Different Gut Microbiomes of Developmental Stages of Field-Collected Native and Invasive Western Bean Cutworm, Striacosta albicosta, in Western Nebraska
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing, DNA Extraction, and Illumina Sequencing
2.3. Processing Data and Statistical Analyses
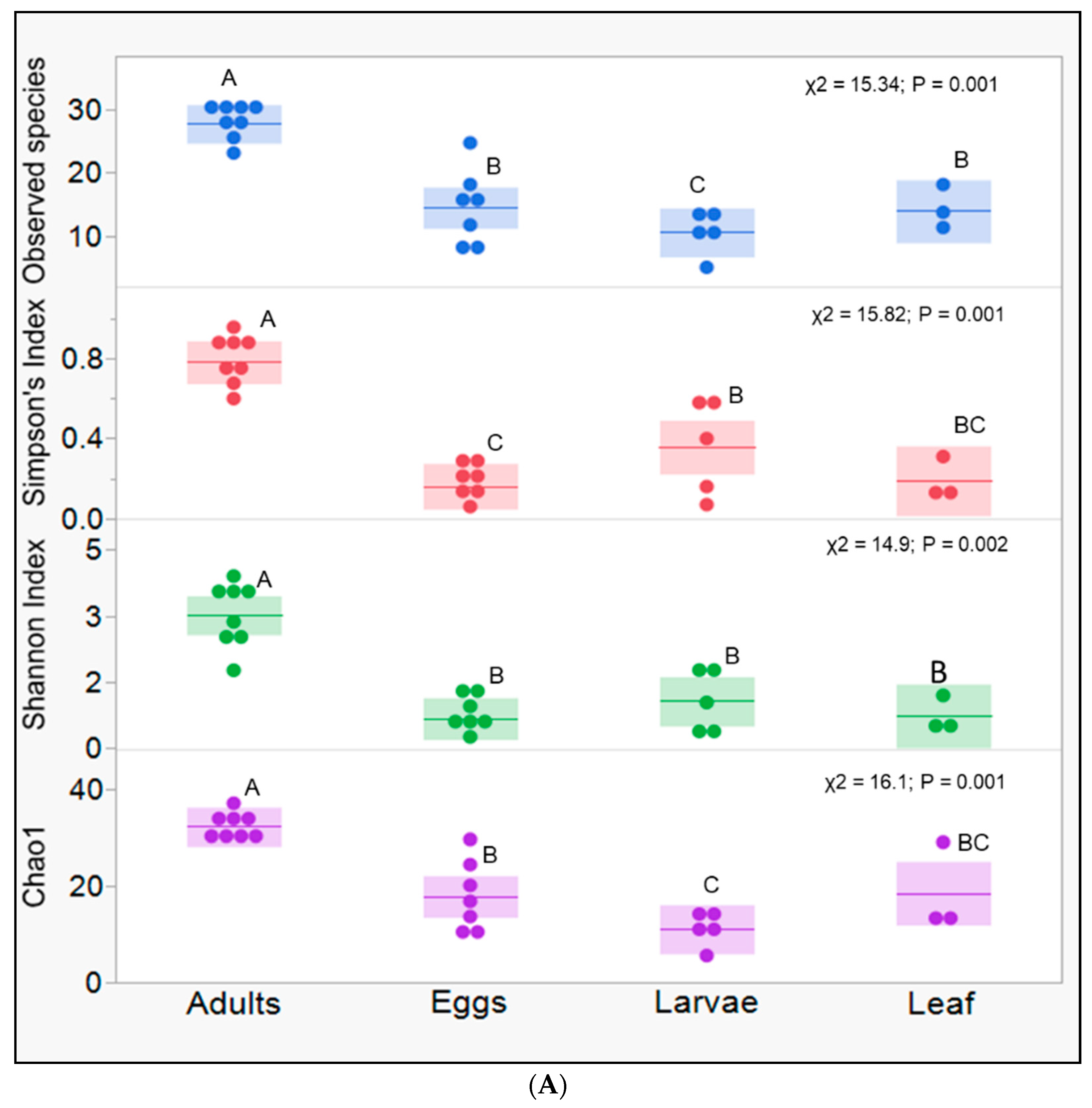
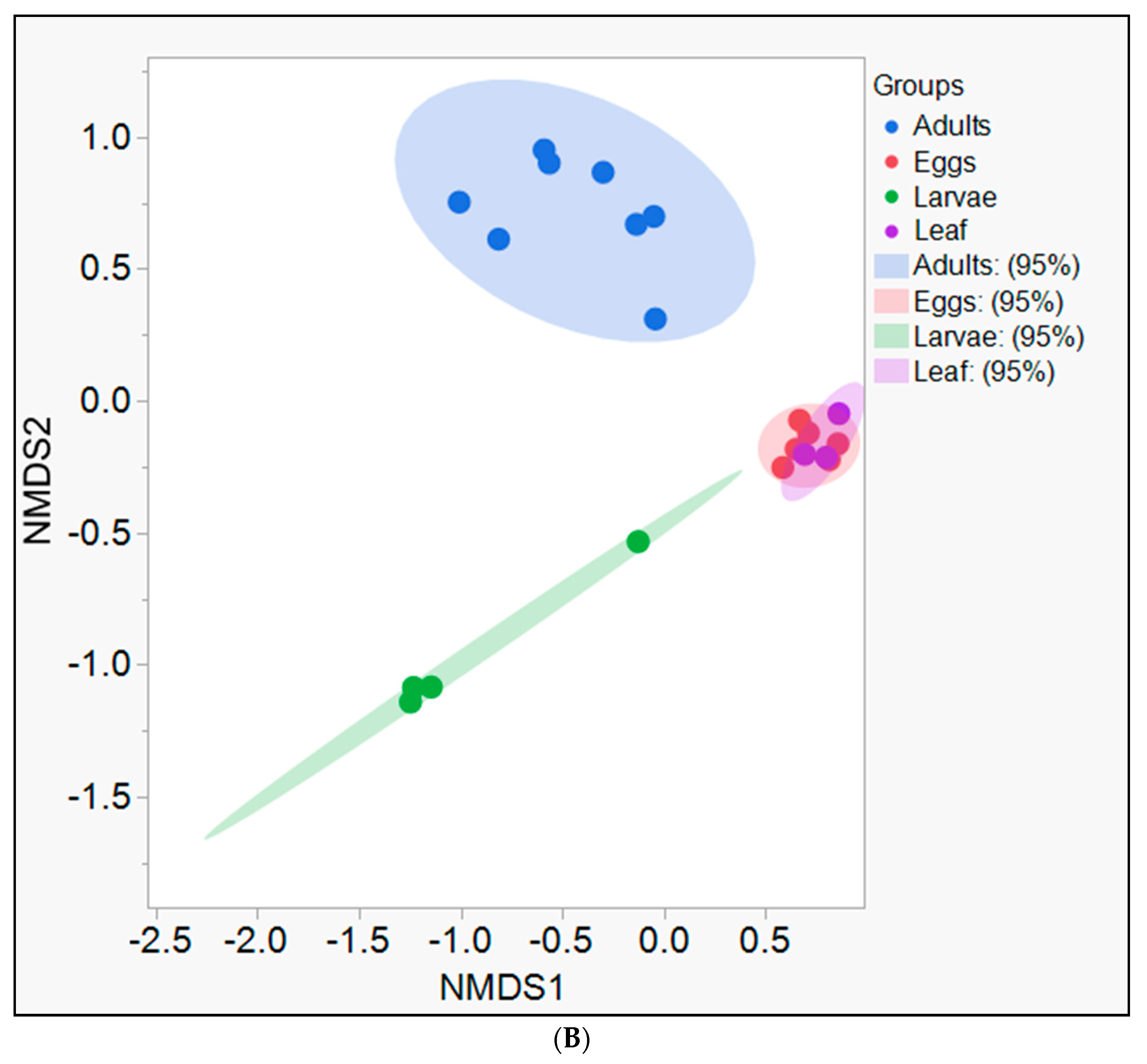
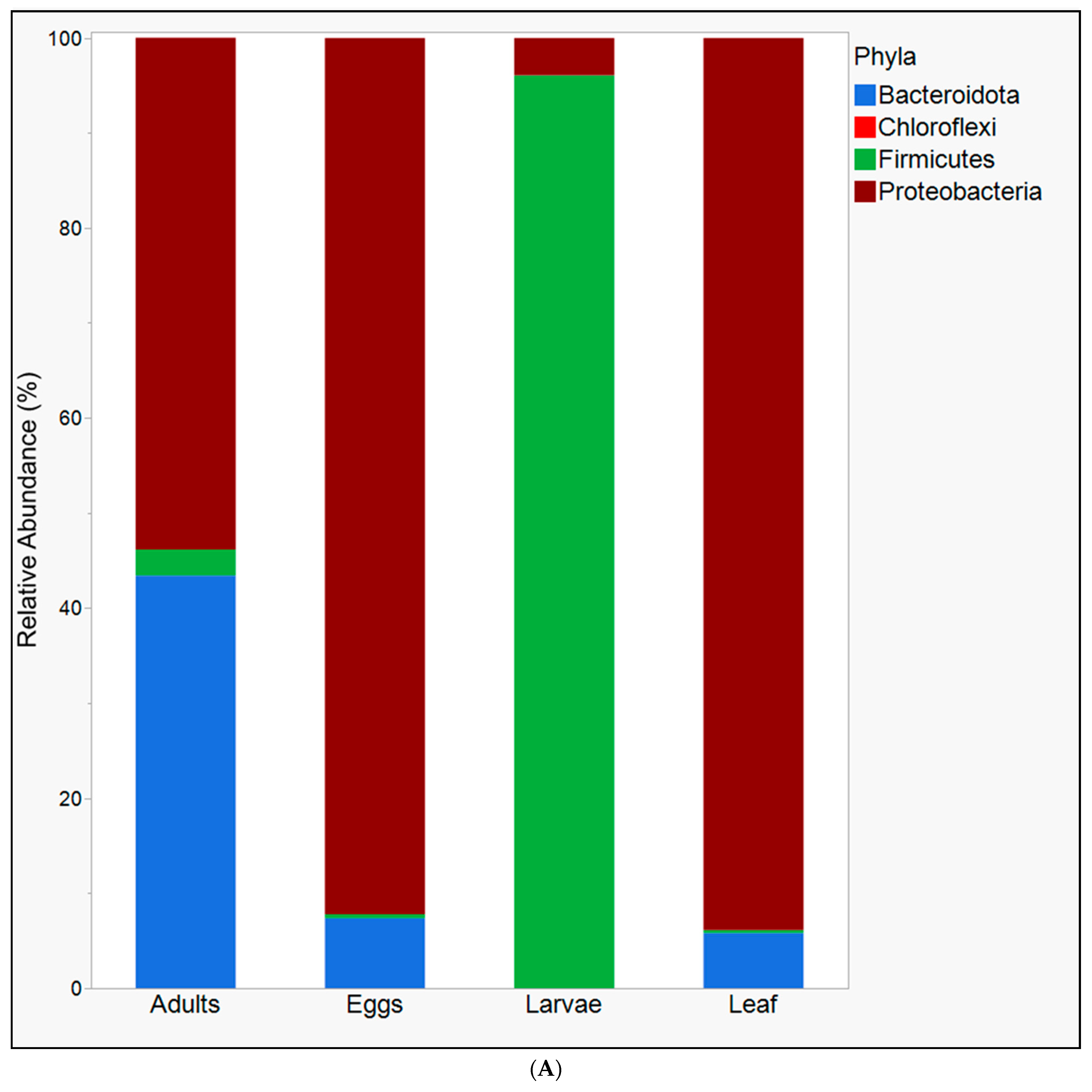
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dillon, R.J.; Dillon, V.M. THE Gut Bacteria of Insects: Nonpathogenic Interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef]
- Gupta, A.; Nair, S. Dynamics of Insect–Microbiome Interaction Influence Host and Microbial Symbiont. Front. Microbiol. 2020, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- Ayayee, P.; Ferry, J.G.; Hoover, K.; Saunders, M.; Felton, G.; Rosa, C. Gut Microbes Contribute to Nitrogen Provisioning in a Wood-Feeding Cerambycid. Environ. Entomol. 2014, 43, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Ayayee, P.A.; Kinney, G.; Yarnes, C.; Larsen, T.; Custer, G.F.; van Diepen, L.T.A.; Muñoz-Garcia, A. Role of the gut microbiome in mediating standard metabolic rate after dietary shifts in the viviparous cockroach, Diploptera punctata. J. Exp. Biol. 2020, 223, jeb218271. [Google Scholar] [CrossRef] [PubMed]
- Ayayee, P.A.; Larsen, T.; Rosa, C.; Felton, G.W.; Ferry, J.G.; Hoover, K. Essential amino acid supplementation by gut microbes of a wood-feeding cerambycid. Environ. Entomol. 2016, 45, 66–73. [Google Scholar] [CrossRef]
- Ayayee, P.A.; Larsen, T.; Sabree, Z. Symbiotic essential amino acids provisioning in the American cockroach, Periplaneta americana (Linnaeus) under various dietary conditions. PeerJ 2016, 4, e2046. [Google Scholar] [CrossRef]
- Sannino, D.R.; Dobson, A.J.; Edwards, K.; Angert, E.R.; Buchon, N.; McFall-Ngai, M.J. The Drosophila melanogaster Gut Microbiota Provisions Thiamine to Its Host. MBio 2018, 9, e00155-18. [Google Scholar] [CrossRef]
