Mining Biosynthetic Gene Clusters in Carnobacterium maltaromaticum by Interference Competition Network and Genome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Competition Assays and Data Processing
2.3. Genome Sequencing and Analysis
2.4. Data Analysis
2.5. Accession Numbers
3. Results
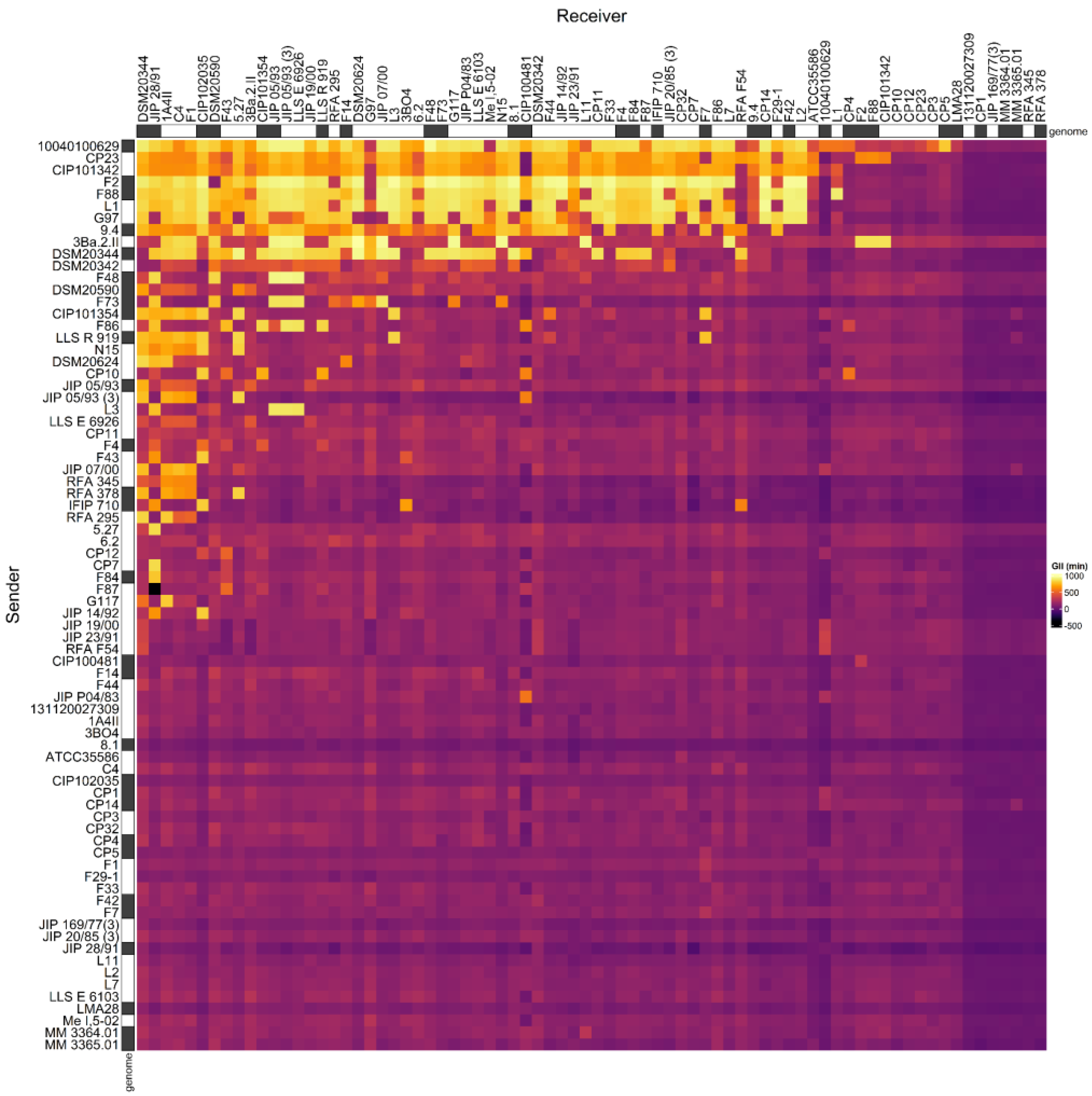
3.1. Adjacency Matrix of Inhibition
3.2. Bacteriocin Gene Clusters
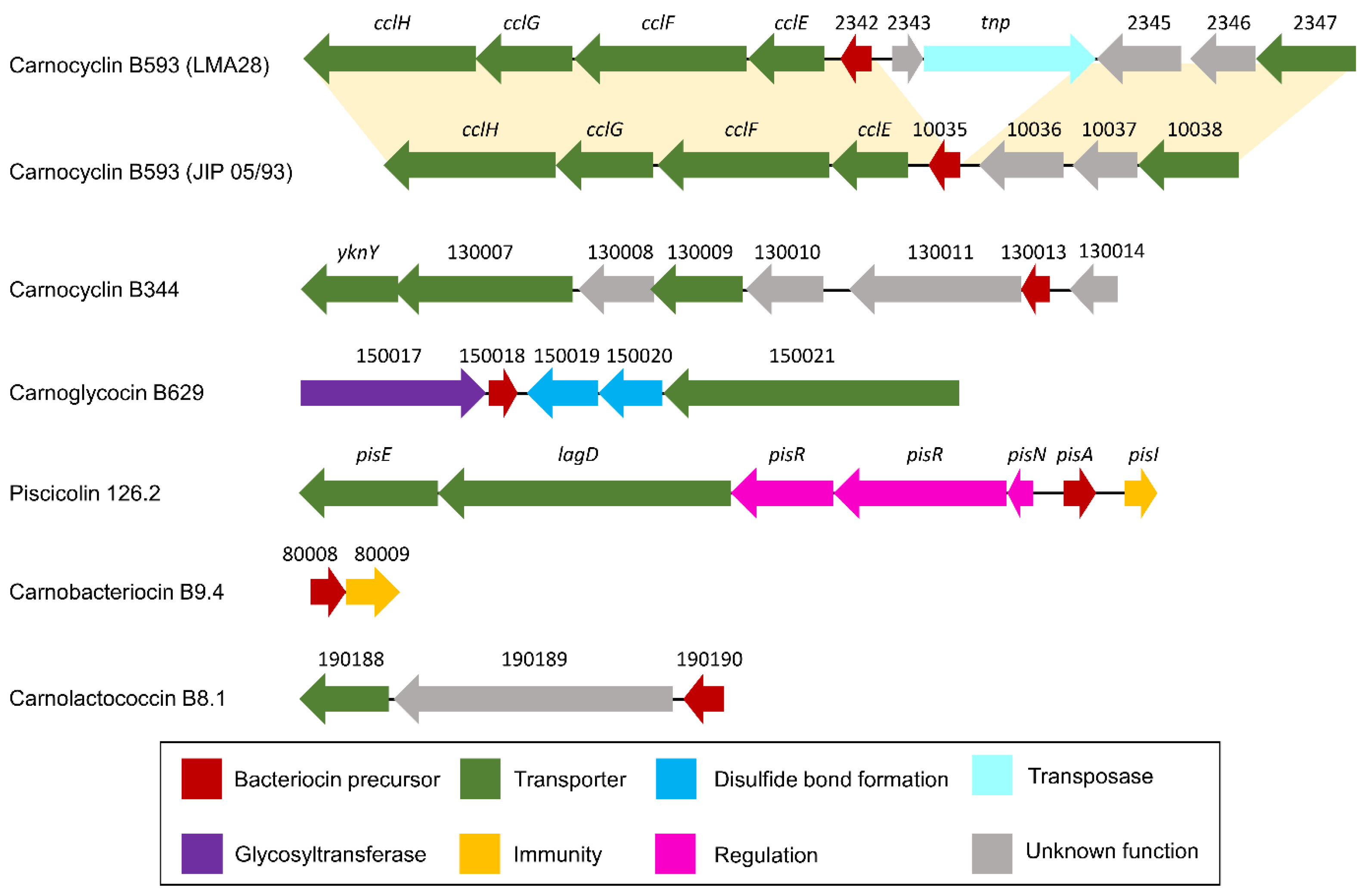
3.2.1. Class I
3.2.2. Class IIa
3.2.3. Class IIc
3.3. NRPS-BGC
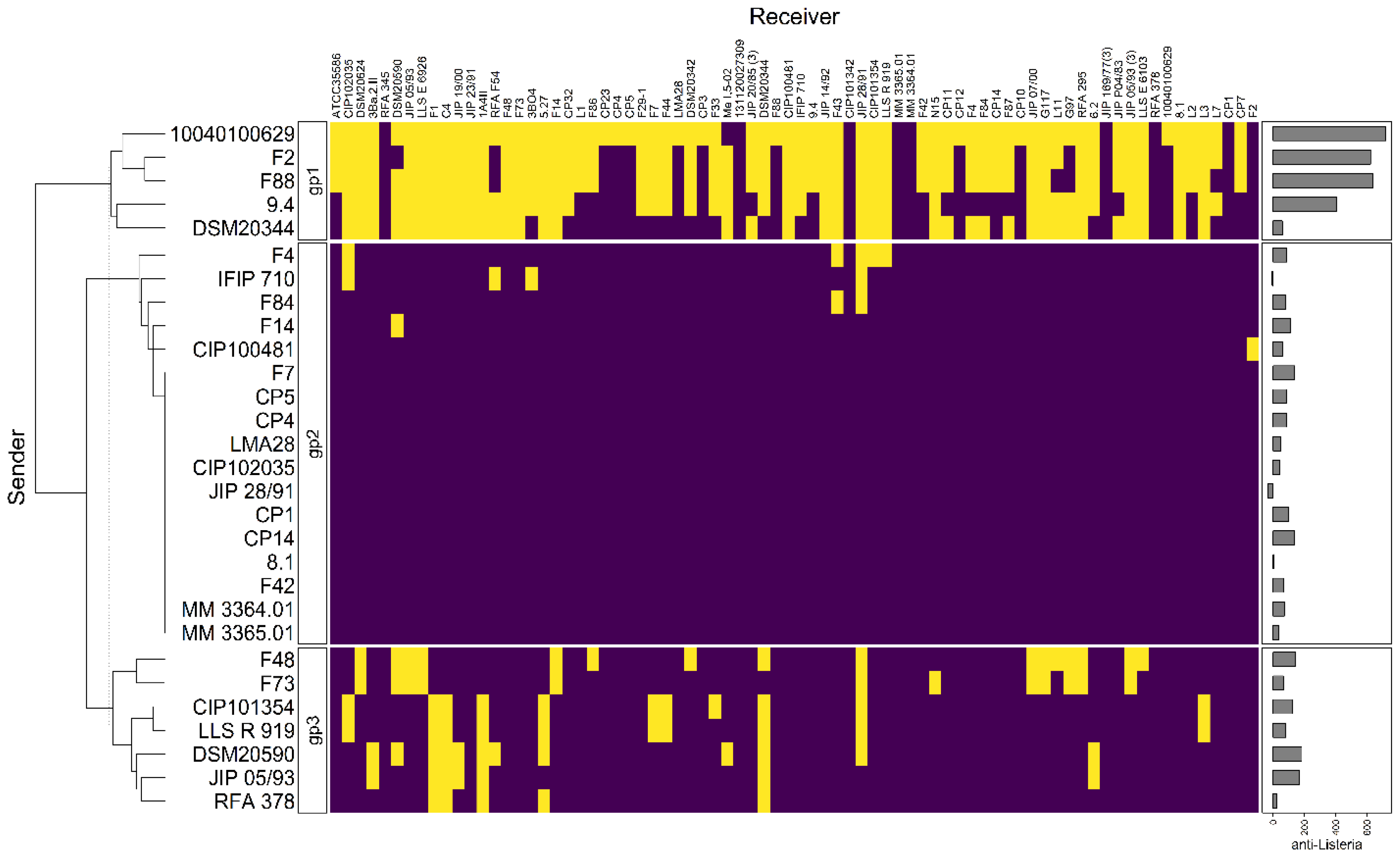
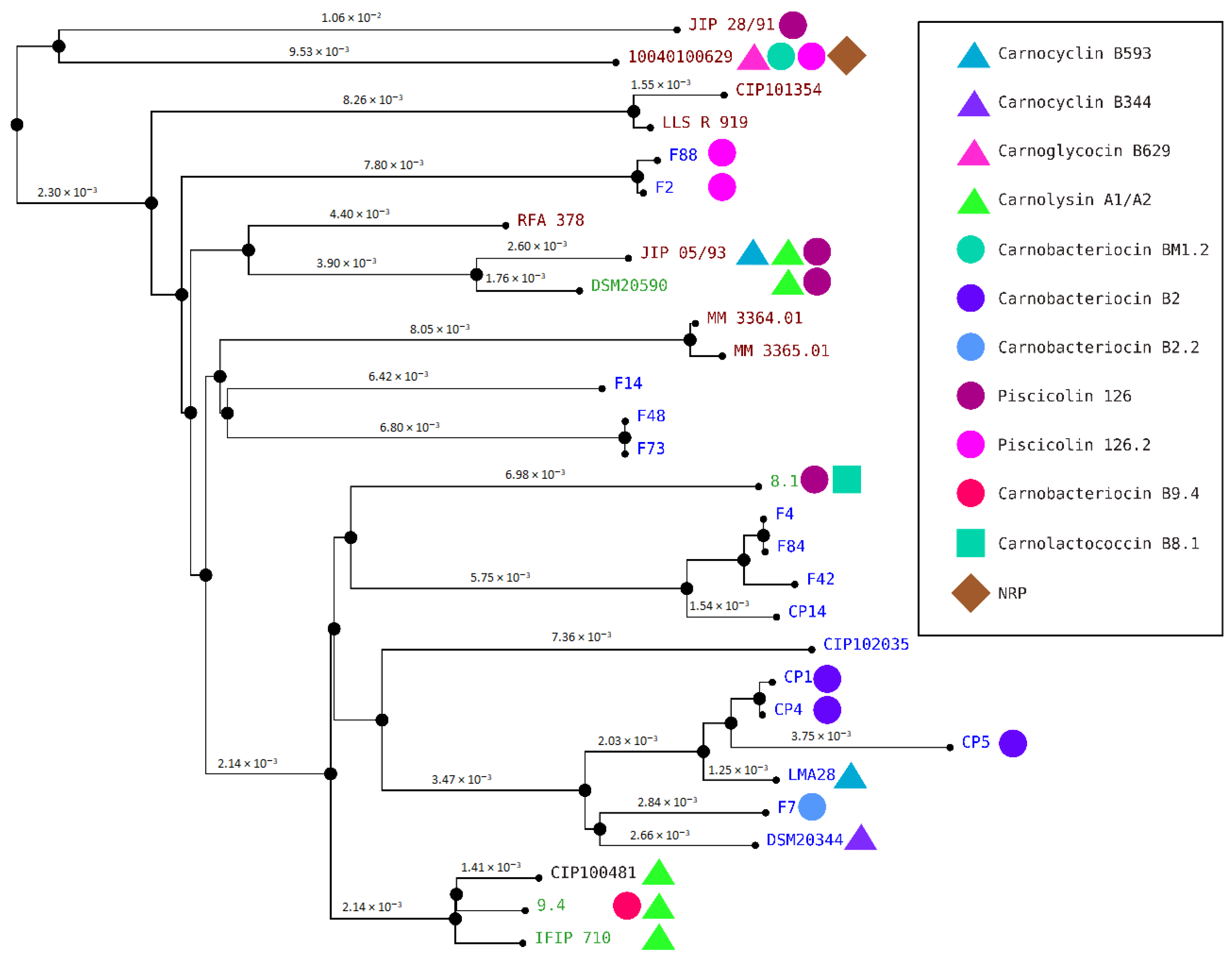
3.4. Genome Clustering
3.5. Comparisons of Antimicrobial Phenotypes and Genome Predictions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Borges, F.; Briandet, R.; Callon, C.; Champomier-Vergès, M.-C.; Christieans, S.; Chuzeville, S.; Denis, C.; Desmasures, N.; Desmonts, M.-H.; Feurer, C.; et al. Contribution of Omics to Biopreservation: Toward Food Microbiome Engineering. Front. Microbiol. 2022, 13, 951182. [Google Scholar] [CrossRef] [PubMed]
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin Production Augments Niche Competition by Enterococci in the Mammalian Gastrointestinal Tract. Nature 2015, 526, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.A.; Wertz, J.E. Bacteriocin Diversity: Ecological and Evolutionary Perspectives. Biochimie 2002, 84, 357–364. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Remenant, B.; Borges, F.; Cailliez-Grimal, C.; Revol-Junelles, A.-M.; Marché, L.; Lajus, A.; Médigue, C.; Pilet, M.-F.; Prévost, H.; Zagorec, M. Draft Genome Sequence of Carnobacterium divergens V41, a Bacteriocin-Producing Strain. Genome Announc. 2016, 4, e01109-16. [Google Scholar] [CrossRef]
- Golomb, B.L.; Yu, A.O.; Coates, L.C.; Marco, M.L. The Lactococcus Lactis KF147 Nonribosomal Peptide Synthetase/Polyketide Synthase System Confers Resistance to Oxidative Stress during Growth on Plant Leaf Tissue Lysate. MicrobiologyOpen 2018, 7, e00531. [Google Scholar] [CrossRef]
- Khayatt, B.I.; van Noort, V.; Siezen, R.J. The Genome of the Plant-Associated Lactic Acid Bacterium Lactococcus Lactis KF147 Harbors a Hybrid NRPS-PKS System Conserved in Strains of the Dental Cariogenic Streptococcus mutans. Curr. Microbiol. 2020, 77, 136–145. [Google Scholar] [CrossRef]
