Polystyrene Degradation by Exiguobacterium sp. RIT 594: Preliminary Evidence for a Pathway Containing an Atypical Oxygenase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media, Chemicals, Bacterial Strains, and Consumables
2.2. Microbial Culture and Isolation of Strains
2.3. PCR Amplification and Nucleotide Sequencing of the 16S V3–V4 Regions
2.4. Whole Genome Sequencing, Strain Identification, and Other Bioinformatics Analysis
2.5. Detection of Polymer Degradation
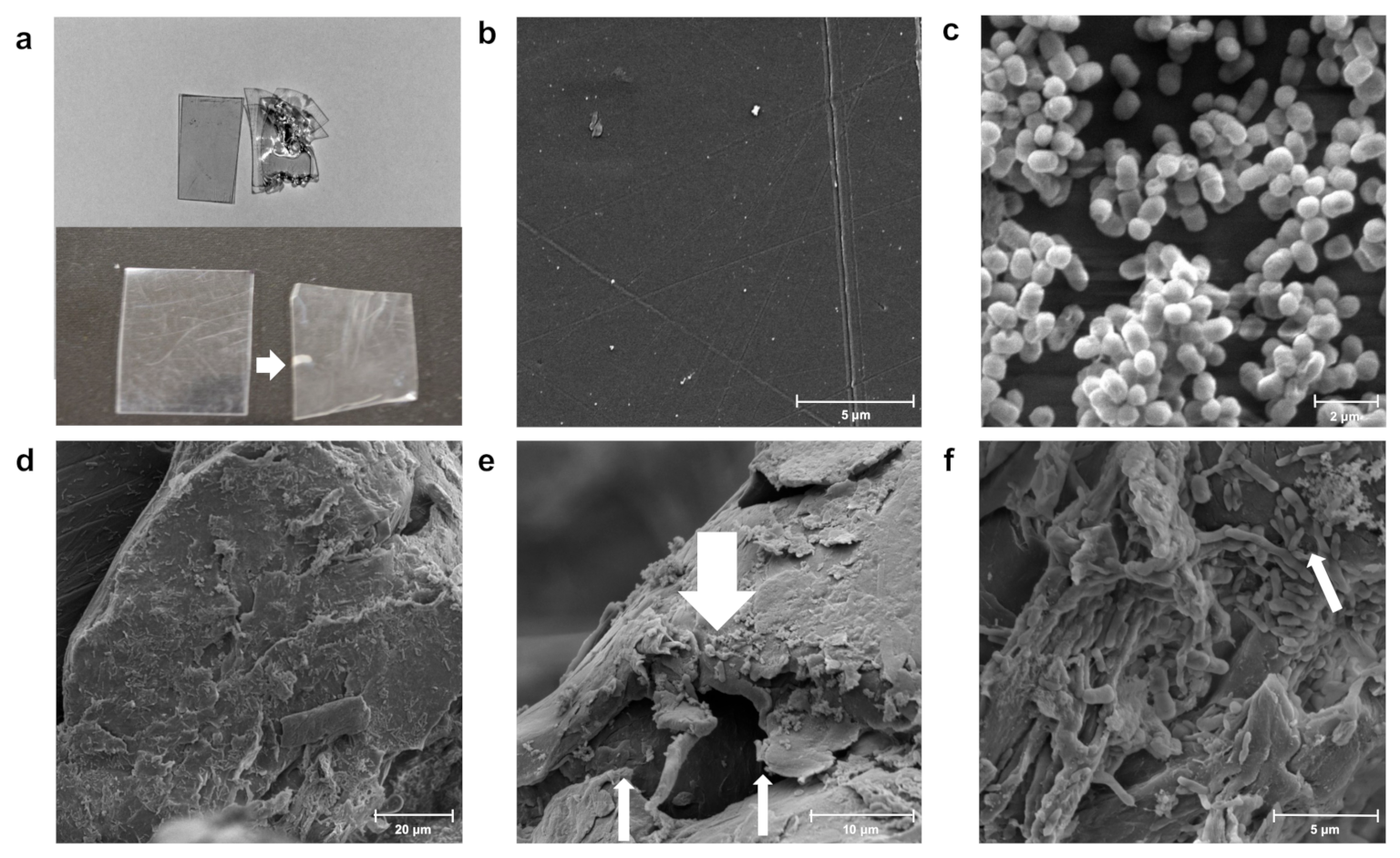
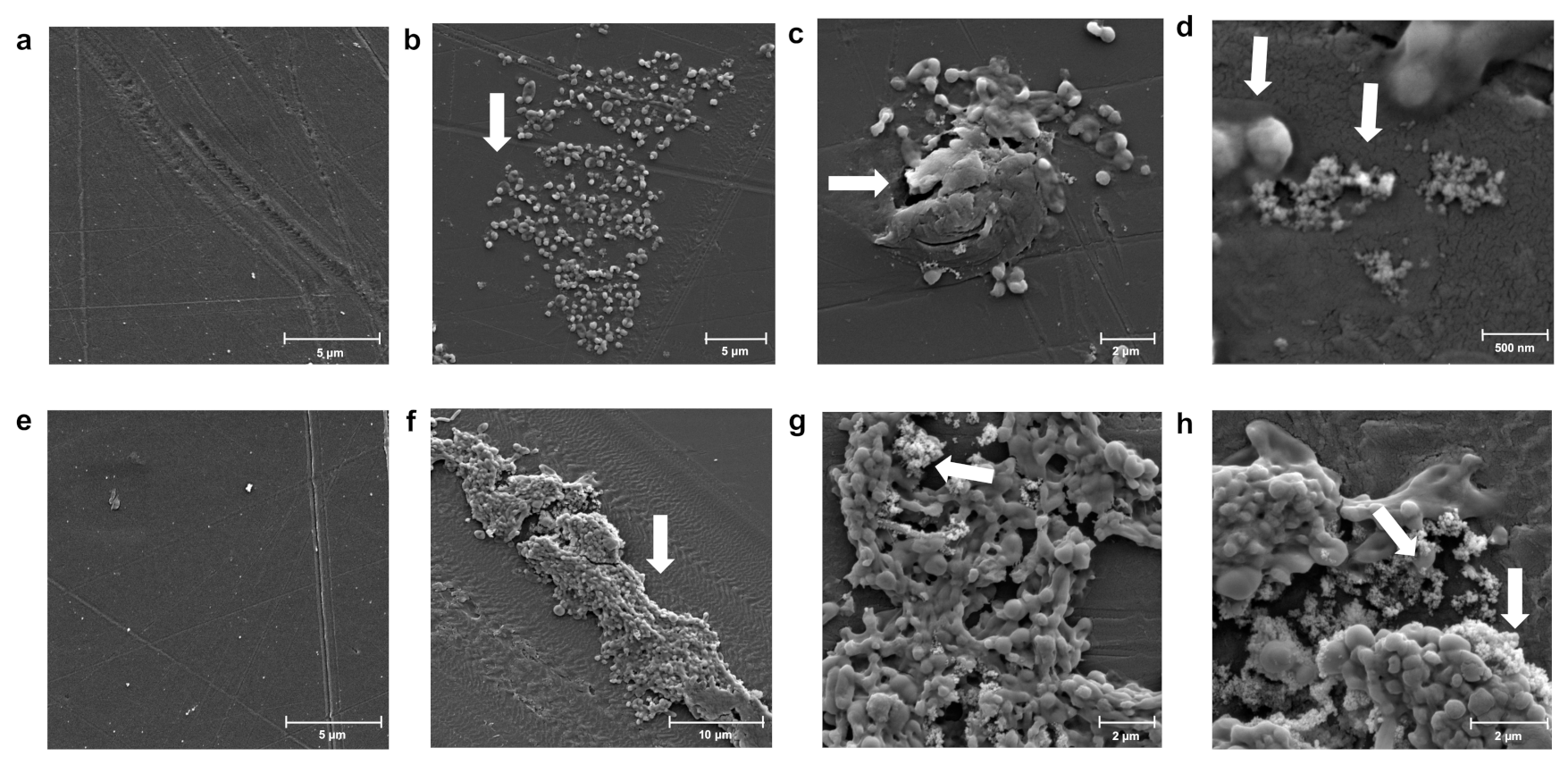
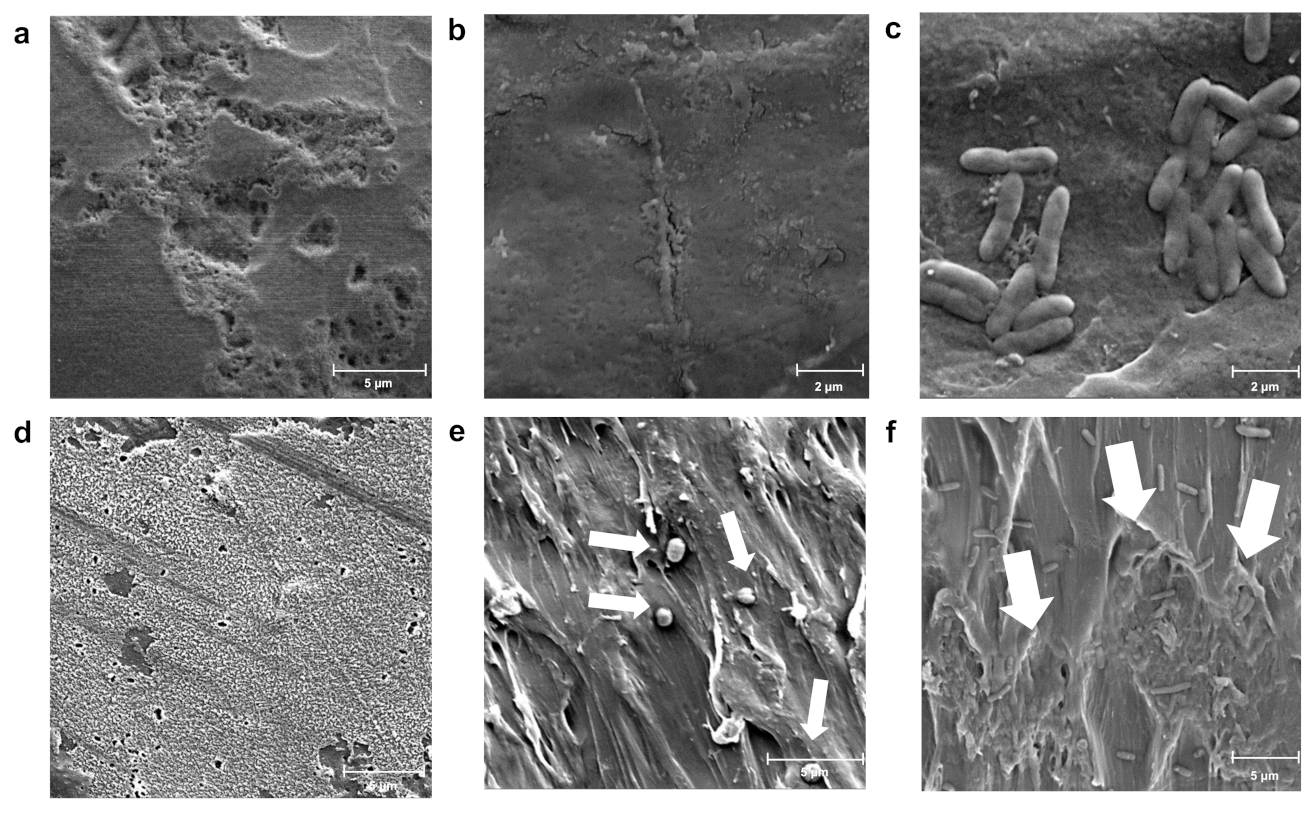
2.6. Scanning Electron Microscopy (SEM)
2.7. Fourier Transform Infrared (FTIR) Spectroscopy
3. Results and Discussion
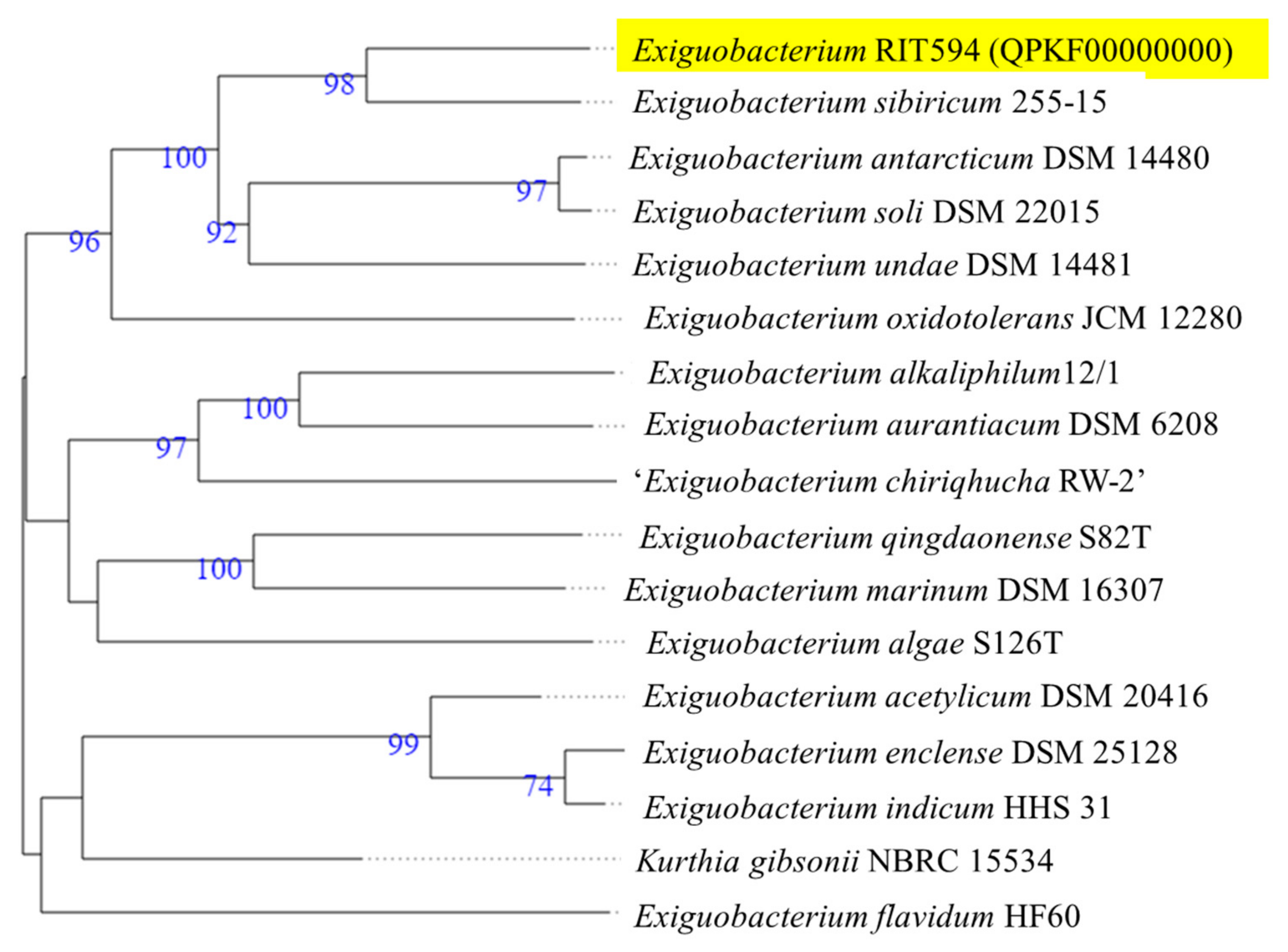
3.1. Strain Identification Using 16S rRNA and Whole Genome-Based Phylogeny
3.2. Detection of Polystyrene (PS) Degradation
3.3. Changes in Morphology during Colonization of Different Polymers
3.4. Chemical Analysis of Biodegraded PS
3.5. The Effects of Oxygen Deprivation
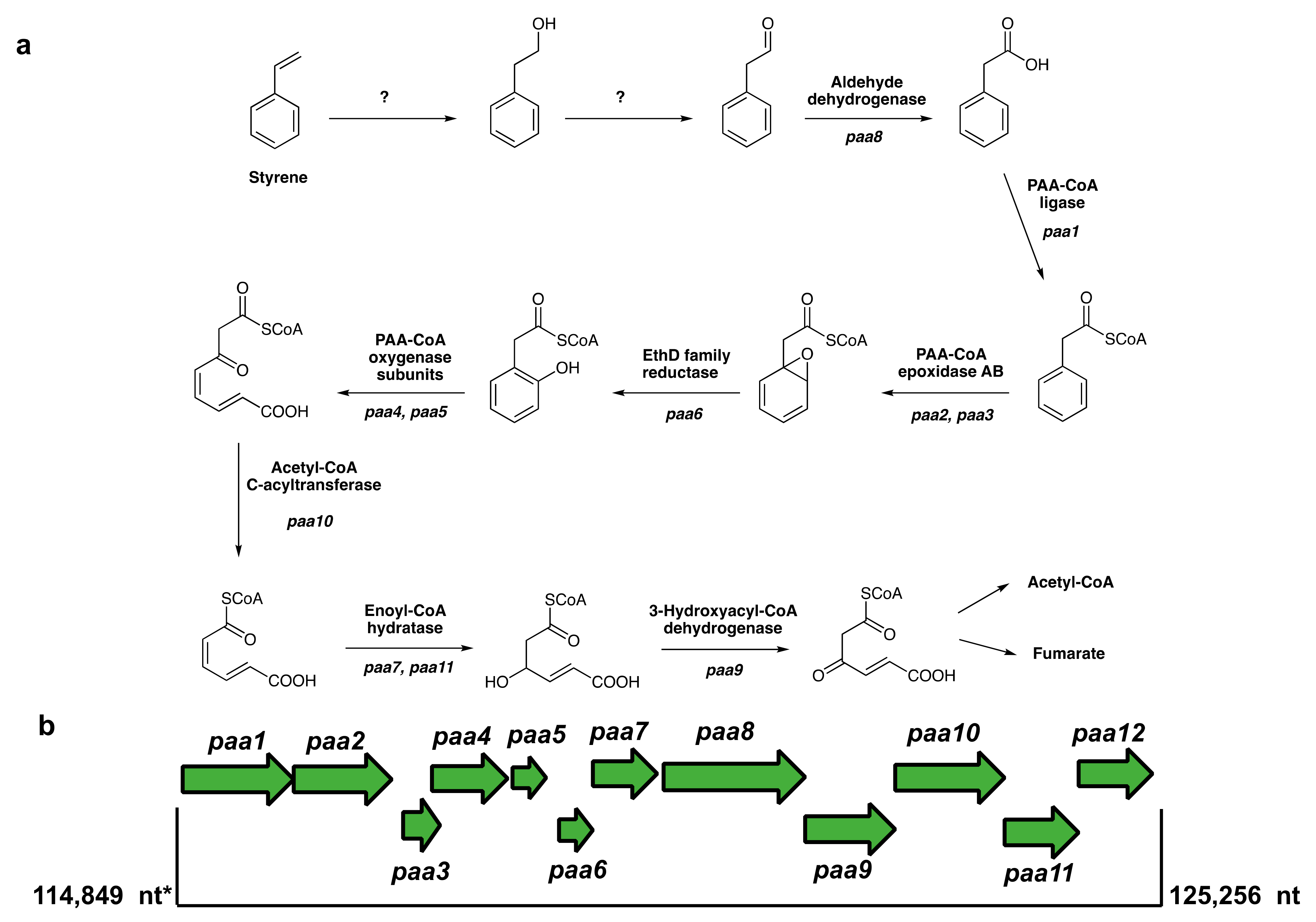
3.6. Putative Pathways for PS Degradation
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A Bacterium That Degrades and Assimilates Poly(Ethylene Terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, R.A.; Aristilde, L. Degradation and Metabolism of Synthetic Plastics and Associated Products by Pseudomonas sp.: Capabilities and Challenges. J. Appl. Microbiol. 2017, 123, 582–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta Lwanga, E.; Thapa, B.; Yang, X.; Gertsen, H.; Salánki, T.; Geissen, V.; Garbeva, P. Decay of Low-Density Polyethylene by Bacteria Extracted from Earthworm’s Guts: A Potential for Soil Restoration. Sci. Total Environ. 2018, 624, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current Status and Application Aspects. ACS Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Chen, C.C.; Dai, L.; Ma, L.; Guo, R.T. Enzymatic Degradation of Plant Biomass and Synthetic Polymers. Nat. Rev. Chem. 2020, 4, 114–126. [Google Scholar] [CrossRef]
- Inderthal, H.; Tai, S.L.; Harrison, S.T.L. Non-Hydrolyzable Plastics—An Interdisciplinary Look at Plastic Bio-Oxidation. Trends Biotechnol. 2021, 39, 12–23. [Google Scholar] [CrossRef]
- Qin, Z.H.; Mou, J.H.; Chao, C.Y.H.; Chopra, S.S.; Daoud, W.; Leu, S.Y.; Ning, Z.; Tso, C.Y.; Chan, C.K.; Tang, S.; et al. Biotechnology of Plastic Waste Degradation, Recycling, and Valorization: Current Advances and Future Perspectives. ChemSusChem 2021, 14, 4103–4114. [Google Scholar] [CrossRef]