- Jing, T.-Z.; Qi, F.-H.; Wang, Z.-Y. Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision? Microbiome 2020, 8, 38. [Google Scholar] [CrossRef]
- Appel, L.L.; Wright, R.J.; Campbell, J.B. Economic Injury Levels for Western Bean Cutworm, Loxagrotis albicosta (Smith) (Lepidoptera: Noctuidae), Eggs and Larvae in Field Corn. J. Kansas Entomol. Soc. 1993, 66, 434–438. [Google Scholar]
- Paula-Moraes, S.; Hunt, T.E.; Wright, R.J.; Hein, G.L.; Blankenship, E.E. Western Bean Cutworm Survival and the Development of Economic Injury Levels and Economic Thresholds in Field Corn. J. Econ. Entomol. 2013, 106, 1274–1285. [Google Scholar] [CrossRef]
- Archibald, W.R.; Bradshaw, J.D.; Golick, D.A.; Wright, R.J.; Peterson, J.A. Nebraska Growers’ and Crop Consultants’ Knowledge and Implementation of Integrated Pest Management of Western Bean Cutworm. J. Integr. Pest Manag. 2018, 9, 1. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.L.; Difonzo, C.D.; Baute, T.S.; Michel, A.P.; Krupke, C.H. Ecology and Management of the Western Bean Cutworm (Lepidoptera: Noctuidae) in Corn and Dry Beans—Revision With Focus on the Great Lakes Region. J. Integr. Pest Manag. 2019, 10, 27. [Google Scholar] [CrossRef]
- Saguez, J.; Neau, M.; Rieux, C.; Vallières-Murray, M.; Petrauskas, P.; Mathieu, S.; Duval, B.; Auger, Y.; Fréchette, I. First Evidence of Western Bean Cutworm (Lepidoptera: Noctuidae) Overwintering in the Province of Québec (Canada). J. Econ. Entomol. 2020, 114, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Baute, T.S.; Sebright, M.M.; Schaafsma, A.W.; DiFonzo, C.D. Establishment of Striacosta albicosta (Lepidoptera: Noctuidae) as a Primary Pest of Corn in the Great Lakes Region. J. Econ. Entomol. 2018, 111, 1732–1744. [Google Scholar] [CrossRef]
- Michel, A.P.; Krupke, C.H.; Baute, T.S.; Difonzo, C.D. Ecology and Management of the Western Bean Cutworm (Lepidoptera: Noctuidae) in Corn and Dry Beans. J. Integr. Pest Manag. 2010, 1, A1–A10. [Google Scholar] [CrossRef]
- Paniagua Voirol, L.R.; Frago, E.; Kaltenpoth, M.; Hilker, M.; Fatouros, N.E. Bacterial Symbionts in Lepidoptera: Their Diversity, Transmission, and Impact on the Host. Front. Microbiol. 2018, 9, 556. [Google Scholar] [CrossRef]