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- Miyanaga, A. Structure and Function of Polyketide Biosynthetic Enzymes: Various Strategies for Production of Structurally Diverse Polyketides. Biosci. Biotechnol. Biochem. 2017, 81, 2227–2236. [Google Scholar] [CrossRef]
- Skiba, M.A.; Maloney, F.P.; Dan, Q.; Fraley, A.E.; Aldrich, C.C.; Smith, J.L.; Brown, W.C. PKS-NRPS Enzymology and Structural Biology: Considerations in Protein Production. Methods Enzymol. 2018, 604, 45–88. [Google Scholar] [CrossRef]
- Fisch, K.M. Biosynthesis of Natural Products by Microbial Iterative Hybrid PKS–NRPS. RSC Adv. 2013, 3, 18228–18247. [Google Scholar] [CrossRef]
- Afzal, M.I.; Jacquet, T.; Delaunay, S.; Borges, F.; Milliere, J.B.; Revol-Junelles, A.M.; Cailliez-Grimal, C. Carnobacterium maltaromaticum: Identification, Isolation Tools, Ecology and Technological Aspects in Dairy Products. Food Microbiol. 2010, 27, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Leisner, J.J.; Laursen, B.G.; Prevost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and Negative Effects in the Environment and in Foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef]
- Afzal, M.I.; Gonzalez Ariceaga, C.C.; Lhomme, E.; Kamel Ali, N.; Payot, S.; Burgain, J.; Gaiani, C.; Borges, F.; Revol-Junelles, A.M.; Delaunay, S.; et al. Characterization of Carnobacterium maltaromaticum LMA 28 for Its Positive Technological Role in Soft Cheese Making. Food Microbiol. 2013, 36, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Hammi, I.; Delalande, F.; Belkhou, R.; Marchioni, E.; Cianferani, S.; Ennahar, S. Maltaricin CPN, a New Class IIa Bacteriocin Produced by Carnobacterium maltaromaticum CPN Isolated from Mould-Ripened Cheese. J. Appl. Microbiol. 2016, 121, 1268–1274. [Google Scholar] [CrossRef]
- Spanu, C.; Piras, F.; Mocci, A.M.; Nieddu, G.; De Santis, E.P.L.; Scarano, C. Use of Carnobacterium spp. Protective Culture in MAP Packed Ricotta Fresca Cheese to Control Pseudomonas spp. Food Microbiol. 2018, 74, 50–56. [Google Scholar] [CrossRef]
- Danielski, G.M.; Imazaki, P.H.; de Andrade Cavalari, C.M.; Daube, G.; Clinquart, A.; de Macedo, R.E.F. Carnobacterium maltaromaticum as Bioprotective Culture in Vitro and in Cooked Ham. Meat Sci. 2020, 162, 108035. [Google Scholar] [CrossRef] [PubMed]
