Abstract
Two bacteria belonging to the Pseudomonas and Pantoea genera were isolated from olive knots. Both bacterial strains were omnipresent in this study’s olive orchard with high susceptibility of the autochthonous olive genotypes indicating coevolution of bacteria with host plants. Genomes of two endemic bacteria show conserved core genomes and genome plasticity. The Pseudomonas ST1 genome has conserved virulence-related genes including genes for quorum sensing, pilus, and flagella biosynthesis, two copies of indole acetic acid biosynthesis (IAA) operons, type I-VI secretions systems, and genes for alginate and levan biosynthesis. Development of knots depends only on the presence of the Pseudomonas ST1 strain which then allows Pantoea paga strain co-infection and cohabitation in developed knots. The two bacteria are sensitive to a large number of antimicrobials, antibiotics, H2O2, and Cu (II) salts that can be efficiently used in propagation of bacterial free olive cultivars.
1. Introduction
Olive knot disease caused by the Gram-negative bacterium Pseudomonas savastanoi pv. savastanoi is recognized as one of the most important diseases that affect olive trees (Olea europaea L.) throughout various regions of the world, in particular the Mediterranean region [1]. Pseudomonas savastanoi belongs to the Pseudomonas syringae complex and includes three pathogenic lineages that cause tumorous overgrowths (tumorous galls or knots) in diverse trees and shrubs of economic relevance. In addition to Pseudomonas savastanoi pv. savastanoi, other pathovars such as P. savastanoi pv. nerii and P. savastanoi pv. fraxini have been isolated from oleander and ash, respectively.
Pseudomonas savastanoi pv. savastanoi (Psv, according to [2]) lives mainly as an epiphyte on the surface of olive organs [3]. Under favorable weather conditions, its population increases and the bacteria may enter into olive tissues and survive as an endophyte. Psv may experience uncontrolled proliferation at the site of infection and form knots trig-gered by phytohormones synthesized by the bacterium [1,4,5]. The knots occur mainly on the stems and branches of the host plants, and more rarely on leaves and fruits [6]. However, there is limited information on the impacts of this disease on tree vigor and yield [7].
At short distances, the bacterium can spread by rain, windblown aerosols, insects and agricultural practices. Processes such as the falling of leaves, budding, harvesting and pruning create wounds on the plant through which infection occurs [8]. Weather conditions favorable for both bacterium epiphytic development and its entry into the host determine the extent of damages caused by Psv. Olive knots are optimal niches for Psv growth and also for other bacteria. For example, many bacterial species reside inside the knots together with Pseudomonas savastanoi pv. savastanoi, commonly of the genera Pantoea, Pectobacterium, Erwinia and Curtobacterium [9]. Previous studies have reported that the microbe interactions can result in enhanced or decreased pathogenicity of one bacterium towards their plant host. Likewise, the interaction of Psv with other bacterial species has shown to mediate the disease potential [10,11]. Several non-pathogenic bacterial species, namely Erwinia toletana, Pantoea agglomerans, and Erwinia oleae, can form a stable bacterial consortium with the pathogen Psv and increase disease severity by boosting the proliferation of olive tissues [9,12,13]. The structure of plant-associated microbiota is genotype dependent [14,15] and may lead to different pathobiomes [16].
The pathogenicity and virulence of bacteria from the Pseudomonas syringae group and olive isolates of P. savastanoi are dependent on a type III secretion system (T3SS) that has been identified as a key virulence determinant among many Gram-negative plant and animal pathogenic bacteria [17,18,19,20]. Bacteria of the P. syringae group cause disease by suppressing plant defense responses after delivering effector proteins into the host cell cytoplasm using a type III secretion system (T3SS) [21]. Sisto et al. [17] reported that T3SS is encoded by hrp/hrc gene clusters. The T3SS is required for the hypersensitive response and for pathogenicity [22]. In addition to effector proteins, phytotoxins and hormones are recognized as molecules that increase virulence or microbial fitness [18,23,24,25,26]. The phytohormones indoleacetic acid (IAA) and cytokinins (CK) are the first and best-characterized Psv determinants that have been shown to be involved in olive knot formation [5,27].