- Schilling, C.; Weiss, S. A Roadmap for Industry to Harness Biotechnology for a More Circular Economy. New Biotechnol. 2021, 60, 9–11. [Google Scholar] [CrossRef]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine Learning-Aided Engineering of Hydrolases for PET Depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef]
- Ho, B.T.; Roberts, T.K.; Lucas, S. An Overview on Biodegradation of Polystyrene and Modified Polystyrene: The Microbial Approach. Crit. Rev. Biotechnol. 2018, 38, 308–320. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.; Wu, W.M.; Zhao, J.; Song, Y.; Gao, L.; Yang, R.; Jiang, L. Biodegradation and Mineralization of Polystyrene by Plastic-Eating Mealworms: Part 2. Role of Gut Microorganisms. Environ. Sci. Technol. 2015, 49, 12087–12093. [Google Scholar] [CrossRef]
- Hou, L.; Majumder, E.L.W. Potential for and Distribution of Enzymatic Biodegradation of Polystyrene by Environmental Microorganisms. Materials 2021, 14, 503. [Google Scholar] [CrossRef]
- Tischler, D.; Eulberg, D.; Lakner, S.; Kaschabek, S.R.; Van Berkel, W.J.H.; Schlömann, M. Identification of a Novel Self-Sufficient Styrene Monooxygenase from Rhodococcus Opacus 1CP. J. Bacteriol. 2009, 191, 4996–5009. [Google Scholar] [CrossRef] [Green Version]
- Warhurst, A.M.; Fewson, C.A. Microbial Metabolism and Biotransformations of Styrene. J. Appl. Bacteriol. 1994, 77, 597–606. [Google Scholar] [CrossRef]
- Grbić-Galić, D.; Churchman-Eisel, N.; Aković, I. Microbial Transformation of Styrene by Anaerobic Consortia. J. Appl. Bacteriol. 1990, 69, 247–260. [Google Scholar] [CrossRef]
- O’Leary, N.D.; O’Connor, K.E.; Dobson, A.D.W. Biochemistry, Genetics and Physiology of Microbial Styrene Degradation. FEMS Microbiol. Rev. 2002, 26, 403–417. [Google Scholar] [CrossRef] [Green Version]
- Warhurst, A.M.; Clarke, K.F.; Hill, R.A.; Holt, R.A.; Fewson, C.A. Metabolism of Styrene by Rhodococcus Rhodochrous NCIMB 13259. Appl. Environ. Microbiol. 1994, 60, 1137–1145. [Google Scholar] [CrossRef] [Green Version]
- Ward, P.G.; Goff, M.; Donner, M.; Kaminsky, W.; O’Connor, K.E. A Two Step Chemo-Biotechnological Conversion of Polystyrene to a Biodegradable Thermoplastic. Environ. Sci. Technol. 2006, 40, 2433–2437. [Google Scholar] [CrossRef]
- Johnston, B.; Radecka, I.; Hill, D.; Chiellini, E.; Ilieva, V.I.; Sikorska, W.; Musioł, M.; Ziȩba, M.; Marek, A.A.; Keddie, D.; et al. The Microbial Production of Polyhydroxyalkanoates from Waste Polystyrene Fragments Attained Using Oxidative Degradation. Polymers 2018, 10, 957. [Google Scholar] [CrossRef] [Green Version]
