Abstract
Programmed cell death (PCD) is the result of an intracellular program and is accomplished by a regulated process in both prokaryotic and eukaryotic organisms. Here, we report a programed cell death process in Mycobacterium smegmatis, an Actinobacteria species which involves a transcription factor and a DNase of the HNH family. We found that over-expression of an ArsR family member of the transcription factor, MSMEG_6762, leads to cell death. Transcriptome analysis revealed an increase in the genes’ transcripts involved in DNA repair and homologous recombination, and in three members of HNH family DNases. Knockout of one of the DNase genes, MSMEG_1275, alleviated cell death and its over-expression of programmed cell death. Purified MSMEG_1275 cleaved the M. smegmatis DNA at multiple sites. Overall, our results indicate that the MSMEG_6762 affects cell death and is mediated, at least partially, by activation of the HNH nuclease expression under a stress condition.
1. Introduction
Programmed cell death (PCD) refers to a genetically regulated process that leads to cell suicide [1]. It is an essential mechanism in the development and homeostasis of multicellular organisms, and is beneficial to bacterial populations and genomes [2]. Recently, PCD systems have also been found in eubacteria, which play a key role in the survival of the population under environmental stresses, such as nutrients deprivation and antibiotics treatments [2,3,4].
A toxin–antitoxin (TA) system has been studied extensively as a mechanism for bacterial PCD [5,6]. TA systems usually consist of two genes encoding a toxin and an antitoxin that counteracts the lethal action of toxin [7,8,9]. A well-characterized chromosomal TA system involved in bacterial PCD is MazEF of Escherichia coli [10,11]. MazF, the toxin, is a sequence-specific endoribonuclease that cleaves mRNAs at ACA or ACU sites in E. coli [12]. The cleavage of mRNAs blocks protein synthesis for metabolism and survival and halts cell proliferation. YihE Kinase was identified as a central regulator of bacterial cell death mediated by the MazEF [13]. Several investigations have revealed that some stress conditions trigger the mazEF PCD system, including starvation [14,15], antibiotics [16,17], high temperature [17], DNA damage [17,18], and oxidative stress [17]. However, the MazEF-mediated PCD in E. coli was controversial due to its reproducibility [19,20,21]. PezAT, a member of type II TA system, and the toxin protein PezT can phosphorylate the ubiquitous peptidoglycan precursor uridine diphosphate-N-acetylglucosamine (UNAG), which inhibits the peptidoglycan synthesis and leads to cell death eventually [22]. Programmed bacterial deaths could also be induced by a variety of restriction–modification (RM) systems. RM systems commonly contain a modification enzyme capable of methylating specific DNA sequences in genomes and a restriction endonuclease capable of cleaving DNA lacking those methylations [23]. The PCD was also found in the sporulating bacteria, Bacillus subtilis. Under nutrient-limited conditions, the spore formation-related regulatory protein Spo0A regulates the sporulating killing factor skfA-H and sporulating delay protein sdpABC operons, which decided the fate of these bacteria—to live or to die [24,25].
MSMEG_6762 is an ArsR family transcriptional regulator abundant in mycobacteria and other bacterial species, such as Staphylococcus aureus, Shigella sonnei, Weissela cibaria, and Klebsiella pneumoniae. The N-terminus of the ArsR family of transcription regulators contains a DNA-binding domain, which binds downstream promoters of target genes to regulate transcription [26]. Generally, ArsR family transcriptional regulators act as metal sensors and modulate the transcription of genes related to metal ion stress [26,27]. Additionally, some ArsR-type regulators, such as HlyU [28,29], SloR [30] and PagR [31], are involved in bacterial pathogenesis.
In this work, we found that the overexpression of MSMEG_6762 leads to cell death. MSMEG_6762 regulates the expression of HNH nuclease MSMEG_1275, which degrades DNA and eventually causes cell death. Knocking out MSMEG_1275 relieved the bactericidal activity of MSMEG_6762. The study found a new PCD in M. smegmatis, which is associated with an ArsR family regulator and HNH nuclease cascade, and which constitutes a live-or-die response decision.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Growth Conditions
The M. smegmatis and E. coli strains and plasmids used in this study are shown in Table S1. The E. coli strain DH5α was used for cloning. E. coli strains were grown on LB broth agar or in LB broth, 37 °C, 200 rpm. M. smegmatis mc2 155 was grown in 7H9 liquid medium (Difco) supplemented with 0.05% w/v Tween 80, 0.5% glycerol, and 0.5% glucose or was grown on 7H10 agar supplemented with 1% glycerol and 0.5% glucose. Restriction enzymes, T4 DNA ligases, and DNA polymerases were purchased from Takara. Ampicillin, kanamycin, hygromycin were bought from Sangon Biotech Co., whose stock solutions were freshly prepared and filter sterilized. When required, the following antibiotics were used at the final concentration: ampicillin, 100 μg/mL; kanamycin, 500 μg/mL for E. coli or 200 μg/mL for M. smegmatis; hygromycin, 50 μg/mL. All cultures were incubated at 37 °C.
2.2. Plasmids and Expression Strains Construction
Mycobacterial expression vector pALACE used in this study has been described previously [32], and all primers used in this study are shown in Supplemental Table S2. The coding sequence of MSMEG_6760 was amplified with primer pair 6760-F/-R, the coding sequence of MSMEG_6762 was amplified with primer pair 6762-F/R, and the coding sequence of MSMEG_6760-MSMEG_6762 was amplified with primer pair 6760-6762-F/-R. All were cloned as BamH I/EcoR I fragments into correspondingly digested pALACE to form pALACE-6760, pALACE-6762 and pALACE-6760-6762. MSMEG_5583, MSMEG_1756, MSMEG_5876, MSMEG_3404, MSMEG_1275, and MSMEG_2148 were amplified with their own primers, and subsequently cloned as Afl II/Nde I fragments into correspondingly digested pALACE to form pALACE-5583, pALACE-1756, pALACE-5876, pALACE-3404, pALACE-1275, and pALACE-2148, respectively. These plasmids were then electroporated into M. smegmatis mc2 155 to generate overexpression strains, respectively.
2.3. Effect of Conditional Expression of Genes on Mycobacterial Growth and Viability
Both solid medium and liquid culture were used to test the toxicity of target genes in M. smegmatis. To assay the effect of expression on solid medium, strains were grown in 7H9 media to an OD600 of approximately 1.0. Each strain was streaked onto an agar plate supplemented with hygromycin, with or without 1% acetamide to induce expression of either the target genes. After 3 days of growth at 37 °C, pictures of the plates were taken by an image analysis system (Furi science & technology Co., Ltd., Shanghai, China). For toxicity assessment in liquid culture, strains were grown in 7H9 media to an OD600 = 0.1 as the expression was induced with 1% acetamide, with OD600 and CFU measured over time. Each experiment was performed in triplicate at each time point.
For the Western blot detection of His-tagged MSMEG_6762 and MSMEG_6760, bacterial pellets were harvested and disrupted by ultrasonication. Samples were subjected to SDS-PAGE, and the His-tagged proteins were detected by the mouse anti-His antibody (TIANGEN, Beijing, China).