Due to the economic impact of olive knot disease, growers need effective control methods to overcome its negative effects on plant growth and fruit yield [28]. Traditional control strategies are mainly based on reducing the endophytic and epiphytic P. savastanoi populations with practices that include pruning of affected branches and treating with copper compounds [28]. The use of resistant olive cultivars is considered one of the most efficient methods of disease control but limited information is available from these comparative studies [29,30,31]. Although different olive cultivars have shown different susceptibility to olive knot disease, there exists no report of olive tree genotypes completely resistant to disease [11]. To avoid the spread of P. savastanoi or other pathogens, sanitary certification programs for olive propagation materials and mother plants have been already launched worldwide [32]. However, the measures for the production of certified olive plants are sometimes based on the visual inspection of the knots, without considering the possible asymptomatic presence of P. savastanoi.
The plant genotype has been increasingly recognized to be a key determinant of phyllosphere microbiota composition [14]. The aim of this work was to evaluate the susceptibility of olive germplasm to olive knot disease caused by Pseudomonas and Pantoea, including isolation of the causative agent of knot formation and confirmation of pathogenicity by inoculation of healthy plants with isolated bacterial strains.
The genus Pantoea is a member of the Enterobacteriaceae and includes several species [33]. Most of them are associated with many plants living as a part of plants epiphyte or endophyte microbiome. Some strains are considered as plant growth promoting bacteria (PGPB). Pantoea aglomerans strains evolved into pathogens that cause gall formation on various plants [34,35] and another prevents root development in pruned cuttings as a limiting factor in plant propagation [36]. In this work we describe the effects of two bacteria in forming galls on olive plants and determine the genomic potential, phylogeny, and virulence of each strain.
2. Materials and Methods
2.1. The Incidence of Olive Knot Disease in Different Olive Cultivars
The study was carried out in 2019 in the collection orchard of olive cultivars from the main olive producing countries of the world and of the most common nationally planted cultivars of Croatia. The collection orchard is situated in Kaštela (43°3′22″ N, 16°20′56″ E). It consists of four Croatian cultivars, ‘Oblica‘, ‘Lastovka’, ‘Levantinka’ and ‘Drobnica’. In addition to the aforementioned national cultivars, the collection preserves cultivars from Italy (Canino, Coratina, Fasolina, Favarol, Frantoio, Grignan, Leccino, Maurino, Moraiolo, Pendolino, Rosciola, Santa Catarina, Taggiasca), Greece (Koroneiki), Algeria (Sigoise), Tunisia (Chemlali) and Morocco (Picholine marocaine).
In November 2019, the symptoms caused by olive knot in different olive cultivars were evaluated, five months after a hailstorm caused great damage to the trees of all cultivars in the collection. The evaluation of the symptoms was carried out according to the proposed methodology for the secondary characterization of olive varieties held in collections. The incidence of the disease was assessed by visual inspection of at least four trees per cultivar. The percentage of area in which symptoms appear was evaluated and cultivars were classified into six different categories of tolerance corresponding to percentages (1. Nil: 0; 2. Very low: 1–20%; 3. Low: 20–40%; 4. Medium: 40–60%; 5. High: 60–80%; 6. Very high: 80–100%).
2.2. Genetic and Phylogenetic Analysis of Pseudomonas ST1 and Pantoea Paga
Two genomes, Pseudomonas ST1 and Pantoea paga strains have been sequenced earlier and deposited under the accession numbers VKOF00000000.1 [37] and VLTF00000000 [38], respectively. Genomes of Pseudomonas ST1 and Pantoea paga strains were analyzed on the Mage platform [39] to compute clusters of orthologous genes (COGs) [40]. Using antiSMASH [41] genes clusters involved in the biosynthesis of secondary metabolites and virulence factors [42] were identified.