- Krueger, M.C.; Seiwert, B.; Prager, A.; Zhang, S.; Abel, B.; Harms, H.; Schlosser, D. Degradation of Polystyrene and Selected Analogues by Biological Fenton Chemistry Approaches: Opportunities and Limitations. Chemosphere 2017, 173, 520–528. [Google Scholar] [CrossRef]
- Mohan, A.J.; Sekhar, V.C.; Bhaskar, T.; Nampoothiri, K.M. Microbial Assisted High Impact Polystyrene (HIPS) Degradation. Bioresour. Technol. 2016, 213, 204–207. [Google Scholar] [CrossRef]
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Screening of Bacillus Strains Isolated from Mangrove Ecosystems in Peninsular Malaysia for Microplastic Degradation. Environ. Pollut. 2017, 231, 1552–1559. [Google Scholar] [CrossRef]
- Yang, S.S.; Brandon, A.M.; Andrew Flanagan, J.C.; Yang, J.; Ning, D.; Cai, S.Y.; Fan, H.Q.; Wang, Z.Y.; Ren, J.; Benbow, E.; et al. Biodegradation of Polystyrene Wastes in Yellow Mealworms (Larvae of Tenebrio Molitor Linnaeus): Factors Affecting Biodegradation Rates and the Ability of Polystyrene-Fed Larvae to Complete Their Life Cycle. Chemosphere 2018, 191, 979–989. [Google Scholar] [CrossRef]
- Chauhan, D.; Agrawal, G.; Deshmukh, S.; Roy, S.S.; Priyadarshini, R. Biofilm Formation by Exiguobacterium Sp. DR11 and DR14 Alter Polystyrene Surface Properties and Initiate Biodegradation. RSC Adv. 2018, 8, 37590–37599. [Google Scholar] [CrossRef] [Green Version]
- Bruckner, J.; Osman, S.; Venkateswaran, K.; Conley, C. Space Microbiology: Planetary Protection, Burden, Diversity and Significance of Spacecraft Associated Microbes. Encycl. Microbiol. 2009, 3, 52–65. [Google Scholar] [CrossRef]
- Chauhan, H.; Bagyaraj, D.J.; Selvakumar, G.; Sundaram, S.P. Novel Plant Growth Promoting Rhizobacteria-Prospects and Potential. Appl. Soil Ecol. 2015, 95, 38–53. [Google Scholar] [CrossRef]
- Kumar, V.; Kumari, A.; Pandey, M.; Sharma, M. Molecular Mechanism of Radio-Resistance and Heavy Metal Tolerance Adaptation in Microbes. In Microbial Extremozymes; Academic Press: Cambridge, MA, USA, 2022; pp. 275–293. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Chauhan, A.; Layton, A.C.; Pfiffner, S.M.; Huntemann, M.; Copeland, A.; Chen, A.; Kyrpides, N.C.; Markowitz, V.M.; Palaniappan, K.; et al. Draft Genome Sequences of 10 Strains of the Genus Exiguobacterium. Genome Announc. 2014, 2, e01058-14. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.R.; Cook, G.M. Isolation and Characterization of Arsenate-Reducing Bacteria from Arsenic-Contaminated Sites in New Zealand. Curr. Microbiol. 2004, 48, 341–347. [Google Scholar] [CrossRef]