2.4. Site-Directed Mutagenesis
The sequences of all primers used in site-directed mutagenesis are shown in Table S2. Four conserved sites were introduced into the MSMEG_6762 by site-directed mutagenesis [33] with the recombinant vectors T-MSMEG_6762 isolated from E. coli DH5α as a template. The mutagenic primers are shown in Table S2. Mutation of 18L (CTC) to A (GTA) used primer pair 6762 L18A-F/-R, 24R (AGG) to A (GTA) used primer pair 6762 R24A-F/-R, 54H (CAT) to A (GTA) used primer pair 6762 H54A, and 58 L (CTC) to A (GTA) used primer pair 6762 L58A-F/-R, respectively. The mutations were confirmed by DNA sequencing using primers 6762-F and 6762-R. For MSMEG_1275, mutation of 258H (CAT) to A (GTA) used primer pair 1275 H258A-F/-R, 272N (AAC) to A (GTA) used primer pair 1275 N272A-F/-R, and 281N (AAC) to A (GTA) used primer pair 1275 N281A-F/-R, respectively.
2.5. RNA-Seq Analysis
For RNA-Seq analysis, MS-VEC and MS-6762 were grown to a turbidity of 0.4, and then the final concentration of 1% acetamide was added to induce MSMEG_6762. After induction for 9 h, cells were harvested. The total amount of RNA was extracted using RNeasy Mini Kit (Qiagen, GmBH, Hilden, Germany) following the manufacturer’s instructions and was checked for a RIN number to inspect RNA integrity by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, USA). Qualified RNA from the previous steps was further purified by Rneasy micro kit (Qiagen, GmBH, Hilden, Germany) and Rnase-Free Dnase Set (Qiagen, GmBH, Hilden, Germany). RNA-Seq was performed by Shanghai Biochip Inc. Results were analyzed in edgeR [34] with Significance Analysis of Microarrays considered significant at q < 0.05.
2.6. Terminal Deoxynucleotidyl Transferase dUTP Nick end Labeling (TUNEL) Assay
Stationary cultures of MS-VEC, MS-6762 and MS-1275 were reinoculated into 7H9 medium, and acetamide was added into the cultures while the OD600 reached 0.1. Aliquots of mycobacterial cells were collected from M. smegmatis cultures induced for 8 h, and the TUNEL assay was performed according to the in situ cell death detection kit (Roche Diagnostics, Indianapolis, IN, USA) instruction. Samples were analyzed by FACS; the FITC signal was analyzed with an emitting laser at 488 nm and bandpass filter of 525/15 nm using a BD Aria II flow cytometer (BD Biosciences, San Jose, CA, USA) with a 70-μm nozzle. For each sample, 10,000 events were acquired, with TUNEL staining gradations expressed as percentages of total gated cells.
2.7. Protein Expression and Purification
The sequences of all primers used in protein expression and purification are shown in Table S2. Recombinant MSMEG_6762 and MSMEG_1275 were expressed in E. coli according to a published protocol [35]. Briefly, the MSMEG_6762 and MSMEG_1275 coding region were amplified by PCR from the genomic DNA of M. smegmatis using pET6762F and pET6762R, or pET1275F and pET1275R. The gene was cloned into pET28a expression vector, E. coli BL21 cells carrying recombinant plasmids were induced with 1 mM IPTG (isopropyl β-D-thiogalactopyranoside), and the bacteria were incubated for 4 h, at 37 °C. Cell lysates were prepared by sonication, and His-MSMEG_6762 was purified by binding to Ni-NTA (GenScript, Tokyo, Japan) equilibrated with wash Buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0), and eluted into the same buffer but containing 250 mM imidazole. His-MSMEG_1275 was dissolved in Buffer A (100 mM NaH2PO4, 300 mM NaCl, 9 M Urea, 5 mM imidazole, 10mMTris-HCl, 1 mM β-mercaptoethanol, pH 7.4). His-MSMEG_1275 was purified by binding to Ni-NTA (GenScript, Nanjing, China) equilibrated with wash Buffer (100 mM NaH2PO4, 300 mM NaCl, 9 M Urea, 20 mM imidazole, 10 mM Tris-HCl, 1 mM β-mercaptoethanol, pH 7.4) and eluted into the same buffer but containing 250 mM imidazole. The elution fractions containing His-MSMEG_1275 were diluted in Buffer B (100 mM NaH2PO4, 300 mM NaCl, 0.1 mM EDTA, 0.01% Triton X-100, 10 mM Tris-HCl, 20% Glycerol, pH 7.4), and concentrated by Millipore Amicon® Ultra-4. Protein concentration was detected by Bicinchoninic Acid (BCA) Assay (TIANGEN, Shanghai, China).
2.8. DNA Digestion Assay
The digestion assays were based on those used by Moodley et al. [36] with some modification. Briefly, the assays were performed with 10 μg/mL of MSMEG_1275 and 6.25 μg/mL M. smegmatis genome DNA. Digestion experiments were conducted in 10 mM HEPES, pH 7.0. Then, 5 mM β-mercaptoethanol was added and the final concentration of β-mercaptoethanol was 0.05 mM. Divalent metal ions (Ni2+, Mg2+, Zn2+, and Cu2+) were tested at concentrations of 1 mM as cofactors. After 5 h, 10 μL of sample was removed, and the reaction was stopped by addition of EDTA to a final concentration of 0.025 mM and DNA loading buffer (Takara, Kusatsu, Japan). Samples were analyzed by 1% agarose gel electrophoresis.
2.9. Electrophoretic Mobility Shift Assays (EMSA)
To evaluate the binding of His-MSMEG_6762 to the operator promoter regions, specific primers (Supplemental Table S2) were used to amplify the genomic DNA of M. smegmatis. DNA substrate and increasing concentrations of protein (as indicated in the legend of the corresponding figure) were incubated for 20 min in EMSA buffer (100 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT and 10% glycerol), at room temperature. Products were separated on native 5% polyacrylamide gels (PAGE) in 0.5 × TBE buffer, at 4 °C, stained with GoldView, and visualized under UV-transmitting light.
2.10. Construction of Deletion Mutant Strains
The sequences of all primers used in knockout and overexpression are shown in Table S2. The genes of M. smegmatismc2 155 was disrupted using recombineering approach previously described [37]. The regions near the deletions were verified by PCR followed by DNA sequencing.
2.11. Statistical Analysis
Data from at least three biological replicates were used to calculate means and standard deviation (SD) for graphing purposes. Statistical analysis employed the unpaired Student’s t test, asterisks indicate statistically significant difference (* p < 0.05; ** p < 0.01; *** p < 0.001).