- Laursen, B.G.; Bay, L.; Cleenwerck, I.; Vancanneyt, M.; Swings, J.; Dalgaard, P.; Leisner, J.J. Carnobacterium Divergens and Carnobacterium maltaromaticum as Spoilers or Protective Cultures in Meat and Seafood: Phenotypic and Genotypic Characterization. Syst. Appl. Microbiol. 2005, 28, 151–164. [Google Scholar] [CrossRef]
- Dos Reis, F.B.; de Souza, V.M.; Thomaz, M.R.S.; Fernandes, L.P.; de Oliveira, W.P.; De Martinis, E.C.P. Use of Carnobacterium maltaromaticum Cultures and Hydroalcoholic Extract of Lippia Sidoides Cham. against Listeria monocytogenes in Fish Model Systems. Int. J. Food Microbiol. 2011, 146, 228–234. [Google Scholar] [CrossRef]
- Kim, D.H.; Austin, B. Innate Immune Responses in Rainbow Trout (Oncorhynchus mykiss, Walbaum) Induced by Probiotics. Fish Shellfish Immunol. 2006, 21, 513–524. [Google Scholar] [CrossRef]
- Koné, A.P.; Zea, J.M.V.; Gagné, D.; Cinq-Mars, D.; Guay, F.; Saucier, L. Application of Carnobacterium maltaromaticum as a Feed Additive for Weaned Rabbits to Improve Meat Microbial Quality and Safety. Meat Sci. 2018, 135, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Stoffels, G.; Nes, I.F.; Guthmundsdottir, A. Isolation and Properties of a Bacteriocin-Producing Carnobacterium piscicola Isolated from Fish. J. Appl. Bacteriol. 1992, 73, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Tulini, F.L.; Lohans, C.T.; Bordon, K.C.F.; Zheng, J.; Arantes, E.C.; Vederas, J.C.; De Martinis, E.C.P. Purification and Characterization of Antimicrobial Peptides from Fish Isolate Carnobacterium maltaromaticum C2: Carnobacteriocin X and Carnolysins A1 and A2. Int. J. Food Microbiol. 2014, 173, 81–88. [Google Scholar] [CrossRef]
- Martin-Visscher, L.A.; van Belkum, M.J.; Garneau-Tsodikova, S.; Whittal, R.M.; Zheng, J.; McMullen, L.M.; Vederas, J.C. Isolation and Characterization of Carnocyclin a, a Novel Circular Bacteriocin Produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 2008, 74, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Quadri, L.E.; Sailer, M.; Roy, K.L.; Vederas, J.C.; Stiles, M.E. Chemical and Genetic Characterization of Bacteriocins Produced by Carnobacterium piscicola LV17B. J. Biol. Chem. 1994, 269, 12204–12211. [Google Scholar] [CrossRef]