- Tang, X.; Freitak, D.; Vogel, H.; Ping, L.; Shao, Y.; Cordero, E.A.; Andersen, G.; Westermann, M.; Heckel, D.G.; Boland, W. Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae. PLoS ONE 2012, 7, e36978. [Google Scholar] [CrossRef]
- Chen, B.; Teh, B.-S.; Sun, C.; Hu, S.; Lu, X.; Boland, W.; Shao, Y. Biodiversity and Activity of the Gut Microbiota across the Life History of the Insect Herbivore Spodoptera littoralis. Sci. Rep. 2016, 6, 29505. [Google Scholar] [CrossRef]
- Visôtto, L.E.; Oliveira, M.G.A.; Guedes, R.N.C.; Ribon, A.O.B.; Good-God, P.I.V. Contribution of gut bacteria to digestion and development of the velvetbean caterpillar, Anticarsia gemmatalis. J. Insect Physiol. 2009, 55, 185–191. [Google Scholar] [CrossRef]
- Chen, B.; Du, K.; Sun, C.; Vimalanathan, A.; Liang, X.; Li, Y.; Wang, B.; Lu, X.; Li, L.; Shao, Y. Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 2018, 12, 2252–2262. [Google Scholar] [CrossRef]
- Dong, H.-L.; Zhang, S.-X.; Chen, Z.-H.; Tao, H.; Li, X.; Qiu, J.-F.; Cui, W.-Z.; Sima, Y.-H.; Cui, W.-Z.; Xu, S.-Q. Differences in gut microbiota between silkworms (Bombyx mori) reared on fresh mulberry (Morus alba var. multicaulis) leaves or an artificial diet. RSC Adv. 2018, 8, 26188–26200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, T.J.; McMillan, W.O.; Fierer, N. Metamorphosis of a Butterfly-Associated Bacterial Community. PLoS ONE 2014, 9, e86995. [Google Scholar] [CrossRef] [PubMed]
- Belda, E.; Pedrola, L.; Peretó, J.; Martínez-Blanch, J.F.; Montagud, A.; Navarro, E.; Urchueguía, J.; Ramón, D.; Moya, A.; Porcar, M. Microbial diversity in the midguts of field and lab-reared populations of the European corn borer Ostrinia nubilalis. PLoS ONE 2011, 6, e21751. [Google Scholar] [CrossRef]
- Montezano, D.G.; Hunt, T.E.; Souza, D.; Vieira, B.C.; Vélez, A.M.; Kruger, G.R.; Zukoff, S.N.; Bradshaw, J.D.; Peterson, J.A. Bifenthrin Baseline Susceptibility and Evaluation of Simulated Aerial Applications in Striacosta albicosta (Lepidoptera: Noctuidae). J. Econ. Entomol. 2019, 112, 2915–2922. [Google Scholar] [CrossRef] [PubMed]