3. Results
3.1. Expression of MSMEG_6762 Causes the Cell Death of M. smegmatis
MSMEG_6762-MSMEG_6760 was predicted as a toxin–antitoxin pair system in M. smegmatis (https://db-mml.sjtu.edu.cn/TAfinder/index.php, accessed on 1 December 2013). MSMEG_6760 is a predicted toxin protein, and MSMEG_6762 is the predicted antitoxin protein [38,39]. In order to confirm this perdition, we performed a co-transcription analysis of MSMEG_6762-MSMEG_6760. As shown by RT-PCR, a single band of ~500 bp was detected using a forward primer that bound to MSMEG_6760 and a reverse primer that bound to MSMEG_6762 using cDNA synthesized from the total RNA as template, indicating that these two genes are co-transcribed. No bands were obtained using total RNA as a template (Figure S1). This result revealed that MSMEG_6762-MSMEG_6760 are co-translated and form an operon. We then study this toxin–antitoxin pair module as in Frampton et al. [40]. The putative toxin gene was inserted into pALACE under the control of an acetamide-inducible promoter. To ensure the effect was observed due to the production of MSMEG_6760 alone, the operon MSMEG_6762-MSMEG_6760 was also cloned in the same manner, thus preventing the effect of the toxin protein by co-expressing the cognate antitoxin. The empty vector pALACE (MS-VEC) was used as a control. However, in the presence of acetamide, cells expressing MSMEG_6760 were able to grow on a 7H10 agar plate, while cells co-expressing toxin and antitoxin failed to grow (Figure 1A, right panel). All three strains could grow normally on a 7H10 agar plate without acetamide (Figure 1A, left panel). A toxic effect of co-expressing MSMEG_6762-MSMEG_6760 was also exhibited in the liquid culture, as shown by the reduction in turbidity (OD600) and colony forming units (CFU) (Figure 1B,C). As shown in Figure 1A–C, this result was exactly the opposite of expectation. Since the expression of putative toxin protein MSMEG_6760 did not affect cell growth, should the putative antitoxin protein MSMEG_6762 be a potent toxin protein? To test this hypothesis, we cloned the region of MSMEG_6762 into pALACE plasmid, and then transferred it into M. smegmatis host. In the presence of acetamide, cells expressing MSMEG_6762 could not grow on a 7H10 agar plate (Figure 1D) and exhibited a notable decrease in cell growth, as shown by the reduction in turbidity (OD600) and colony-forming units (CFU) (Figure 1E,F). The expression of MSMEG_6760 and MSMEG_6762 were verified by Western blot (Figure S2A,B). These results suggest that overexpression of MSMEG_6762 is lethal for M. smegmatis.
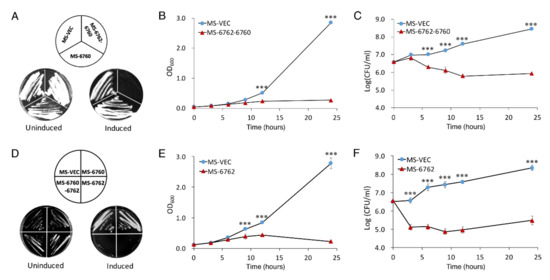
Figure 1.
Effect of MSMEG_6760 and MSMEG_6762 on the growth and viability of M. smegmatis. Growth on 7H10 plates with 50 μg/mL hygromycin without (left) and with (right) 1% acetamide (A,D), were incubated for 3 days. M. smegmatis hosts containing pALACE-based constructs were cultured in 7H9 medium supplemented with 50 μg/μL hygromycin without (left) and with (added at OD600 = 0.1). Cell growth of MS-6762-6760 (B), MS-6762 (E) and viability (CFU/mL) of MS-6762-6760 (C), MS-6762 (F) were tested at indicated intervals. MS-VEC, M. smegmatis with pALACE plasmid. MS-6062, M. smegmatis with pALACE-MSMEG_6760-MSMEG_6762 plasmid. MS-6762: with pALACE-MSMEG_6762 plasmid. Experiments were performed in triplicate. Data are represented as mean +/− SEM. Significance of MS-VEC strain compared to MS-6762 strain was determined using a Student’s t test: *** p < 0.001.
3.2. L18, R24, H54, and L58 Residues Are Critical for the Toxicity of MSMEG_6762
MSMEG_6762 was identified as an ArsR transcriptional factor [41] with four amino acid residues conserved among the ArsR family regulator (Figure 2A). The 3D structure predicted by Phyre2 protein homology recognition engine [42] indicated that the conserved domain is situated at the end of one helix and R54, close to L58. The four amino acid residues might be crucial for DNA binding (Figure 2B). Then, we performed site-directed mutagenesis on L18, R24, H54, and L58 to explore the significance of the conserved residues in determining toxicity of MSMEG_6762. Substitution of any one of these amino acid residues with alanine abolished the toxicity of MSMEG_6762, both in liquid and solid medium (Figure 2C,D). The results indicate that residues L18, R24, H54, and L58 are critical in the toxicity of MSMEG_6762.
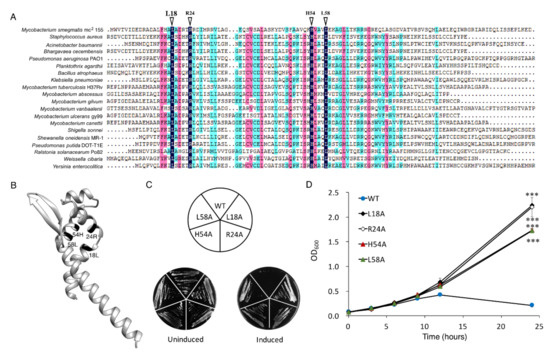
Figure 2.
Key residues for MSMEG_6762 toxicity. (A) Conserved amino acid residues of MSMEG_6762. (B) Predicted 3D structure of MSMEG_6762. Toxicity results of single-site mutagenesis of 18L, 24R, 54H, and 58L of MSMEG_6762 in solid (C) and liquid medium (D). MS-VEC: M. smegmatis with pALACE plasmid. MS-6762: with pALACE-MSMEG_6762 plasmid. WT indicates the wild-type MSMEG_6762 protein; the remainders are mutated proteins. The number in the mutated protein indicates the position of the amino acid in MSMEG_6762. Log-phase cultures were streaked on 50 μg/mL hygromycin 7H10 plates, with or without 1% acetamide. Experiments were performed in triplicate. Data are represented as mean +/− SEM. Significance of mutant strains compared to MS-6762 WT strain was determined using a Student’s t test: *** p < 0.001.
3.3. Overexpression of MSMEG_6762 Induces the DNA Damage in M. smegmatis
MSMEG_6762 is a transcriptional regulator governing the expression of target genes [41]. To find the genes underlying the lethal effect of MSMEG_6762, RNA-Seq based transcriptome analysis was performed. Upon MSMEG_6762 overexpression, at least 580 genes were upregulated, and 1127 genes were downregulated using log2 fold change (greater than 1 or less than 1) as a threshold (Table S3). The genes involved in mismatch repair, nucleotide excision repair, base excision repair, and homologous recombination were upregulated at least two-fold (log2 fold change greater than 1) (Table 1) (Figure 3A). Bacterial SOS is a global response to DNA damage to arrest the cell cycle, and initiate DNA repair. RecA-lexA modulates the SOS response. During normal growth, LexA encoded by the lexA gene acts as a repressor by binding to an operator DNA of a specific sequence, the SOS box, and prevents their expression [43,44,45]. Upon DNA damage, single-stranded DNA occurs [46]. RecA binds to these single-stranded regions and is converted to an active form to stimulate the self-cleavage of LexA [47]. The recB, recC, and recD gene-encoded proteins comprise a RecBCD complex, which is required for recombinational DNA repair of double-stranded DNA (dsDNA) breaks in bacteria [48,49]. The RuvA and RuvB proteins form a complex that catalyzes branch migration, and RuvC catalyzes resolution of Holliday junctions [50,51,52]. Real-time analyses were performed for selected three genes to confirm the RNA-seq results, including MSMEG_1620 (the most highly upregulated gene), lexA and recA (two genes involved in DNA repair pathway). The real-time PCR results are as follows: up-regulated (5.11 times), up-regulated (4.27 times), and up-regulated (2.20 times) (Figure S3), which are in good agreement with the RNA-seq data. These data suggest that the expression of MSMEG_6762 resulted in DNA damage in M. smegmatis.