- Ordoñez, O.F.; Rasuk, M.C.; Soria, M.N.; Contreras, M.; Farías, M.E. Haloarchaea from the Andean Puna: Biological Role in the Energy Metabolism of Arsenic. Microb. Ecol. 2018, 76, 695–705. [Google Scholar] [CrossRef]
- Belfiore, C.; Ordoñez, O.F.; Farías, M.E. Proteomic Approach of Adaptive Response to Arsenic Stress in Exiguobacterium Sp. S17, an Extremophile Strain Isolated from a High-Altitude Andean Lake Stromatolite. Extremophiles 2013, 17, 421–431. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, V.; Kumar, R. Characterization of an Alkaliphile, Exiguobacterium Sp. and It’s Application in Bioremediation. In Proceedings of the International Conference on Extremophiles, Brest, France, 17–21 September 2006. [Google Scholar]
- López, L.; Pozo, C.; Rodelas, B.; Calvo, C.; Juárez, B.; Martínez-Toledo, M.V.; González-López, J. Identification of Bacteria Isolated from an Oligotrophic Lake with Pesticide Removal Capacities. Ecotoxicology 2005, 14, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Pattanapipitpaisal, P.; Mabbett, A.N.; Finlay, J.A.; Beswick, A.J.; Paterson-Beedle, M.; Essa, A.; Wright, J.; Tolley, M.R.; Badar, U.; Ahmed, N.; et al. Reduction of Cr(VI) and Bioaccumulation of Chromium by Gram Positive and Gram Negative Microorganisms Not Previously Exposed to Cr-Stress. Environ. Technol. 2002, 23, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Okeke, B.C. Bioremoval of Hexavalent Chromium from Water by a Salt Tolerant Bacterium, Exiguobacterium sp. GS1. J. Ind. Microbiol. Biotechnol. 2008, 35, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Petrova, M.A.; Mindlin, S.Z.; Gorlenko, Z.M.; Kaliaeva, E.S.; Soina, V.S.; Bogdanova, E.S. Mercury-Resistant Bacteria from Permafrost Sediments and Prospects for Their Use in Comparative Studies of Mercury Resistance Determinants. Genetika 2002, 38, 1569–1574. [Google Scholar]
- Amaresan, N.; Senthil Kumar, M.; Annapurna, K.; Kumar, K.; Sankaranarayanan, A. Beneficial Microbes in Agro-Ecology; Academic Press: Cambridge, MA, USA, 2020; ISBN 9780128234143. [Google Scholar]
- Yumoto, I.; Hishinuma-Narisawa, M.; Hirota, K.; Shingyo, T.; Takebe, F.; Nodasaka, Y.; Matsuyama, H.; Hara, I. Exiguobacterium oxidotolerans sp. nov., a Novel Alkaliphile Exhibiting High Catalase Activity. Int. J. Syst. Evol. Microbiol. 2004, 54, 2013–2017. [Google Scholar] [CrossRef] [Green Version]