- Jack, R.W.; Wan, J.; Gordon, J.; Harmark, K.; Davidson, B.E.; Hillier, A.J.; Wettenhall, R.E.; Hickey, M.W.; Coventry, M.J. Characterization of the Chemical and Antimicrobial Properties of Piscicolin 126, a Bacteriocin Produced by Carnobacterium piscicola JG126. Appl. Environ. Microbiol. 1996, 62, 2897–2903. [Google Scholar] [CrossRef]
- Yamazaki, K.; Suzuki, M.; Kawai, Y.; Inoue, N.; Montville, T.J. Purification and Characterization of a Novel Class IIa Bacteriocin, Piscicocin CS526, from Surimi-Associated Carnobacterium piscicola CS526. Appl. Environ. Microbiol. 2005, 71, 554–557. [Google Scholar] [CrossRef]
- Bhugaloo-Vial, P.; Dousset, X.; Metivier, A.; Sorokine, O.; Anglade, P.; Boyaval, P.; Marion, D. Purification and Amino Acid Sequences of Piscicocins V1a and V1b, Two Class IIa Bacteriocins Secreted by Carnobacterium piscicola V1 That Display Significantly Different Levels of Specific Inhibitory Activity. Appl Environ Microbiol 1996, 62, 4410–4416. [Google Scholar] [CrossRef]
- Holck, A.L.; Axelsson, L.; Schillinger, U. Purification and Cloning of Piscicolin 61, a Bacteriocin from Carnobacterium Piscicola LV61. Curr. Microbiol. 1994, 29, 63–68. [Google Scholar] [CrossRef]
- Worobo, R.W.; Henkel, T.; Sailer, M.; Roy, K.L.; Vederas, J.C.; Stiles, M.E. Characteristics and Genetic Determinant of a Hydrophobic Peptide Bacteriocin, Carnobacteriocin A, Produced by Carnobacterium piscicola LV17A. Microbiology 1994, 140 Pt 3, 517–526. [Google Scholar] [CrossRef][Green Version]
- Stiles, M.E.; Calrson, D.; Smith, D.C. Enhanced Preservation of Processed Food. U.S. Patent 9,066,521 B2, 30 June 2015. [Google Scholar]
- Borges, F.; Revol-Junelles, A.-M. Nouvelles Souches de Carnobacterium maltaromaticum et Leurs Utilisations. International Patent WO2021078612A1, 29 April 2021. [Google Scholar]
- Ramia, N.E.; Mangavel, C.; Gaiani, C.; Muller-Gueudin, A.; Taha, S.; Revol-Junelles, A.-M.; Borges, F. Nested Structure of Intraspecific Competition Network in Carnobacterium maltaromaticum. Sci. Rep. 2020, 10, 7335. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing: Vienna, Austria. 2021. Available online: https://www.R-project.org/ (accessed on 28 August 2022).