Table 1.
Transcriptional profile of genes in the response to MEMSG_6762 expression reveals the DNA damage in M. smegmatis.
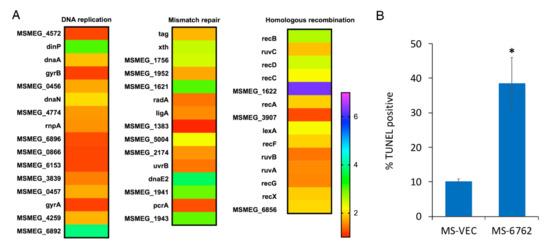
Figure 3.
DNA damage induced by MSMEG_6762. (A) Heat maps of DNA repair related pathway. The relative fold change in expression level in several pathways was calculated and visualized over time using Excel; heat map was made with GraphPad Prism 6.0. (B) The percentage of TUNEL-positive of MS-VEC and MS-6762 (mean ± SD at 12 h after induction). Data are represented as mean +/− SEM. Significance of MS-6762 strain compared to MS-VEC strain was determined using a Student’s t test: * p < 0.05.
To assess DNA double-strand break potentially caused by MSMEG_6762 expression, we measured the DNA fragmentation in M. smegmatis by the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (Figure 3B). The percentage of cells with DNA breaks in MSMEG_6762 overexpression strain reaches to 38% after 12 h induction, 3.82-fold higher than the MS-VEC strain. The data indicates that the expression of MSMEG_6762 induces DNA damage in M. smegmatis.
3.4. MSMEG_6762 Causes Cell Death by an Unregulated HNH Nuclease MSMEG_1275
HNH motif is a small DNA binding and cleavage module characterized by two tightly conserved histidine residues separated by an asparagine residue [53]. To date, more than 1000 HNH motif-containing proteins have been identified from bacteria, archaea, and eukaryotes [54]. The largest subgroup of HNH motif-containing proteins with known function is the site-specific homing endonucleases [55], such as Cpc [56], I-TevIII [57], and I-BasI [58]. Bacterial toxins with HNH motifs include E7 [59], E9 [60], colicins, and pyocins S1, S2 [61]. Both E7 and E9 are endonucleases active on single- and double-stranded DNA, but with no clear specificity, and result in cell death [62,63]. Pyocin S1 and S2 exhibit DNase activity, which can degrade cellular DNA in susceptible cells [61]. HNH motif has also been identified in restriction or repair enzymes, such as MnlI [64] and McrA [65]. RNA-Seq was used to explore the relationship between MSMEG_6762 expression and DNA damage. The data showed that five HNH motif-containing proteins were upregulated during the expression of MSMEG_6762, including MSMEG_5583, MSMEG_5876, MSMEG_3404, MSMEG_1275, and MSMEG_2148 (Table 2 and Figure 4A). To the extent that MSMEG_6762 is a transcriptional factor, HNH motif-containing proteins might cause such DNA damage. To test this hypothesis, we overexpressed these five genes in M. smegmatis solely. The result shows that only overexpression of MSMEG_1275 greatly inhibited cell growth both on 7H10 agar (Figure 4B) and 7H9 liquid culture medium (Figure 4C).

Table 2.
Upregulated HNH family genes in response to MEMSG_6762 expression in M. smegmatis.
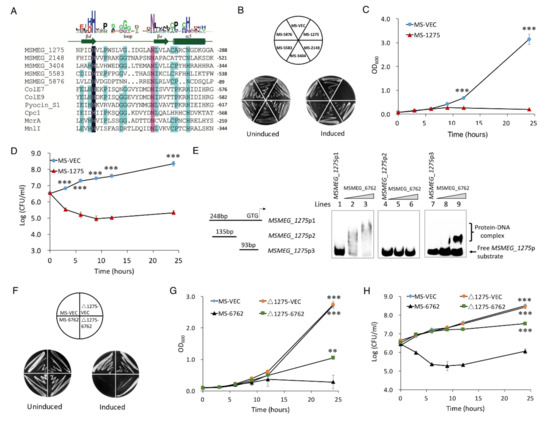
Figure 4.
MSMEG_6762 causes cell death by an unregulated HNH nuclease MSMEG_1275. (A) Sequence alignment of HNH proteins. (B) Overexpression of HNH domain genes on 7H10 plates. (C) The effect of MSMEG_1275 on the cell growth of MS-1275. (D) The bactericidal activity of HNH nuclease MSMEG_1275 on cell viability of MS-1275. Log-phase cultures were streaked on 50 μg/mL hygromycin 7H10 plates with or without 1% acetamide. Significance of MS-VEC strain compared to MS-1275 strain was determined using a Student’s t test: *** p < 0.001. (E) EMSA assays for the binding of MSMEG_6762 to MSMEG_1275 promoter DNA fragments. The MSMEG_1275 promoter DNA substrates were co-incubated with gradually increasing concentrations of MSMEG_6762 protein (0, 2 and 4 μM). (F) The effect of HNH nuclease MSMEG_1275 on MSMEG_6762 mediated cell death in solid culture medium. Log-phase cultures were streaked on 50 μg/mL hygromycin 7H10 plates with or without 1% acetamide. (G,H) The effect of HNH nuclease MSMEG_1275 on MSMEG_6762 mediated cell death in liquid culture medium. Experiments were performed in triplicate. Data are represented as mean +/− SEM. Significance of tested strains compared to MS-6762 strain was determined using a Student’s t test: ** p < 0.01 and *** p < 0.001.
Since MSMEG_1275 was upregulated in response to the expression of MSMEG_6762, the expression level of MSMEG_1275 might be regulated by MSMEG_6762 directly. To test whether recombinant MSMEG_6762 (Figure S4A) can interact with MSMEG_1275 promoter, an EMSA assay was performed. As shown in Figure 4E (lines 1–4), when the MSMEG_1275p1 DNA substrate (100 bp) was co-incubated with increasing concentrations of recombinant His-MSMEG_6762 (0, 2, and 4 μM, respectively), clear shifted bands were observed. MSMEG_6762 specifically bound to the MSMEG_1275p2 DNA substrate (Figure 4, lines 5–8); however, no band shifted by the shorter MSMEG_1275p3 DNA substrate (Figure 4E, lines 9–12). This result indicates that MSMEG_6762 can specifically bind to the MSMEG_1275 promoter region.
To confirm MSMEG_1275 is the downstream regulation target of MSMEG_6762, we knocked out MSMEG_1275 and expressed MSMEG_6762 in the ΔMSMEG_1275 strain. If MSMEG_1275 is the downstream regulation target of MSMEG_6762, the lethal effect of MSMEG_6762 will be abolished or relieved. As we expected, knockout MSMEG_1275 relieved the bactericidal activity of MSMEG_6762 on a 7H10 agar plate, as well as in the liquid culture medium (the OD600 of Δ1275-6762 reached 0.946 after 24 h induction) (Figure 4F, G). Knockout MSMEG_1275 aborted the bacteria killing activity of MSMEG_6762, but caused a delay in growth (Figure 4H). These results demonstrate that MSMEG_1275 is the downstream target of MSMEG_6762.