Using GBDP distances calculated from 16S rDNA gene sequences, phylogenetic trees were created with FastME 2.1.6.1. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 52.6%. The tree was rooted at the midpoint [43,44].
2.3. Confirmation of Pathogenicity of Pseudomonas ST1 and Pantoea Paga Isolated from the Trees in a Collection Orchard
The experiment was performed in an unshaded greenhouse at the Institute for Adriatic Crops and Karst Reclamation, Split in October 2020. Two olive cultivars Oblica and Lastovka that showed significantly different susceptibilities to olive knot disease in the first experiment were included in the study. Plants were planted in 5 L pots with a mixture of substrates including “Brill TYPical 4” and eutric brown soil (pH 5.5–6.8; 2–6% humus content). With a sterile scalpel, lateral wounds, approximately 5 to 10 mm in diameter, were introduced to the shoots of the two-year-old plants. The cambial tissue was exposed to artificial infection performed by inoculation with Pseudomonas ST1 strain [37], Pantoea paga strain [38], and combination of these two bacteria. Replicates of five plants per cultivar with ten lateral wounds was used (in total, fifty wounds per treatment condition and cultivar). In control plants no inoculation with bacterial strains was performed. Olive trees were assessed 6 months after inoculation and the percentage of knot formation was calculated for each treatment (number of wounds with symptoms/total number of wounds × 100). Treatments were evaluated based on the incidence of disease. The data for knot formation in different treatments were subjected to an analysis of variance using generalised linear models in SAS software (SAS Institute Inc., Cary, NC, USA). Differences between groups were determined by the LSD test at p ≤ 0.05.
To confirm the presence of bacteria in lateral wounds, bacteria from the olive knots and plant tissue around the wounds were cultured from the plants that showed symptoms of infection as well as from the plants that did not develop symptoms including control plants. The samples were sterilized using 75% ethanol, sliced with a sterile scalpel, and put onto King’s medium agar plates, kept in the dark [45]. The bacterial cells developed on medium were transferred onto a new King’s medium agar plates and kept at 37 °C. An isolated single colony from each treatment was transferred into 5 ml of liquid lysogeny broth (LB) medium and grown at 28 °C. DNA was isolated from bacterial cells in the precipitate using a microbial DNA kit (Macherey-Nagel, Hoerdt, France). 16S RNA gene was amplified in each sample using PCR reaction and sequenced.
The Efficiency of the Primers in Detection of the Bacteria
‘Lastovka’, the cultivar with the highest intensity symptoms, was used for virulence determination. Olive knots and plant tissue around the inoculation sites for the two bacterial strains were taken from plant shoots and disassociated with a sterile scalpel. The control plants were also treated similarly. The DNA isolation from disease knots and control plant tissue was purified with a microbial DNA kit (Macherey-Nagel, Hoerdt, France). The concentration of the DNA was measured spectrophotometrically by UV absorbance at 260–280 nm. The PCR amplifications were performed using sets of specific primers. Pantoea primers were: paga20853f 5′-TTACCCAGCGTGAAACCCG, paga20853r 5′-TCGGCTGGGTTCAGGCTC, paga20492f 5′-GGCCAAAGAAGACAATATTGAA, paga20492r 5′-AGCGACTACGGAAGACAATG, paga20832f 5′-TGGTACTGATTGCAGCGGG, paga20832r 5′-GCCCTTTGCCATCCTCCG. Pseudomonas ST1 primers were: PSST1-1220002f 5′-TCGGGGACCTGCACGTCC, PSST1-1220002r 5′-AGCGCCCTGGCCTACTGG, PSST1-1220003f 5′-ACTACCAGCTCCCAGGGC, PSST1-1220003r 5′-GGCGCCATCAGAGCCATAC. For 16sRNA primers were: 16S27f 5′-AGAGTTTGATCMTGGCTCAG, 16S1492r 5′-GGYTWCCTTGTTACGACTT [46,47], pA 5′-AGAGTTTGATCCTGGCTCAG, pH 5′-AAGGAGGTGATCCAGCCGCA [48], com1 CAGCAGCCGCGGTAATAC and com2 CCGTCAATTCCTTTGAGTTT [49]. DNA amplification was done in 25 µL volume of reaction mixture containing specific primers, 1× reaction buffer, dNTPs, DNA template and Taq polymerase (NEB) in accordance with manufacturer’s instructions with the following conditions: 95 °C for 5 min; 35 cycles of 95 °C for 20 s, 55 °C for 45 s and 72 °C for 1 min.