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 28 August 2022).
- Vallenet, D.; Engelen, S.; Mornico, D.; Cruveiller, S.; Fleury, L.; Lajus, A.; Rouy, Z.; Roche, D.; Salvignol, G.; Scarpelli, C.; et al. MicroScope: A Platform for Microbial Genome Annotation and Comparative Genomics. Database 2009, 2009, bap021. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fevre, F.; Longin, C.; Mornico, D.; Roche, D.; et al. MicroScope—An Integrated Microbial Resource for the Curation and Comparative Analysis of Genomic and Metabolic Data. Nucleic Acids Res. 2013, 41, D636–D647. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Calteau, A.; Cruveiller, S.; Gachet, M.; Lajus, A.; Josso, A.; Mercier, J.; Renaux, A.; Rollin, J.; Rouy, Z.; et al. MicroScope in 2017: An Expanding and Evolving Integrated Resource for Community Expertise of Microbial Genomes. Nucleic Acids Res. 2017, 45, D517–D528. [Google Scholar] [CrossRef]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An Integrated Platform for the Annotation and Exploration of Microbial Gene Functions through Genomic, Pangenomic and Metabolic Comparative Analysis. Nucleic Acids Res. 2020, 48, D579–D589. [Google Scholar] [CrossRef]
- De Jong, A.; van Hijum, S.A.F.T.; Bijlsma, J.J.E.; Kok, J.; Kuipers, O.P. BAGEL: A Web-Based Bacteriocin Genome Mining Tool. Nucleic Acids Res. 2006, 34, W273–W279. [Google Scholar] [CrossRef]
- De Jong, A.; van Heel, A.J.; Kok, J.; Kuipers, O.P. BAGEL2: Mining for Bacteriocins in Genomic Data. Nucleic Acids Res. 2010, 38, W647–W651. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Montalbán-López, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated Identification of Genes Encoding Bacteriocins and (Non-)Bactericidal Posttranslationally Modified Peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. AntiSMASH 4.0—Improvements in Chemistry Prediction and Gene Cluster Boundary Identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0—A Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid Identification, Annotation and Analysis of Secondary Metabolite Biosynthesis Gene Clusters in Bacterial and Fungal Genome Sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. AntiSMASH 2.0—A Versatile Platform for Genome Mining of Secondary Metabolite Producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Dawson, C. Ggprism: A “ggplot2” Extension Inspired by “GraphPad Prism”. 2021. Available online: https://cran.r-project.org/web/packages/ggprism/index.html (accessed on 28 August 2022).
- Slowikowski, K. Ggrepel: Automatically Position Non-Overlapping Text Labels with “Ggplot2”. 2021. Available online: https://cran.r-project.org/web/packages/ggrepel/ (accessed on 28 August 2022).
- Garnier, S.; Ross, N.; Rudis, B.; Sciaini, M.; Camargo, A.P.; Scherer, C. Viridis-Colorblind-Friendly Color Maps for R. 2021. Available online: https://cran.r-project.org/web/packages/viridis/ (accessed on 28 August 2022).
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and Leaderless Bacteriocins: Biosynthesis, Mode of Action, Applications, and Prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef]
- Van Belkum, M.J.; Martin-Visscher, L.A.; Vederas, J.C. Cloning and Characterization of the Gene Cluster Involved in the Production of the Circular Bacteriocin Carnocyclin A. Probiotics Antimicrob. Proteins 2010, 2, 218–225. [Google Scholar] [CrossRef]
- Kaunietis, A.; Buivydas, A.; Čitavičius, D.J.; Kuipers, O.P. Heterologous Biosynthesis and Characterization of a Glycocin from a Thermophilic Bacterium. Nat. Commun. 2019, 10, 1115. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa Bacteriocins: Diversity and New Developments. Int. J. Mol. Sci. 2012, 13, 16668. [Google Scholar] [CrossRef]
- Lohans, C.T.; Li, J.L.; Vederas, J.C. Structure and Biosynthesis of Carnolysin, a Homologue of Enterococcal Cytolysin with D-Amino Acids. J. Am. Chem. Soc. 2014, 136, 13150–13153. [Google Scholar] [CrossRef] [PubMed]
- Drider, D.; Fimland, G.; Hechard, Y.; McMullen, L.M.; Prevost, H. The Continuing Story of Class IIa Bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Balciunas, E.M.; Castillo Martinez, F.A.; Todorov, S.D.; de Melo Franco, B.D.G.; Converti, A.; de Souza Oliveira, R.P. Novel Biotechnological Applications of Bacteriocins: A Review. Food Control 2013, 32, 134–142. [Google Scholar] [CrossRef]