3.5. MSMEG_1275 Mediates Double-Stranded Digestion of M. smegmatis Chromosome DNA
To determine whether MSMEG_1275 possesses nuclease activity, we examined its ability to cleave the M. smegmatis chromosome. Purified MSMEG_1275 (Figure S4B) was tested for nuclease activity in the presence of a variety of divalent metal ions. Without metal ions, MSMEG_1275 cleaves the chromosome at multiple sites, as evidenced by the formation of a continuum of DNA fragments with varying lengths (Figure 5A). The enzyme exhibited high activity with 1 mM Mg2+ in the reaction buffer, less activity with 1 mM Ni2+, Zn2+, and very low activity with 1 mM Cu2+ (Figure 5A). In the second subfamily of HNH superfamily, the second His residue is usually substituted by a conserved Asn residue and forms a HNN motif [66]. To check whether these conserved residues constitute the HNN motif, we constructed three MSMEG_1275 mutants, H258A, N272A and N281A, and we found that the mutation of H258 or N272 abolished the MSMEG_1275 bacteria-killing activity both in liquid and solid medium in vivo, and mutation of N281 decreased the activity of MSMEG_1275 (the OD600 of N281A reached 0.417 after 24 induction) (Figure 5B,C). These results indicate that MSMEG_1275 has nuclease activity, and the conserved residues H258 and N272 are essential for its nuclease activity. Next, we measured the DNA fragmentation in M. smegmatis by the TUNEL assay. The results showed that the percentage of cells with DNA breaks in MSMEG_1275 overexpression strain reached 55%, 5.72-fold higher than the MS-VEC strain (Figure 5D). The data indicates that the HNH nuclease MSMEG_1275 cleaves M. smegmatis chromosome DNA.
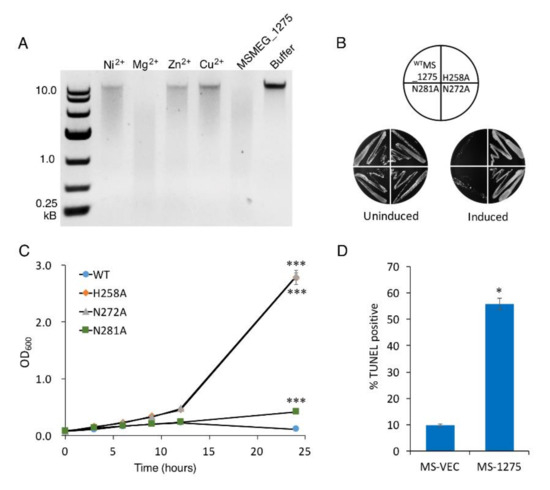
Figure 5.
MSMEG_1275 is an HNH nuclease and cleaves double-stranded DNA of M. smegmatis. (A) Effect of divalent cations on the DNase activity. Toxicity results of single-site mutagenesis of 258H, 272N and 281N of MSMEG_1275 in solid (B) and liquid medium (C). (D) The percentage of TUNEL-positive of MS-VEC and MS-1275 after 12 h induction. Data are represented as mean +/− SEM. Significance of mutant strains compared to MS-1275 WT strain was determined using a Student’s t test: * p < 0.05 and *** p < 0.001.
4. Discussion
PCD in bacteria is a form of active suicide phenomenon that is controlled by related genes. Within the group of bacteria, a part of these bacteria that has programmed cell death is important to the entire bacterial population. Here, we report for the first time that the ArsR family transcriptional regulator MSMEG_6762 is involved in programmed cell death. We demonstrated that the overexpression of MSMEG_6762 causes bacterial death in M. smegmatis cells and identified that residues L18, R24, H54, and L58 are crucial for its activity. We further showed that MSMEG_6762 regulates the expression of HNH nuclease MSMEG_1275, which cleaves the double-stranded DNA in an iron-independent manner and eventually causes bacterial cell death. Moreover, we found that mutation of H258 or N272 aborted the nuclease activity of MSMEG_1275. Together, our findings unveiled a novel programmed cell death pathway in M. smegmatis which is consistent with an ArsR family transcription factor, MSMEG_6762, and an HNH nuclease, MSMEG_1275. Upon the treatment of amikacin, MSMEG_6762 was expressed and binds the promoter of MSMEG_1275, and then MSMEG_1275 degrades the DNA of the host, which leads to cell death.
Zhang et al. [67] and Gao et al. [41] conducted a similar overexpression experiment of MSMEG_6762 in M. smegmatis, but no bacteria-killing activity of MSMEG_6762 was documented, which might be related to the plasmid and promoters used in the different expression vectors in M. smegmatis. Zhang et al. [67] found that the overexpression of MSMEG_6760 and MSMEG_6762-6760 did not affect the growth of M. smegmatis. However, the expression of target proteins has not been confirmed by Western blot. In this study, we assessed the expression level of MSMEG_6762 and MSMEG_6760 by Western blot (Figure S1) and confirmed that overexpression of MSMEG_6762 causes cell death in M. smegmatis. We also tested the toxicity of MSMEG_6762 in another mycobacteria expression, plasmid pNIT [68], overexpression of MSMEG_6762 by pNIT was toxic to M. smegmatis as well (Figure S5). Hence, we confirm that the expression of MSMEG_6762 was toxic to M. smegmatis.
The ArsR family transcriptional regulators have been found to be involved in various important cellular events, such as metal ion homeostasis, biofilm formation, and virulence [69]. phoPR is one of the two-component systems in mycobacteria and plays an important role in cell wall biosynthesis [70], virulence, hypoxic response [71], and pH response [72]. The expression of phoP was positive regulated by MSMEG_6762 [41]. There is a zero-fold change in information in our transcriptome data. MSMEG_3932 (hspX), encoding a small heat shock protein in M. smegmatis, also positively regulated by MSMEG_6762. However, hspX was the most down-regulated gene upon MSMEG_6762 overexpression in our study. Thus, we believe that phoP and hspX might not be involved in MSMEG_6762-mediated cell death in M. smegmatis. Since the expression of MSMEG_6762 resulted in cell death in M. smegmatis, this means MSMEG_6762 might be induced under lethal conditions, such as antibiotic, heat or heavy metal treatment. It is interesting to know the physiological role of MSMEG_6762 in M. smegmatis.
HNH motif is a small nucleic-acid binding and cleavage module, including site-specific homing endonucleases, non-specific endonuclease [73], DNA fragmentation during cell apoptosis [74] and repair of enzymes [75]. We overexpressed five HNH motif nucleases in this study and found only one was lethal for bacteria. HNH endonuclease/nuclease motif-containing proteins have been shown to play an important role in the competition between rival bacteria [76]. Furthermore, knockout MSMEG_1275 only can abolish a part of the killing activity of MSMEG_6762 (as the OD600 of Δ1275-6762 was 4.27-fold higher than MS-6762 after 24 h induction), which means there is more than one pathway involved in the MSMEG_6762-induced cell death. It would be interesting to isolate the surviving mutants and identify the possible pathways which are involved in MSMEG_6762-caused cell death.