2.4. Susceptibility of Bacteria to Antibiotic and Chemical Agents Tested by a Standardized Single Disk Method
2.4.1. Bacteria Strains
The strains of two bacterial species used in this study were from our culture collection. Pseudomonas ST1 and Pantoea paga cells were isolated from olive knots from olive plants grown in the central region of Dalmatia, Croatia. The complete genome sequence was described and has been deposited in NCBI/GenBank [37,38].
2.4.2. Growth Media, Antibiotics and Chemical Agents
King’s A media for a single-disc diffusion technique was prepared according to King et al. [45]. The medium was used for antibiotic and chemical agents’ susceptibility testing. A single-disc diffusion technique was used for determination of the antibiotic susceptibility of bacteria. Eighteen antibiotics were tested including: Ciprofloxacin, Novobiocin, Streptomycin, Hygromycin, Chloramphenicol, Tetracycline, Trimethoprim, Carbenicillin, G418, Neomycin, Ampicillin, Vancomycin, Geneticin, Rifampicin, Spectinomycin, Apramycin, Ticarcillin and Kanamycin.
Antibiotics and chemical agents were diluted in sterile distilled water to prepare the solutions at the specified working concentrations. The antibiotics (Sigma Aldrich, Saint-Quentin-Fallavier, France) were used at the working concentrations of the following ranges: Ciprofloxacin (from 0.05 to 2.5 mg/mL), Novobiocin (from 0.748 to 37.4 mg/mL), Streptomycin (from 0.6 to 30 mg/mL), Hygromycin, Carbenicillin, G418, Neomycin, Ampicillin (from 2 to 100 mg/mL), Chloramphenicol (from 0.4 to 20 mg/mL), Tetracycline, Trimethoprim (from 0.5 to 25 mg/mL), Vancomycin (from 0.58 to 28.86 mg/mL), Geneticin, Kanamycin (from 0.8 to 40 mg/mL), Rifampicin (from 1.8 to 90 mg/mL), Spectinomycin (from 6 to 300 mg/mL), Apramycin (from 1 to 50 mg/mL) and Ticarcillin (250 mg/mL).
Chemical agents used in the experiment were: hydrogen peroxide (H2O2), magnesium carbonate (MgCO3), benzoic acid (C7H6O2), manganese sulphate monohydrate (MnSO4*H2O), sodium molybdate (Na2MoO4), zinc sulfate heptahydrate (ZnSO4*7H2O), iron (III) chloride (FeCl3), copper (II) sulfate pentahydrate (CuSO4*5H2O), ethylenediaminetetraacetic acid (EDTA; C10H16N2O8), iron (II) citrate (C12H30Fe3O24), potassium iodide (KI), potassium sulfate (K2SO4), p-hydroxybenzoic acid (C7H6O3), polyoxyethylenesorbitan monolaurate (tween; C58H114O26), mineral oil, and sodium dodecyl sulfate (SDS; C12H25NaO4S). Solutions of different chemical agents were prepared at the concentrations from 1 mg/ml to 50 mg/ml with the exception of hydrogen peroxide and dodecyl sulfate, which were used at the concentration from 0.6 to 30% and 0.004 to 20%, respectively. Antibiotic and chemical agents were added on the discs. The discs were placed on the King’s A media with growing bacterial cultures. To determine the efficiency of an individual antibiotic and chemical agent and their effective concentrations, the zone diameters were measured after overnight incubation at 37 °C. We measured the diameter of the growth inhibition zone [circular zone around an antibiotic disc with no visible bacterial growth (Figure S1)]. The diameter of the zone of inhibition of the test isolates were recorded. Some strains exhibited an inner area of light growth immediately around the antibiotic disc with an obvious area of inhibition outside this hazy growth. In these instances, the outer zone of inhibition was measured. The most obvious zone of inhibition was measured in those instances when several concentric zones were observed around an antibiotic disc.