- Sánchez, J.; Diep, D.B.; Herranz, C.; Nes, I.F.; Cintas, L.M.; Hernández, P.E. Amino Acid and Nucleotide Sequence, Adjacent Genes, and Heterologous Expression of Hiracin JM79, a Sec-Dependent Bacteriocin Produced by Enterococcus Hirae DCH5, Isolated from Mallard Ducks (Anas platyrhynchos). FEMS Microbiol. Lett. 2007, 270, 227–236. [Google Scholar] [CrossRef]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guédon, G. Conjugative and Mobilizable Genomic Islands in Bacteria: Evolution and Diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef]
- Quadri, L.E.N. Regulation of Class II Bacteriocin Production by Cell-Cell Signaling. J. Microbiol. 2003, 41, 175–182. [Google Scholar]
- Giello, M.; La Storia, A.; De Filippis, F.; Ercolini, D.; Villani, F. Impact of Lactobacillus curvatus 54M16 on Microbiota Composition and Growth of Listeria Monocytogenes in Fermented Sausages. Food Microbiol. 2018, 72, 1–15. [Google Scholar] [CrossRef]
- Revol-Junelles, A.M.; Mathis, R.; Krier, F.; Fleury, Y.; Delfour, A.; Lefebvre, G. Leuconostoc Mesenteroides Subsp. Mesenteroides FR52 Synthesizes Two Distinct Bacteriocins. Lett. Appl. Microbiol. 1996, 23, 120–124. [Google Scholar]
- Begrem, S.; Ivaniuk, F.; Gigout-Chevalier, F.; Kolypczuk, L.; Bonnetot, S.; Leroi, F.; Grovel, O.; Delbarre-Ladrat, C.; Passerini, D. New Insight into Antimicrobial Compounds from Food and Marine-Sourced Carnobacterium Species through Phenotype and Genome Analyses. Microorganisms 2020, 8, 1093. [Google Scholar] [CrossRef]
- Reis, J.A.; Paula, A.T.; Casarotti, S.N.; Penna, A.L.B. Lactic Acid Bacteria Antimicrobial Compounds: Characteristics and Applications. Food Eng. Rev. 2012, 4, 124–140. [Google Scholar] [CrossRef]
- Walsh, C.J.; Guinane, C.M.; Hill, C.; Ross, R.P.; O’Toole, P.W.; Cotter, P.D. In Silico Identification of Bacteriocin Gene Clusters in the Gastrointestinal Tract, Based on the Human Microbiome Project’s Reference Genome Database. BMC Microbiol. 2015, 15, 183. [Google Scholar] [CrossRef] [PubMed]
- Borsetto, C.; Amos, G.C.A.; da Rocha, U.N.; Mitchell, A.L.; Finn, R.D.; Laidi, R.F.; Vallin, C.; Pearce, D.A.; Newsham, K.K.; Wellington, E.M.H. Microbial Community Drivers of PK/NRP Gene Diversity in Selected Global Soils. Microbiome 2019, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Aleti, G.; Sessitsch, A.; Brader, G. Genome Mining: Prediction of Lipopeptides and Polyketides from Bacillus and Related Firmicutes. Comput. Struct. Biotechnol. J. 2015, 13, 192–203. [Google Scholar] [CrossRef]
- Bloudoff, K.; Schmeing, T.M. Structural and Functional Aspects of the Nonribosomal Peptide Synthetase Condensation Domain Superfamily: Discovery, Dissection and Diversity. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1587–1604. [Google Scholar] [CrossRef]
- Martínez-Núñez, M.; López, V.E. Nonribosomal Peptides Synthetases and Their Applications in Industry. Sustain. Chem. Process. 2016, 4, 13. [Google Scholar] [CrossRef]
- Agrawal, S.; Acharya, D.; Adholeya, A.; Barrow, C.J.; Deshmukh, S.K. Nonribosomal Peptides from Marine Microbes and Their Antimicrobial and Anticancer Potential. Front. Pharmacol. 2017, 8, 828. [Google Scholar] [CrossRef] [PubMed]
- Lohans, C.T.; Vederas, J.C. Development of Class IIa Bacteriocins as Therapeutic Agents. Int. J. Microbiol. 2012, 2012, 386410. [Google Scholar] [CrossRef]
- Kazazic, M.; Nissen-Meyer, J.; Fimland, G. Mutational Analysis of the Role of Charged Residues in Target-Cell Binding, Potency and Specificity of the Pediocin-like Bacteriocin Sakacin P. Microbiology 2002, 148, 2019–2027. [Google Scholar] [CrossRef][Green Version]
- Zheng, Y.; Du, Y.; Qiu, Z.; Liu, Z.; Qiao, J.; Li, Y.; Caiyin, Q. Nisin Variants Generated by Protein Engineering and Their Properties. Bioengineering 2022, 9, 251. [Google Scholar] [CrossRef]
- Gursky, L.J.; Martin, N.I.; Derksen, D.J.; van Belkum, M.J.; Kaur, K.; Vederas, J.C.; Stiles, M.E.; McMullen, L.M. Production of Piscicolin 126 by Carnobacterium maltaromaticum UAL26 Is Controlled by Temperature and Induction Peptide Concentration. Arch. Microbiol. 2006, 186, 317–325. [Google Scholar] [CrossRef]