In conclusion, we have shown that a novel PCD pathway has been identified in M. smegmatis. In this PCD pathway, MSMEG_6762 works as a regulator, and MSMEG_1275 works as an executor which controls the cell fate of bacteria. This finding has expanded the understanding of ArsR family transcriptional regulators, because this is the first report of this family member serving as a death regulator in bacterial PCD.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10081535/s1, Figure S1. Genetic organization and co-transcription analysis of MSMEG_6762-MSMEG_6760. Figure S2. The expression of MSMEG_6760 and MSMEG_6762 in M. smegmatis. Figure S3. Verification of RNA-seq results by real-time PCR. Numbers means the numbering of gene in M. smegmatis mc2 155. Figure S4. SDS-PAGE gel of recombinant M. smegmatis protein expressed and purified from E. coli. Figure S5. The effect of MSMEG_6762 on M. smegmatis growth when overexpressed by pNIT plasmid. Table S1. Bacterial strains and plasmids in this study. Table S2. Oligonucleotides used for RT-PCR, cloning (restriction sites are underlined and in bold), site directed mutagenesis (target mutated nucleotides are underlined), knockout and verification (restriction sites are underlined), and EMSA. Table S3: Upregulated and downregulated genes upon MSMEG_6762 overexpression. References [77,78,79] are cited in the supplementary materials.
Author Contributions
X.D. (Xiangke Duan), X.H., J.X. (Junqi Xu), X.L., J.N., X.D. (Xiaoli Du), X.W., J.L., M.K., J.G., K.Z. and Y.H. performed the experiments. X.D. (Xiangke Duan), X.H. and J.X. (Junqi Xu) analyzed the data. J.X. (Jianping Xie) contributed with reagents and materials. X.D. (Xiangke Duan), B.K. and J.X. (Jianping Xie) designed the study and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation (grant numbers 82072246 and 81871182), the National key R & D plan (2016YFC0502304), and the Chongqing Graduate Research and Innovation Foundation (CYB20091).
Data Availability Statement
Not applicable.
Conflicts of Interest
All authors declare that they have no conflict of interest.
Abbreviations
| PCD | programmed cell death |
| TA | toxin–antitoxin |
| CFU | colony-forming units |
| dsDNA | double-stranded DNA |
| TUNEL assay | Terminal deoxynucleotidyl transferase dUTP nick end labeling assay |
| LB | Luria-Bertani |
| EMSA | electrophoretic mobility shift assay |
References
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wide-Ranging Implications in Tissue Kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Programmed Death in Bacteria. Microbiol. Mol. Biol. Rev. 2000, 64, 503–514. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Diggle, S.P.; Buckling, A.; Gardner, A.; Griffins, A.S. The Social Lives of Microbes. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 53–77. [Google Scholar] [CrossRef]
- Rice, K.C.; Bayles, K.W. Death’s Toolbox: Examining the Molecular Components of Bacterial Programmed Cell Death. Mol. Microbiol. 2003, 50, 729–738. [Google Scholar] [CrossRef]
- Hu, M.-X.; Zhang, X.; Li, E.-L.; Feng, Y.-J. Recent Advancements in Toxin and Antitoxin Systems Involved in Bacterial Programmed Cell Death. Int. J. Microbiol. 2010, 2010, 781430. [Google Scholar] [CrossRef]
- Tanouchi, Y.; Lee, A.J.; Meredith, H.; You, L. Programmed cell death in bacteria and implications for antibiotic therapy. Trends Microbiol. 2013, 21, 265–270. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-Antitoxin Systems: Biology, Identification, and Application. Mob. Genet. Elem. 2013, 3, e26219. [Google Scholar] [CrossRef]
- Gerdes, K.; Christensen, S.K.; Lobner-Olesen, A. Prokaryotic Toxin-Antitoxin Stress Response Loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Park, J.-H.; Inouye, M. Toxin-Antitoxin Systems in Bacteria and Archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef]
- Ramisetty, B.C.; Natarajan, B.; Santhosh, R.S. Mazef-Mediated Programmed Cell Death in Bacteria: “What Is This?”. Crit. Rev. Microbiol. 2015, 41, 89–100. [Google Scholar] [CrossRef]
- Engelberg-Kulka, H.; Hazan, R.; Amitai, S. mazEF: A chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 2005, 118, 4327–4332. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Hoeflich, K.P.; Ikura, M.; Qing, G.; Inouye, M. MazF Cleaves Cellular mRNAs Specifically at ACA to Block Protein Synthesis in Escherichia coli. Mol. Cell 2003, 12, 913–923. [Google Scholar] [CrossRef]
- Dorsey-Oresto, A.; Lu, T.; Mosel, M.; Wang, X.; Salz, T.; Drlica, K.; Zhao, X. YihE Kinase Is a Central Regulator of Programmed Cell Death in Bacteria. Cell Rep. 2013, 3, 528–537. [Google Scholar] [CrossRef]
- Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. An Escherichia Coli Chromosomal” Addiction Module” Regulated by Guanosine [Corrected] 3′, 5′-Bispyrophosphate: A Model for Programmed Bacterial Cell Death. Proc. Natl. Acad. Sci. USA 1996, 93, 6059–6063. [Google Scholar] [CrossRef]
- Sat, B.; Reches, M.; Engelberg-Kulka, H. The Escherichia coli mazEF Suicide Module Mediates Thymineless Death. J. Bacteriol. 2003, 185, 1803–1807. [Google Scholar] [CrossRef]
- Engelberg-Kulka, H.; Sat, B.; Reches, M.; Amitai, S.; Hazan, R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004, 12, 66–71. [Google Scholar] [CrossRef]
- Hazan, R.; Sat, B.; Engelberg-Kulka, H. Escherichia coli mazEF-Mediated Cell Death Is Triggered by Various Stressful Conditions. J. Bacteriol. 2004, 186, 3663–3669. [Google Scholar] [CrossRef]
- Erental, A.; Sharon, I.; Engelberg-Kulka, H. Two Programmed Cell Death Systems in Escherichia Coli: An Apoptotic-Like Death Is Inhibited by the Mazef-Mediated Death Pathway. PLoS Biol. 2012, 10, e1001281. [Google Scholar] [CrossRef]
- Ramisetty BC, M.; Raj, S.; Ghosh, D. Escherichia Coli Mazef Toxin-Antitoxin System Does Not Mediate Programmed Cell Death. J. Basic Microbiol. 2016, 56, 1398–1402. [Google Scholar] [CrossRef]
- Ramisetty, B.C.M.; Ghosh, D.; Chowdhury, M.R.; Santhosh, R.S. What is the link between stringent response, endoribonuclease encoding type II toxin–antitoxin systems and persistence? Front. Microbiol. 2016, 7, 1882. [Google Scholar] [CrossRef]