3. Results
3.1. Susceptibility of Different Olive Cultivars to Olive Knot Disease
Different cultivars showed diverse intensities of symptoms for olive knot disease and were classified in six categories according to percentage of area affected (Table 1). We found no symptoms of disease in three Italian cultivars, Coratina, Favarol, and Leccino. The highest percentages of area covered by tumorous overgrowths ranging from 60–80% (category 5) and from 80–100% (category 6) were found for Frantoio, Lastovka, Santa Catarina and Chemlali. The Tunisian cultivar Chemlali was the most affected cultivar with the highest incidence of symptoms (category 6). Together with cultivars Fasolina, Grignan, Moraiolo and Sigoise, trees of Oblica, which is the most widespread cultivar in Croatia, showed very low percentage of area covered by symptoms ranging from 1-20% (category 2).

Table 1.
The classification of 21 olive cultivars in six categories according to their tolerance to olive knot disease (from category 1–the highest tolerance to category 6–the lowest tolerance). The categories correspond to percentages of area affected by disease (1. Nil: 0; 2. Very low: 1–20%; 3. Low: 20–40%; 4. Medium: 40–60%; 5. High: 60–80%; 6. Very high: 80–100% of area affected).
3.2. Genome Analysis of the Pseudomonas ST1 and Pantoea Paga
Plants of Lastovka and Oblica, the most representative olive cultivars grown along the eastern coast of the Adriatic area were used to isolate and identify causal agent of gall formation that could coevolve with olive plants. Fresh galls from different plants where surface sterilized in ethanol, sliced by sterile scalpel and placed on King’s agar plates. We cultured two visually different types of colonies after two days of incubation at room temperature. Total DNA was isolated from twelve colonies and part of the 16sRNA gene was amplified and sequenced. Blast results of sequence analysis of the 16sRNA genes from twelve random isolated colonies show presence of two groups of bacteria belonging to Pseudomonas and Pantoea genera.
Genomes of Pantoea paga and Pseudomonas ST1 isolates were sequenced using ilumina NGS technology end each genome was assembled for genome analysis [37,38].
The genome of Pseudomonas ST1 has 6.1 Mbps, genome coding for 6271 genomic objects and 58% of GC contents (Figure 1). Phylogenetic analysis of the whole genome and 16S rRNA gene sequence of Pseudomonas ST1 show that this strain belongs to known species Pseudomonas amygdali (Figure 2A,B).
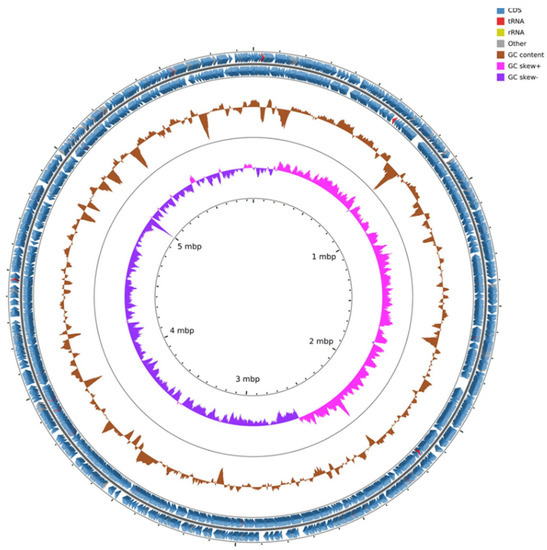
Figure 1.