- Mathieu, F.; Michel, M.; Lebrihi, A.; Lefebvre, G. Effect of the Bacteriocin Carnocin CP5 and of the Producing Strain Carnobacterium piscicola CP5 on the Viability of Listeria monocytogenes ATCC 15313 in Salt Solution, Broth and Skimmed Milk, at Various Incubation Temperatures. Int J Food Microbiol 1994, 22, 155–172. [Google Scholar] [CrossRef]
- Mørtvedt, C.; Nes, I. Plasmid-Associated Bacteriocin Production by a Lactobacillus sake Strain. Microbiology 1990, 136, 1601–1607. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Cailliez-Grimal, C.; Bontemps, C.; Payot, S.; Chaillou, S.; Revol-Junelles, A.-M.; Borges, F. High Genetic Diversity among Strains of the Unindustrialized Lactic Acid Bacterium Carnobacterium maltaromaticum in Dairy Products as Revealed by Multilocus Sequence Typing. Appl. Environ. Microbiol. 2014, 80, 3920–3929. [Google Scholar] [CrossRef]
- Millière, J.B.; Michel, M.; Mathieu, F.; Lefebvre, G. Presence of Carnobacterium Spp. in French Surface Mould-Ripened Soft-Cheese. J. Appl. Bacteriol. 1994, 76, 264–269. [Google Scholar] [CrossRef]
- Miller, A.; Morgan, M.E.; Libbey, L.M. Lactobacillus maltaromicus, a New Species Producing a Malty Aroma. Int. J. Syst. Bacteriol. 1974, 24, 346–354. [Google Scholar] [CrossRef]
- Ramia, N.E.; El Kheir, S.M.; Taha, S.; Mangavel, C.; Revol-Junelles, A.M.; Borges, F. Multilocus Sequence Typing of Carnobacterium maltaromaticum Strains Associated with Fish Disease and Dairy Products. J. Appl. Microbiol. 2019, 126, 377–387. [Google Scholar] [CrossRef]
- Cailliez-Grimal, C.; Edima, H.C.; Revol-Junelles, A.M.; Millière, J.B. Short Communication: Carnobacterium maltaromaticum: The Only Carnobacterium Species in French Ripened Soft Cheeses as Revealed by Polymerase Chain Reaction Detection. J. Dairy Sci. 2007, 90, 1133–1138. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Culturable Psychrotrophic Bacterial Communities in Raw Milk and Their Proteolytic and Lipolytic Traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef]
- Walker, V.K.; Palmer, G.R.; Voordouw, G. Freeze-Thaw Tolerance and Clues to the Winter Survival of a Soil Community. Appl. Environ. Microbiol. 2006, 72, 1784–1792. [Google Scholar] [CrossRef]
- Leisner, J.J.; Vogensen, F.K.; Kollmann, J.; Aideh, B.; Vandamme, P.; Vancanneyt, M.; Ingmer, H. α-Chitinase Activity among Lactic Acid Bacteria. Syst. Appl. Microbiol. 2008, 31, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Hiu, S.F.; Holt, R.A.; Sriranganathan, N.; Seidler, R.J.; Fryer, J.L. Lactobacillus piscicola, a New Species from Salmonid Fish. Int. J. Syst. Bacteriol. 1984, 34, 393–400. [Google Scholar] [CrossRef]
- Holzapfel, W.H.; Gerber, E.S. Lactobacillus divergens Sp. Nov., a New Heterofermentative Lactobacillus Species Producing L(+)-Lactate. Syst. Appl. Microbiol. 1983, 4, 522–534. [Google Scholar] [CrossRef]
- Feurer, C.; Ellouze, M. Utilisation Raisonnée de La Flore Naturelle de Viande de Porc Fraîche Pour Améliorer Sa Qualité Sanitaire. IFIP 2010, 34, 125. [Google Scholar]


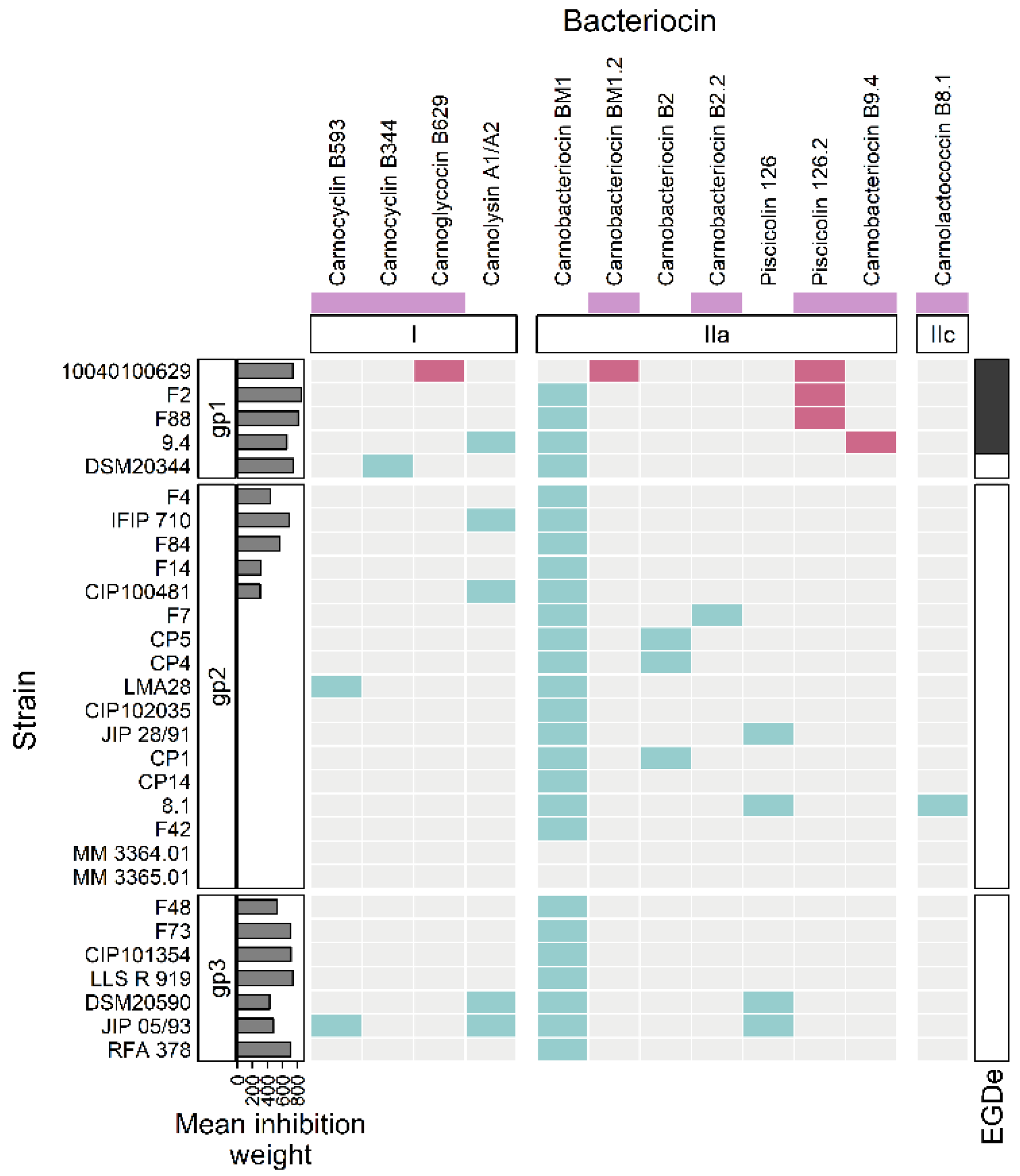


| Class | Family | Name | Strain | |
|---|---|---|---|---|
| I Cyclic peptide | circularin A/uberolysin | Carnocyclin | B593 | JIP 05/93 |
| B344 | DSM20344 | |||
| I Glycocin | Carnoglycocin | B629 | 10040100629 | |
| I Lantibiotic | Two component bacteriocin | Carnolysin | A1 | JIP 05/93; DSM20590; CIP100481; 9.4; and IFIP 710 |
| A2 | ||||
| IIa | pediocin-like | Carnobacteriocin | BM1 | All excluding MM 3364.01; MM 3365.01; and 10040100629 |
| BM1.2 | 10040100629 | |||
| Piscicolin | 126 | DSM20590; JIP 28/91; JIP 05/93; and 8.1 | ||
| 126.2 | 10040100629 | |||
| Carnobacteriocin | B2 | CP1; CP4; and CP5 | ||
| B2.2 | F7 | |||
| Carnobacteriocin | B9.4 | 9.4 | ||
| IIc | Lactococcin 972 | Carnolactococcin | B8.1 | 8.1 |
| Metabolite | Locus Tag | Gene | Putative Function |
|---|---|---|---|
| Carnocyclin B593 | CMALT398_v1_10031/BN424_2338 | cllH | ABC transporter, permease protein |
| CMALT398_v1_10032/BN424_2339 | cclG | Uncharacterized ABC transporter ATP-binding protein YknY | |
| CMALT398_v1_10033/BN424_2340 | cclF | Membrane-fusion protein/RND family efflux transporter | |
| CMALT398_v1_10034/BN424_2341 | cclE | Putative membrane protein | |
| CMALT398_v1_10035/BN424_2342 | Bacteriocin precursor, circularin A/uberolysin family protein | ||
| BN424_2343 | Hypothetical protein | ||
| BN424_2344 | tnp | Transposase | |