- Takebe, F.; Hara, I.; Matsuyama, H.; Yumoto, I. Effects of H2O2 under Low- and High-Aeration-Level Conditions on Growth and Catalase Activity in Exiguobacterium oxidotolerans T-2-2T. J. Biosci. Bioeng. 2007, 104, 464–469. [Google Scholar] [CrossRef]
- Usuda, Y.; Kawasaki, H.; Shimaoka, M.; Utagawa, T. Molecular Characterization of Guanosine Kinase Gene from a Facultative Alkalophile, Exiguobacterium aurantiacum ATCC 35652. Biochim. Biophys. Acta 1998, 1442, 373–379. [Google Scholar] [CrossRef]
- Suga, S.; Koyama, N. Purification and Properties of a Novel Azide-Sensitive ATPase of Exiguobacterium aurantiacum. Arch. Microbiol. 2000, 173, 200–205. [Google Scholar] [CrossRef]
- Kasana, R.C.; Pandey, C.B. Exiguobacterium: An Overview of a Versatile Genus with Potential in Industry and Agriculture. Crit. Rev. Biotechnol. 2017, 38, 141–156. [Google Scholar] [CrossRef]
- Molina, G.; De Lima, E.A.; Borin, G.P.; De Barcelos, M.C.S.; Pastore, G.M. Beta-Glucosidase from Penicillium. In New and Future Developments in Microbial Biotechnology and Bioengineering Penicillum System Properties and Applications; Elsevier: Amsterdam, The Netherlands, 2017; pp. 137–151. [Google Scholar] [CrossRef]
- Koul, B.; Chaudhary, R.; Taak, P. Extremophilic Microbes and Their Application in Bioremediation of Environmental Contaminants. In Microbe Mediated Remediation of Environmental Contaminants; Woodhead Publishing: Sawston, UK, 2021; pp. 115–128. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Hakeem, K.R.; Öztürk, M.; Fujita, M. Arsenic Toxicity in Plants and Possible Remediation. In Soil Remediation and Plants: Prospects and Challenges; Academic Press: Cambridge, MA, USA, 2015; pp. 433–501. [Google Scholar] [CrossRef]
- Etesami, H. Plant-Microbe Interactions in Plants and Stress Tolerance. In Plant Life Under Changing Environment; Academic Press: Cambridge, MA, USA, 2020; pp. 355–396. [Google Scholar] [CrossRef]
- Tripathi, P.; Singh, A. Biofertilizers: “An Ace in the Hole” in Medicinal and Aromatic Plants Cultivation. In Biofertilizers; Woodhead Publishing: Sawston, UK, 2021; pp. 253–263. [Google Scholar] [CrossRef]
- Wu, W.M.; Criddle, C.S. Characterization of Biodegradation of Plastics in Insect Larvae. Methods Enzymol. 2021, 648, 95–120. [Google Scholar] [CrossRef]
- Pandey, N. Exiguobacterium. In Beneficial Microbes in Agro-Ecology; Academic Press: Cambridge, MA, USA, 2020; pp. 169–183. [Google Scholar] [CrossRef]
- López, M.C.; Galán, B.; Carmona, M.; Llorens, J.M.N.; Peretó, J.; Porcar, M.; Getino, L.; Olivera, E.R.; Luengo, J.M.; Castro, L.; et al. Xerotolerance: A New Property in Exiguobacterium Genus. Microorganisms 2021, 9, 2455. [Google Scholar] [CrossRef]