Genome map of Pseudomonas ST1 strain with coding sequence (CDS), transfer RNA (tRNA), ribosomal RNA (rRNA), GC content, and inner cycle scale.
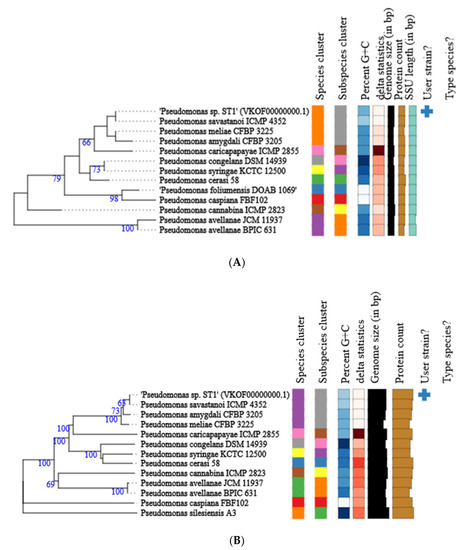
Figure 2.
Phylogenetic tree of Pseudomonas ST1 strain calculated from 16S rDNA gene sequences (A); phylogenetic tree of Pseudomonas ST1 strain calculated from whole genome sequences (B). Different colors within each column indicate different minimum, maximum and intermediate values; and +represents user strain.
Pseudomonas ST1 is a motile bacterium as confirmed under light microscopy observation and presence of flagellum and pili biosynthesis genes in the genome. Genome analysis shows presence of six secretion systems, from TSS1 to TSS6, and an alginate biosynthesis operon related to production of extracellular polysaccharides necessary to host attachment and virulence. The global activator, two component system gacS is also present in the genome and could regulate not only alginate production but also homo serine lactone production, toxin production and other virulence related genes and operons (Figure 3 and Table S1). Presence of two operons with genes (iaaM1, iaaH1, iaaM2, and iaaH2) involved in indole acetic acid (IAA) biosynthesis indicates potential for the strain to produce plant hormone. Eight genome clusters coding for secondary metabolites are present in the genome. Most of them are nonribosomal peptides synthetases (NRPS) involved in biosynthesis of secondary metabolites. Based on homology searches, these clusters can be involved in biosynthesis of antibiotics, signaling molecules, siderophores, toxins, and storage materials (Table S2).
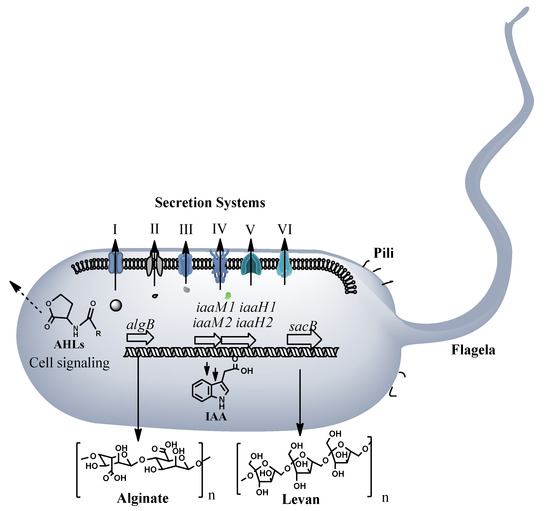
Figure 3.
Pathogenicity and virulence factors of Pseudomonas ST1 strain, secretion system (I–VI), motility factors, pili and flagella, alginate and levan biosynthesis, IAA hormone biosynthesis and signaling molecule biosynthesis. For full list of conserved virulence related genes and proteins see Table S1.
Whole genome homology searches show similarity of Pseudomonas ST1 isolates to the Pseudomonas savastanoi ICMP. Two strains share 5442 common genes and each strain has strain-specific genes. Pseudomonas ST1 has 568 strain-specific genes in comparison and Pseudomonas savastanoi that has 434 specific genes (Table S3 and Figure S2).