| CMALT398_v1_10036 | Membrane protein of unknown function | ||
| CMALT398_v1_10037 | Conserved membrane protein of unknown function | ||
| CMALT398_v1_10038 | ABC-type multidrug transport system, ATPase component/(ABC) transporter | ||
| CMDSM20344_v1_130006 | cclG | Uncharacterized ABC transporter ATP-binding protein YknY | |
| Carnocyclin B344 | CMDSM20344_v1_130007 | Periplasmic component of efflux system | |
| CMDSM20344_v1_130008 | Conserved membrane protein of unknown function | ||
| CMDSM20344_v1_130009 | ABC transporter domain-containing protein | ||
| CMDSM20344_v1_130010 | Conserved membrane protein of unknown function | ||
| CMDSM20344_v1_130011 | Conserved membrane protein of unknown function | ||
| CMDSM20344_v1_130012 | Protein of unknown function | ||
| CMDSM20344_v1_130013 | Bacteriocin precursor, circularin A/uberolysin family protein | ||
| CMDSM20344_v1_130014 | Protein of unknown function | ||
| Carnobacteriocin B9.4 | CMALT94_v1_80008 | Bacteriocin precursor | |
| CMALT94_v1_80009 | putative immunity protein | ||
| Carnoglycocin B629 | CMALT430_v1_150017 | Glycosyltransferases involved in cell wall biogenesis/Glycosyltransferase, family 2 | |
| CMALT430_v1_150018 | Bacteriocin precursor | ||
| CMALT430_v1_150019 | Thiol-disulfide isomerase and thioredoxins, lipoprotein signal peptide | ||
| CMALT430_v1_150020 | Thiol-disulfide isomerase and thioredoxins | ||
| CMALT430_v1_150021 | Peptidase domain-containing ABC transporter | ||
| Carnolactococcin B8.1 | CMALT81_v1_190188 | Bacteriocin ABC transporter ATP-binding protein | |
| CMALT81_v1_190189 | Membrane protein of unknown function | ||
| CMALT81_v1_190190 | Bacteriocin precursor, lipoprotein signal peptide | ||
| NRP-PK compound (ser) + (glu-ser) + (mal) + (X) | CMALT430_v1_70061 | SMCOG1012:4’-phosphopantetheinyl transferase, ACPS | |
| CMALT430_v1_70062 | Protein of unknown function | ||
| CMALT430_v1_70063 | Conserved membrane protein of unknown function, transport | ||
| CMALT430_v1_70064 | putative Surfactin synthase thioesterase subunit, SMCOG1004: thioesterase | ||
| CMALT430_v1_70065 | SMCOG1002: AMP-dependent synthetase and ligase, AMP-binding, PCP, Thioesterase | ||
| CMALT430_v1_70066 | Putative Acyl-CoA dehydrogenase, Acyl-CoA_dh_1 | ||
| CMALT430_v1_70067 | Putative Acyl-CoA dehydrogenase, Acyl-CoA_dh_N; Acyl-CoA_dh_M; Acyl-CoA_dh_1; SMCOG1006:acyl-CoA dehydrogenase | ||
| CMALT430_v1_70068 | Putative predicted aminopeptidases | ||
| CMALT430_v1_70069 | PP-binding, PCP | ||
| CMALT430_v1_70070 | Putative carrier domain-containing protein, SMCOG1022:Beta-ketoacyl synthase, PKS_KS, PCP, Condensation_LCL | ||
| CMALT430_v1_70071 | PP-binding, PCP | ||
| CMALT430_v1_70072 | Condensation_LCL, AMP-binding, PCP, Condensation_LCL, AMP-binding, PCP, Condensation_LCL | ||
| CMALT430_v1_70073 | Condensation_LCL, AMP-binding, PCP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gontijo, M.T.P.; Ramia, N.E.; Dijamentiuk, A.; Elfassy, A.; Taha, S.; Mangavel, C.; Revol-Junelles, A.-M.; Borges, F. Mining Biosynthetic Gene Clusters in Carnobacterium maltaromaticum by Interference Competition Network and Genome Analysis. Microorganisms 2022, 10, 1794. https://doi.org/10.3390/microorganisms10091794
Gontijo MTP, Ramia NE, Dijamentiuk A, Elfassy A, Taha S, Mangavel C, Revol-Junelles A-M, Borges F. Mining Biosynthetic Gene Clusters in Carnobacterium maltaromaticum by Interference Competition Network and Genome Analysis. Microorganisms. 2022; 10(9):1794. https://doi.org/10.3390/microorganisms10091794
Chicago/Turabian StyleGontijo, Marco Túlio Pardini, Nancy E. Ramia, Alexis Dijamentiuk, Annelore Elfassy, Samir Taha, Cécile Mangavel, Anne-Marie Revol-Junelles, and Frédéric Borges. 2022. "Mining Biosynthetic Gene Clusters in Carnobacterium maltaromaticum by Interference Competition Network and Genome Analysis" Microorganisms 10, no. 9: 1794. https://doi.org/10.3390/microorganisms10091794
APA StyleGontijo, M. T. P., Ramia, N. E., Dijamentiuk, A., Elfassy, A., Taha, S., Mangavel, C., Revol-Junelles, A.-M., & Borges, F. (2022). Mining Biosynthetic Gene Clusters in Carnobacterium maltaromaticum by Interference Competition Network and Genome Analysis. Microorganisms, 10(9), 1794. https://doi.org/10.3390/microorganisms10091794