- Przemieniecki, S.W.; Kosewska, A.; Ciesielski, S.; Kosewska, O. Changes in the Gut Microbiome and Enzymatic Profile of Tenebrio Molitor Larvae Biodegrading Cellulose, Polyethylene and Polystyrene Waste. Environ. Pollut. 2020, 256, 113265. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Steiner, K.K.; Parthasarathy, A.; Wong, N.H.; Cavanaugh, N.T.; Chu, J.; Hudson, A.O. Isolation and Whole-Genome Sequencing of Pseudomonas Sp. RIT 623, a Slow-Growing Bacterium Endowed with Antibiotic Properties. BMC Res. Notes 2020, 13, 370. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Type Strain Genome Server. Available online: https://tygs.dsmz.de/ (accessed on 5 July 2022).
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Parthasarathy, A. Scanning Electron Microscopy (SEM) for Microbes—A Simple and Inexpensive Method for Sample Preparation; ResearchGate: Berlin, Germany, 2019. [Google Scholar]
- ASTM G22: Standard Practice for Determining Resistance of Plastics to Bacteria. Available online: https://global.ihs.com/doc_detail.cfm?document_name=ASTMG22&item_s_key=00021438 (accessed on 11 April 2022).
- Lambert, S.; Wagner, M. Formation of Microscopic Particles during the Degradation of Different Polymers. Chemosphere 2016, 161, 510–517. [Google Scholar] [CrossRef]
- Kongpol, A.; Kato, J.; Tajima, T.; Vangnai, A.S. Characterization of Acetonitrile-Tolerant Marine Bacterium Exiguobacterium sp. SBH81 and Its Tolerance Mechanism. Microbes Environ. 2012, 27, 30–35. [Google Scholar] [CrossRef] [Green Version]
- Tischler, D. Pathways for the Degradation of Styrene; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Oelschlägel, M.; Zimmerling, J.; Tischler, D. A Review: The Styrene Metabolizing Cascade of Side-Chain Oxygenation as Biotechnological Basis to Gain Various Valuable Compounds. Front. Microbiol. 2018, 9, 490. [Google Scholar] [CrossRef]
- Otto, K.; Hofstetter, K.; Röthlisberger, M.; Witholt, B.; Schmid, A. Biochemical Characterization of StyAB from Pseudomonas Sp. Strain VLB120 as a Two-Component Flavin-Diffusible Monooxygenase. J. Bacteriol. 2004, 186, 5292–5302. [Google Scholar] [CrossRef] [Green Version]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial Phenylalanine and Phenylacetate Catabolic Pathway Revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef] [Green Version]