The Pantoea paga genome is 5.08 Mbps with 54.58% of G + C contents. The genome has 4777 coding sequences, one ribosomal operon coding for 16s, 23s, and 5s rRNA genes, 70 tRNA genes and 91 misc_RNA (Figure 4).
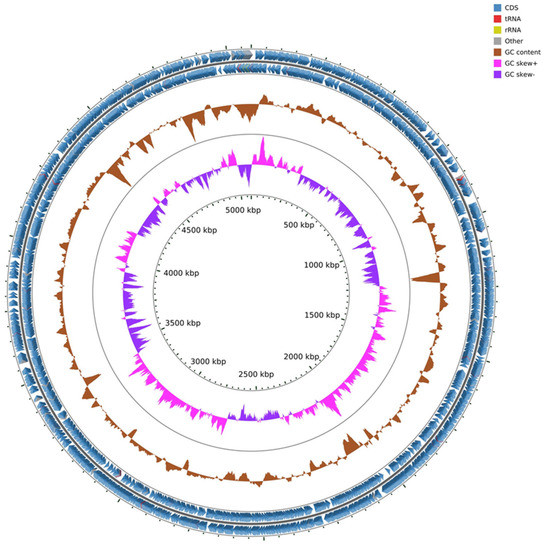
Figure 4.
Genome map of Pantoea paga strain with coding sequence (CDS), transfer RNA (tRNA), ribosomal RNA (rRNA), GC content, and inner cycle scale.
The genome has T1SS, T3SS, and T5SS genes for secretion systems, focA gene for pilin biosynthesis, hlyF gene for hemolysin, and only one hopD1 gene related to pathogenicity. Seven genome regions contain genes for secondary metabolite biosynthesis such as carotenoids, zeaxantinins, siderophores, and homoserine lactone. The Pantoea paga strain has mcb bacteriocin biosynthesis operon containing genes involved in biosynthesis, maturation, modification, and secretion of antimicrobial peptide.
Phylogenetic analysis of the whole genome and 16S rRNA gene sequence of Pantoea paga shows that this strain belongs to the known species Pantoea agglomerans (Figure 5A,B).
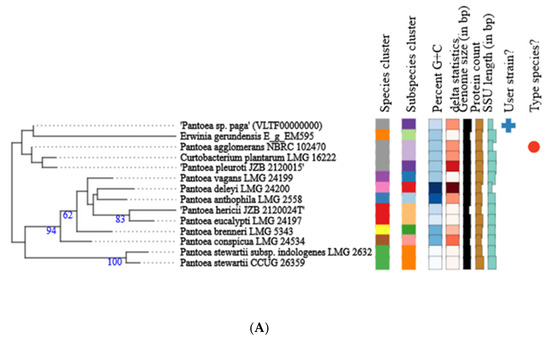
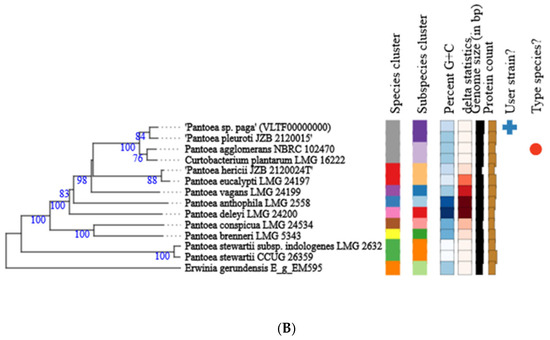
Figure 5.
Phylogenetic tree of Pantoea paga strain calculated from 16S rDNA gene sequences (A); phylogenetic tree of Pantoea paga strain calculated from whole genome sequences (B). Different colors within each column indicate different minimum, maximum and intermediate values; + represents user strain and red dot indicates type species.
The statistic distribution of the protein coding genes of the two genomes within the clusters of orthologous groups of functional categories is shown in Figure 6 [40].