- Coates, B.S.; Abel, C.A.; Swoboda-Bhattarai, K.A.; Palmquist, D.E.; Montezano, D.G.; Zukoff, S.N.; Wang, Y.; Bradshaw, J.D.; DiFonzo, C.D.; Shields, E.; et al. Geographic Distribution of Bacillus thuringiensis Cry1F Toxin Resistance in Western Bean Cutworm (Lepidoptera: Noctuidae) Populations in the United States. J. Econ. Entomol. 2020, 113, 2465–2472. [Google Scholar] [CrossRef] [PubMed]
- Montezano, D.G.; Hunt, T.E.; Specht, A.; Luz, P.M.C.; Peterson, J.A. Life-History Parameters of Striacosta albicosta (Lepidoptera: Noctuidae) Under Laboratory Conditions. J. Insect Sci. 2019, 19, 14. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Keskitalo, A.; Pietilä, S.; Munukka, Ë.; Eerola, E.; Pursiheimo, J.-P.; Laiho, A.; Pekkala, S.; Huovinen, P. Gut Microbiota Analysis Results Are Highly Dependent on the 16S rRNA Gene Target Region, Whereas the Impact of DNA Extraction Is Minor. J. Biomol. Tech. 2017, 28, 19. [Google Scholar] [CrossRef]
- Mukherjee, S.; Huntemann, M.; Ivanova, N.; Kyrpides, N.C.; Pati, A. Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Stand. Genomic Sci. 2015, 10, 18. [Google Scholar] [CrossRef]
- Callahan, B.J.; Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O.; Caporaso, J.G.; et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to Analyze 16S rRNA Gene Sequences from Microbial Communities. In Current Protocols in Microbiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 1E.5.1–1E.5.20. ISBN 9780471729259. [Google Scholar]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Cameron, E.S.; Schmidt, P.J.; Tremblay, B.J.-M.; Emelko, M.B.; Müller, K.M. Enhancing diversity analysis by repeatedly rarefying next generation sequencing data describing microbial communities. Sci. Rep. 2021, 11, 22302. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, H. The Impact of Environmental Heterogeneity and Life Stage on the Hindgut Microbiota of Holotrichia parallela Larvae (Coleoptera: Scarabaeidae). PLoS ONE 2013, 8, 57169. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Shannon, C. E A Mathematical Theory of Communication. Bell Syst. Tech. J. 1957, 27, 379–423. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; Wiley: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Li, J.; Wang, S.; Zhao, J.; Dong, Z.; Shao, T. Gut Microbiota of Ostrinia nubilalis Larvae Degrade Maize Cellulose. Front. Microbiol. 2022, 13, 858. [Google Scholar] [CrossRef]
- Dow, J. pH gradients in Lepidopteran midgut. J. Exp. Biol. 1992, 172, 355–375. [Google Scholar] [CrossRef]
- Funke, M.; Büchler, R.; Mahobia, V.; Schneeberg, A.; Ramm, M.; Boland, W. Rapid hydrolysis of quorum-sensing molecules in the gut of lepidopteran larvae. Chembiochem 2008, 9, 1953–1959. [Google Scholar] [CrossRef] [PubMed]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef] [PubMed]
- Shannon, A.L.; Attwood, G.; Hopcroft, D.H.; Christeller, J.T. Characterization of lactic acid bacteria in the larval midgut of the keratinophagous lepidopteran, Hofmannophila pseudospretella. Lett. Appl. Microbiol. 2001, 32, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Tomás, A.A.; Anderson, M.A.; Suen, G.; Stevenson, D.M.; Chu, F.S.T.; Cleland, W.W.; Weimer, P.J.; Currie, C.R. Symbiotic Nitrogen Fixation in the Fungus Gardens of Leaf-Cutter Ants. Science 2009, 326, 1120–1123. [Google Scholar] [CrossRef] [PubMed]
- Morales-Jiménez, J.; Vera-Ponce de León, A.; García-Domínguez, A.; Martínez-Romero, E.; Zúñiga, G.; Hernández-Rodríguez, C. Nitrogen-Fixing and Uricolytic Bacteria Associated with the Gut of Dendroctonus rhizophagus and Dendroctonus valens (Curculionidae: Scolytinae). Microb. Ecol. 2013, 66, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Ayayee, P.A.; Keeney, G.; Sabree, Z.L.; Muñoz-Garcia, A. Compositional differences among female-associated and embryo-associated microbiota of the viviparous Pacific Beetle cockroach, Diploptera punctata. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Scully, E.D.; Geib, S.M.; Carlson, J.E.; Tien, M.; McKenna, D.; Hoover, K. Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genom. 2014, 15, 1096. [Google Scholar] [CrossRef]