- Luengo, J.M.; García, J.L.; Olivera, E.R. The Phenylacetyl-CoA Catabolon: A Complex Catabolic Unit with Broad Biotechnological Applications. Mol. Microbiol. 2001, 39, 1434–1442. [Google Scholar] [CrossRef]
- Sánchez-Reyez, A.; Batista-García, R.A.; Valdés-García, G.; Ortiz, E.; Perezgasga, L.; Zárate-Romero, A.; Pastor, N.; Folch-Mallol, J.L. A Family 13 Thioesterase Isolated from an Activated Sludge Metagenome: Insights into Aromatic Compounds Metabolism. Proteins Struct. Funct. Bioinf. 2017, 85, 1222–1237. [Google Scholar] [CrossRef]
- Chauvaux, S.; Chevalier, F.; Le Dantec, C.; Fayolle, F.; Miras, I.; Kunst, F.; Beguin, P. Cloning of a Genetically Unstable Cytochrome P-450 Gene Cluster Involved in Degradation of the Pollutant Ethyl Tert-Butyl Ether by Rhodococcus Ruber. J. Bacteriol. 2001, 183, 6551–6557. [Google Scholar] [CrossRef] [Green Version]
- Sielicki, M.; Focht, D.D.; Martin, J.P. Microbial Transformations of Styrene and [14C] Styrene in Soil and Enrichment Cultures. Appl. Environ. Microbiol. 1978, 35, 124. [Google Scholar] [CrossRef] [Green Version]
- Collins, M.D.; Lund, B.M.; Farrow, J.A.E.; Schleifer, K.H. Chemotaxonomic Study of an Alkalophilic Bacterium, Exiguobacterium aurantiacum gen. nov., sp. nov. Microbiology 1983, 129, 2037–2042. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, L.; Zhou, J.; Wu, H.; Li, D.; Cui, Y.; Lu, B. Exiguobacterium sp. A1b/GX59 Isolated from a Patient with Community-Acquired Pneumonia and Bacteremia: Genomic Characterization and Literature Review. BMC Infect. Dis. 2017, 17, 508. [Google Scholar] [CrossRef] [Green Version]



| Enzyme Annotations | Genome Location (nt) | Protein ID | Identity to R. opacus SMO1 (Query Cover) | Identity to R. opacus SMO2 (Query Cover) | Identity to R. opacus SOI (Query Cover) |
|---|---|---|---|---|---|
| Antibiotic biosynthesis monooxygenase | 130,054–130,347 | RDB32576.1 | - | - | - |
| Heme oxygenase | 173,723–174,055 | RDB32997.1 | - | - | - |
| Monooxygenase, FAD-dependent monooxygenase | 851,364–852,938 | RDB33898.1 | 35% (5%) | - | 31.88% (14%) |
| Antibiotic biosynthesis monooxygenase | 1,100,990–1,101,292 | RDB34148.1 | - | - | - |
| Antibiotic biosynthesis monooxygenase | 130,054–130,347 | RDB32576.1 | - | - | - |
| Heme oxygenase | 173,723–174,055 | RDB32997.1 | - | - | - |
| Monooxygenase, FAD-dependent monooxygenase | 851,364–852,938 | RDB33898.1 | 35% (5%) | - | 31.88% (14%) |
| Antibiotic biosynthesis monooxygenase | 1,100,990–1,101,292 | RDB34148.1 | - | - | - |
| Antibiotic biosynthesis monooxygenase | 130,054–130,347 | RDB32576.1 | - | - | - |
| Heme oxygenase | 173,723–174,055 | RDB32997.1 | - | - | - |