- Tsilibaris, V.; Maenhaut-Michel, G.; Mine, N.; van Melderen, L. What Is the Benefit to Escherichia Coli of Having Multiple Toxin-Antitoxin Systems in Its Genome? J. Bacteriol. 2007, 189, 6101–6108. [Google Scholar] [CrossRef] [PubMed]
- Mutschler, H.M.; Gebhardt, R.L.; Shoeman, A.; Meinhart, A. Novel Mechanism of Programmed Cell Death in Bacteria by Toxin-Antitoxin Systems Corrupts Peptidoglycan Synthesis. PLoS Biol. 2011, 9, e1001033. [Google Scholar] [CrossRef] [PubMed]
- Ichige, A.; Kobayashi, I. Stability of Ecori Restriction-Modification Enzymes in Vivo Differentiates the Ecori Restriction-Modification System from Other Postsegregational Cell Killing Systems. J. Bacteriol. 2005, 187, 6612–6621. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pastor, J.E.; Hobbs, E.C.; Losick, R. Cannibalism by Sporulating Bacteria. Science 2003, 301, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Ellermeier, C.D.; Hobbs, E.C.; Gonzalez-Pastor, J.E.; Losick, R. A Three-Protein Signaling Pathway Governing Immunity to a Bacterial Cannibalism Toxin. Cell 2006, 124, 549–559. [Google Scholar] [CrossRef]
- Al Mamun, A.A.M.; Lombardo, M.-J.; Shee, C.; Lisewski, A.M.; Gonzalez, C.; Lin, D.; Nehring, R.B.; Saint-Ruf, C.; Gibson, J.L.; Frisch, R.L.; et al. Identity and Function of a Large Gene Network Underlying Mutagenic Repair of DNA Breaks. Science 2012, 338, 1344–1348. [Google Scholar] [CrossRef]
- OSummers, A. Damage control: Regulating Defenses Against Toxic Metals and Metalloids. Curr. Opin. Microbiol. 2009, 12, 138–144. [Google Scholar] [CrossRef]
- Williams, S.G.; Attridge, S.R.; Manning, P.A. The Transcriptional Activator Hlyu of Vibrio Cholerae: Nucleotide Sequence and Role in Virulence Gene Expression. Mol. Microbiol. 1993, 9, 751–760. [Google Scholar] [CrossRef]
- Liu, M.; Alice, A.F.; Naka, H.; Crosa, J.H. The Hlyu Protein Is a Positive Regulator of Rtxa1, a Gene Responsible for Cytotoxicity and Virulence in the Human Pathogen Vibrio Vulnificus. Infect. Immun. 2007, 75, 3282–3289. [Google Scholar] [CrossRef]
- O'Rourke, K.P.; Shaw, J.D.; Pesesky, M.W.; Cook, B.T.; Roberts, S.M.; Bond, J.P.; Spatafora, G.A. Genome-Wide Characterization of the SloR Metalloregulome in Streptococcus mutans. J. Bacteriol. 2010, 192, 1433–1443. [Google Scholar] [CrossRef]
- Zhao, H.; Volkov, A.; Veldore, V.H.; Hoch, J.A.; Varughese, K.I. Crystal structure of the transcriptional repressor PagR of Bacillus anthracis. Microbiology 2010, 156, 385–391. [Google Scholar] [CrossRef][Green Version]
- Duan, X.; Li, Y.; Du, Q.; Huang, Q.; Guo, S.; Xu, M.; Lin, Y.; Liu, Z.; Xie, J. Mycobacterium Lysine ε-aminotransferase is a novel alarmone metabolism related persister gene via dysregulating the intracellular amino acid level. Sci. Rep. 2016, 6, 19695. [Google Scholar] [CrossRef]
- Zhou, M.; Xie, L.; Yang, Z.; Zhou, J.; Xie, J. Lysine Succinylation of Mycobacterium Tuberculosis Isocitrate Lyase (Icl) Fine-Tunes the Microbial Resistance to Antibiotics. J. Biomol. Struct. Dyn. 2017, 35, 1030–1041. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Lin, Y.; Li, Q.; Xie, L.; Xie, J. Mycobacterium tuberculosis rv1400c encodes functional lipase/esterase. Protein Expr. Purif. 2017, 129, 143–149. [Google Scholar] [CrossRef]
- Moodley, S.; Maxwell, K.; Kanelis, V. The protein gp74 from the bacteriophage HK97 functions as a HNH endonuclease. Protein Sci. 2012, 21, 809–818. [Google Scholar] [CrossRef]
- van Kessel, J.C.; Hatfull, G.F. Mycobacterial Recombineering. Methods Mol. Biol. 2008, 435, 203–215. [Google Scholar]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.-Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin–antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2010, 39, D606–D611. [Google Scholar] [CrossRef]
- Bajaj, R.A.; Arbing, M.A.; Shin, A.; Cascio, D.; Miallau, L. Crystal Structure of the Toxin Msmeg_6760, the Structural Homolog of Mycobacterium Tuberculosis Rv2035, a Novel Type Ii Toxin Involved in the Hypoxic Response. Acta Crystallogr. Sect. F: Struct. Biol. Commun. 2016, 72, 863–869. [Google Scholar] [CrossRef]
- Frampton, R.; Aggio, R.B.; Villas-Boas, S.; Arcus, V.L.; Cook, G.M. Toxin-Antitoxin Systems of Mycobacterium smegmatis Are Essential for Cell Survival. J. Biol. Chem. 2012, 287, 5340–5356. [Google Scholar] [CrossRef]
- Gao, C.H.; Yang, M.; He, Z.G. An Arsr-Like Transcriptional Factor Recognizes a Conserved Sequence Motif and Positively Regulates the Expression of Phop in Mycobacteria. Biochem. Biophys. Res. Commun. 2011, 411, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- De Henestrosa, A.R.F.; Ogi, T.; Aoyagi, S.; Chafin, D.; Hayes, J.J.; Ohmori, H.; Woodgate, R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2002, 35, 1560–1572. [Google Scholar] [CrossRef]
- Courcelle, J.; Khodursky, A.; Peter, B.; Brown, O.P.; Hanawalt, P.C. Comparative Gene Expression Profiles Following UV Exposure in Wild-Type and SOS-Deficient. Escherichia coli. Genetics 2001, 158, 41–64. [Google Scholar] [CrossRef]
- Smollett, K.L.; Smith, K.M.; Kahramanoglou, C.; Arnvig, K.B.; Buxton, R.S.; Davis, E.O. Global Analysis of the Regulon of the Transcriptional Repressor Lexa, a Key Component of Sos Response in Mycobacterium Tuberculosis. J. Biol. Chem. 2012, 287, 22004–22014. [Google Scholar] [CrossRef]
- Sassanfar, M.; Roberts, J.W. Nature of the Sos-Inducing Signal in Escherichia Coli. The Involvement of DNA Replication. J. Mol. Biol. 1990, 212, 79–96. [Google Scholar] [CrossRef]
- Little, J. Mechanism of specific LexA cleavage: Autodigestion and the role of RecA coprotease. Biochimie 1991, 73, 411–421. [Google Scholar] [CrossRef]
- Dillingham, M.S.; Kowalczykowski, S.C. RecBCD Enzyme and the Repair of Double-Stranded DNA Breaks. Microbiol. Mol. Biol. Rev. 2008, 72, 642–671, Table of Contents. [Google Scholar] [CrossRef]