- Scully, E.D.; Hoover, K.; Carlson, J.E.; Tien, M.; Geib, S.M. Midgut transcriptome profiling of Anoplophora glabripennis, a lignocellulose degrading cerambycid beetle. BMC Genom. 2013, 14, 850. [Google Scholar] [CrossRef]
- Hammer, T.J.; Janzen, D.H.; Hallwachs, W.; Jaffe, S.P.; Fierer, N. Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. 2017, 114, 9641–9646. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Cho, G.-S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef] [Green Version]
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H. El Lactic acid bacteria: From starter cultures to producers of chemicals. FEMS Microbiol. Lett. 2018, 365, fny213. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayayee, P.A.; Currie, A.; Peterson, J.A. Different Gut Microbiomes of Developmental Stages of Field-Collected Native and Invasive Western Bean Cutworm, Striacosta albicosta, in Western Nebraska. Microorganisms 2022, 10, 1828. https://doi.org/10.3390/microorganisms10091828
Ayayee PA, Currie A, Peterson JA. Different Gut Microbiomes of Developmental Stages of Field-Collected Native and Invasive Western Bean Cutworm, Striacosta albicosta, in Western Nebraska. Microorganisms. 2022; 10(9):1828. https://doi.org/10.3390/microorganisms10091828
Chicago/Turabian StyleAyayee, Paul A., Austin Currie, and Julie A. Peterson. 2022. "Different Gut Microbiomes of Developmental Stages of Field-Collected Native and Invasive Western Bean Cutworm, Striacosta albicosta, in Western Nebraska" Microorganisms 10, no. 9: 1828. https://doi.org/10.3390/microorganisms10091828
APA StyleAyayee, P. A., Currie, A., & Peterson, J. A. (2022). Different Gut Microbiomes of Developmental Stages of Field-Collected Native and Invasive Western Bean Cutworm, Striacosta albicosta, in Western Nebraska. Microorganisms, 10(9), 1828. https://doi.org/10.3390/microorganisms10091828