| Enzyme Annotations | Genome Location (nt) | Protein ID | Identity to P. putida DO (Query Cover) | Identity to R. rhodocrous CatA (Query Cover) | Identity to R. rhodocrous Extradiol DO (Query Cover) | Identity to C. necator DO (Query Cover) |
|---|---|---|---|---|---|---|
| Ring-cleaving dioxygenase | 126,220–127,185 | RDB32955.1 | - | - | 26.92% (8%) | - |
| Ring-cleaving dioxygenase, VOC family protein | 139,158–140,096 | RDB33200.1 | - | - | 40.74% (8%) | - |
| VOC family protein, Glyoxalase, Extradiol dioxygenase, Bleomycin resistance protein | 1,198,498–1,198,878 | RDB34253.1 | - | - | 23.40% (14%) | - |
| VOC family protein, glyoxalase | 1,244,862–1,245,542 | RDB34313.1 | - | - | - | - |
| Ring-cleaving dioxygenase | 1,423,920–1,424,903 | RDB34501.1 | - | - | 26.92% (15%) | - |
| Alpha/beta hydrolase, Dienelactone hydrolase family protein | 1,066,665–1,067,267 | WP_158537521.1 | ||||
| Esterase/lipase | 1,422,520–1,423,257 | RDB34499.1 | ||||
| Fumarylacetoacetate (FAA) hydrolase family protein, 2-Keto-4-pentenoate hydratase, 2-oxohepta-3-ene-1,7-dioic acid hydratase (catechol pathway) | 1,490,795–1,491,628 | RDB34570.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parthasarathy, A.; Miranda, R.R.; Eddingsaas, N.C.; Chu, J.; Freezman, I.M.; Tyler, A.C.; Hudson, A.O. Polystyrene Degradation by Exiguobacterium sp. RIT 594: Preliminary Evidence for a Pathway Containing an Atypical Oxygenase. Microorganisms 2022, 10, 1619. https://doi.org/10.3390/microorganisms10081619
Parthasarathy A, Miranda RR, Eddingsaas NC, Chu J, Freezman IM, Tyler AC, Hudson AO. Polystyrene Degradation by Exiguobacterium sp. RIT 594: Preliminary Evidence for a Pathway Containing an Atypical Oxygenase. Microorganisms. 2022; 10(8):1619. https://doi.org/10.3390/microorganisms10081619
Chicago/Turabian StyleParthasarathy, Anutthaman, Renata Rezende Miranda, Nathan C. Eddingsaas, Jonathan Chu, Ian M. Freezman, Anna C. Tyler, and André O. Hudson. 2022. "Polystyrene Degradation by Exiguobacterium sp. RIT 594: Preliminary Evidence for a Pathway Containing an Atypical Oxygenase" Microorganisms 10, no. 8: 1619. https://doi.org/10.3390/microorganisms10081619
APA StyleParthasarathy, A., Miranda, R. R., Eddingsaas, N. C., Chu, J., Freezman, I. M., Tyler, A. C., & Hudson, A. O. (2022). Polystyrene Degradation by Exiguobacterium sp. RIT 594: Preliminary Evidence for a Pathway Containing an Atypical Oxygenase. Microorganisms, 10(8), 1619. https://doi.org/10.3390/microorganisms10081619