- Wigley, D.B. Bacterial DNA Repair: Recent Insights into the Mechanism of Recbcd, Addab and Adnab. Nat. Rev. Microbiol. 2013, 11, 9–13. [Google Scholar] [CrossRef]
- Donaldson, J.R.; Courcelle, C.T.; Courcelle, J. RuvABC Is Required to Resolve Holliday Junctions That Accumulate following Replication on Damaged Templates in Escherichia coli. J. Biol. Chem. 2006, 281, 28811–28821. [Google Scholar] [CrossRef]
- Grove, J.I.; Harris, L.; Buckman, C.; Lloyd, R.G. DNA double strand break repair and crossing over mediated by RuvABC resolvase and RecG translocase. DNA Repair 2008, 7, 1517–1530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Mahdi, A.; Briggs, G.S.; Lloyd, R.G. Promoting and Avoiding Recombination: Contrasting Activities of the Escherichia coli RuvABC Holliday Junction Resolvase and RecG DNA Translocase. Genetics 2010, 185, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Keeble Anthony, H.; Maté, M.J.; Kleanthous, C. Hnh Endonucleases. In Homing Endonucleases and Inteins; Springer: Berlin/Heidelberg, Germany, 2005; pp. 49–65. [Google Scholar]
- Bateman, A.; Birney, E.; Cerruti, L.; Durbin, R.; Etwiller, L.; Eddy, S.R.; Griffiths-Jones, S.; Howe, K.L.; Marshall, M.; Sonnhammer, E.L. The Pfam Protein Families Database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, B.L. Homing endonuclease structure and function. Q. Rev. Biophys 2005, 38, 49–95. [Google Scholar] [CrossRef]
- Ferat, J.L.; Michel, F. Group Ii Self-Splicing Introns in Bacteria. Nature 1993, 364, 358–361. [Google Scholar] [CrossRef]
- Eddy, S.R.; Gold, L. The phage T4 nrdB intron: A deletion mutant of a version found in the wild. Genes Dev. 1991, 5, 1032–1041. [Google Scholar] [CrossRef]
- Landthaler, M.; Shub, D.A. The Nicking Homing Endonuclease I-Basi Is Encoded by a Group I Intron in the DNA Polymerase Gene of the Bacillus Thuringiensis Phage Bastille. Nucleic Acids Res. 2003, 31, 3071–3077. [Google Scholar] [CrossRef]
- Chak, K.F.; Kuo, W.S.; Lu, F.M.; James, R. Cloning and Characterization of the Cole7 Plasmid. J. Gen. Microbiol. 1991, 137, 91–100. [Google Scholar] [CrossRef]
- Wallis, R.; Moore, G.R.; Kleanthous, C.; James, R. Molecular Analysis of the Protein-Protein Interaction between the E9-Immunity Protein and Colicin-E9. Eur. J. Biochem. 1992, 210, 923–930. [Google Scholar] [CrossRef]
- Sano, Y.; Matsui, H.; Kobayashi, M.; Kageyama, M. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J. Bacteriol. 1993, 175, 2907–2916. [Google Scholar] [CrossRef]
- Pommer, A.J.; Cal, S.; Keeble, A.; Walker, D.; Evans, S.J.; Kühlmann, U.C.; Cooper, A.; Connolly, A.B.; Hemmings, A.; Moore, G.R.; et al. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 2001, 314, 735–749. [Google Scholar] [CrossRef]
- Hsia, K.C.; Chak, K.F.; Liang, P.H.; Cheng, Y.S.; Ku, W.Y.; Yuan, H.S. DNA Binding and Degradation by the Hnh Protein Cole7. Structure 2004, 12, 205–214. [Google Scholar] [CrossRef]
- Kriukiene, E.; Lubiene, J.; Lagunavicius, A.; Lubys, A. Mnli-the Member of H-N-H Subtype of Type Iis Restriction Endonucleases. Biochim. Biophys. Acta-Proteins Proteom. 2005, 1751, 194–204. [Google Scholar] [CrossRef]
- Hiom, K.; Sedgwick, S.G. Cloning and structural characterization of the mcrA locus of Escherichia coli. J. Bacteriol. 1991, 173, 7368–7373. [Google Scholar] [CrossRef][Green Version]
- Mehta, P.; Katta, K.; Krishnaswamy, S. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Sci. 2004, 13, 295–300. [Google Scholar] [CrossRef]
- Zhang, L.-Y.; Geng, Y.; Jia, H.; Xiao, J.; Li, Z.; Pan, L.; Sun, Y.; Zhang, Z. Preliminary Study on the Gene Function of a Novel Toxin-Antitoxin System Msmeg_3435-3436 in Mycobacterium Smegmatis. Chin. J. Antituberc. 2020, 42, 133. [Google Scholar]
- Pandey, A.K.; Raman, S.; Proff, R.; Joshi, S.; Kang, C.-M.; Rubin, E.J.; Husson, R.N.; Sassetti, C.M. Nitrile-inducible gene expression in mycobacteria. Tuberculosis 2009, 89, 12–16. [Google Scholar] [CrossRef]
- Ren, S.; Li, Q.; Xie, L.; Xie, J.; Sai, R. Molecular Mechanisms Underlying the Function Diversity of ArsR Family Metalloregulator. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 19–35. [Google Scholar] [CrossRef]
- Walters, S.B.; Dubnau, E.; Kolesnikova, I.; Laval, F.; Daffé, M.; Smith, I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006, 60, 312–330. [Google Scholar] [CrossRef]
- Gonzalo-Asensio, J.; Mostowy, S.; Harders-Westerveen, J.; Huygen, K.; Hernández-Pando, R.; Thole, J.; Behr, M.; Gicquel, B.; Martin, C. PhoP: A Missing Piece in the Intricate Puzzle of Mycobacterium tuberculosis Virulence. PLoS ONE 2008, 3, e3496. [Google Scholar] [CrossRef]
- Abramovitch Robert, B.; Rohde, K.H.; Hsu, F.o.; Russell, D.G. Aprabc: A Mycobacterium Tuberculosis Complex-Specific Locus That Modulates Ph-Driven Adaptation to the Macrophage Phagosome. Mol. Microbiol. 2011, 80, 678–694. [Google Scholar] [CrossRef]
- Rangarajan, E.S.; Shankar, V. Sugar non-specific endonucleases. FEMS Microbiol. Rev. 2001, 25, 583–613. [Google Scholar] [CrossRef]
- Widlak, P.; Garrard, W.T. Discovery, Regulation, and Action of the Major Apoptotic Nucleases Dff40/Cad and Endonuclease G. J. Cell. Biochem. 2005, 94, 1078–1087. [Google Scholar] [CrossRef]
- Marti, T.M.; Fleck, O. DNA Repair Nucleases. Cell. Mol. Life Sci. 2004, 61, 336–354. [Google Scholar] [CrossRef]
- Parret, A.H.A.; de Mot, R. Bacteria Killing Their Own Kind: Novel Bacteriocins of Pseudomonas and Other Γ-Proteobacteria. Trends Microbiol. 2002, 10, 107–112. [Google Scholar] [CrossRef]
- Du, Q.; Long, Q.; Mao, J.; Fu, T.; Duan, X.; Xie, J. Characterization of a novel mutation in the overlap of tlyA and ppnK involved in capreomycin resistance in Mycobacterium. IUBMB Life 2014, 66, 405–414. [Google Scholar]
- Kessel, J.C.v.; Hatfull, G.F. Mycobacterial Recombineering Chromosomal Mutagenesis; Springer: Berlin/Heidelberg, Germany, 2008; pp. 203–215. [Google Scholar]
- Snapper, S.B.; Melton, R.E.; Mustafa, S.; Kieser, T.; Jacobs, W.R., Jr. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990, 4, 1911–1919. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).