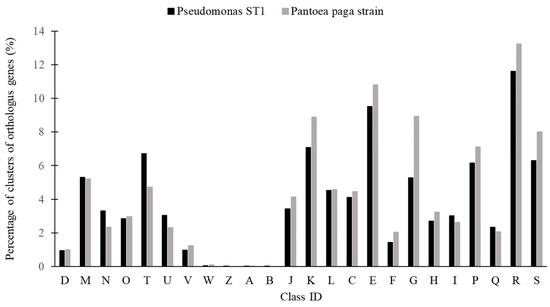
Figure 6.
Percentage of clusters of orthologus genes (%) in Pseudomonas ST1 strain and Pantoea paga strain. Letters on the x-axis correspond to different groups of functional genes: D—cell cycle control, cell division, chromosome partitioning; M—cell wall/membrane/envelope biogenesis; N—cell motility; O—posttranslational modification, protein turnover, chaperones; T—signal transduction mechanisms; U—intracellular trafficking, secretion, and vesicular transport; V—defense mechanisms; W—extracellular structures; Z—cytoskeleton; A—RNA processing and modification; B—chromatin structure and dynamics; J—translation, ribosomal structure, and biogenesis; K—transcription; L—replication, recombination, and repair; C—energy production and conversion; E—amino acid transport and metabolism; F—nucleotide transport and metabolism; G—carbohydrate transport and metabolism; H—coenzyme transport and metabolism; I—lipid transport and metabolism; P—inorganic ion transport and metabolism; Q—secondary metabolites biosynthesis, transport and catabolism; R—general function prediction only; and S—function unknown.
3.3. Pathogenicity of Pseudomonas ST1 and Pantoea Paga
To verify the causative agent of knot formation we inoculated healthy olive plants with isolated bacterial species alone and in a mixture of the two bacterial isolates. Pure culture of two strains were mixed and inoculated on healthy olive plants. Six months post inoculation of bacteria we observed formation of galls on plants tissues and compared symptoms with control plants (Figure 7A–D). We observed formation of galls when Pseudomonas ST1 strain was used as inoculant either alone or in a mixture with Pantoea sp. paga strain (Figure 7C,D). There was no gall formation on the plants where Pantoea paga strain was applied alone (Figure 7B).

Figure 7.
Appearance of symptoms of olive knot disease six months post inoculation by Pseudomonas ST1 and Pantoea paga strains (A) control plants; (B) plants inoculated with Pantoea paga strain; (C) plants inoculated with Pseudomonas ST1 strain; and (D) plants inoculated with a mixture of both bacteria.
Total DNA was isolated from infected areas of plant tissues. Presence of two bacteria was confirmed by PCR amplification of 16s rDNA genes and PCR amplification of strain specific genes using specific pairs of primers followed by amplicon sequencing. Formation of knots and presence of Pseudomonas ST1 strain was confirmed in all tested knots. We conclude that the isolate of Pseudomonas sp. alone is the causative agent of knot formation on olive plants (Figure 8). Pantoea sp. cells inoculated alone on olive plants did not induce formation of galls. Lastovka showed higher level of knot formation compared to Oblica when inoculated with Pseudomonas ST1 strain (Figure 8). A mixture of two bacteria enhanced the development of knots as observed in Oblica.
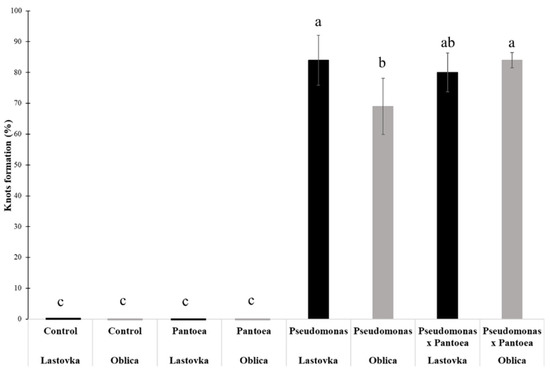
Figure 8.
Percentage of knots formation on Oblica and Lastovka cultivars inoculated with Pantoea paga or Pseudomonas ST1 strains used alone or with mixture of two strains compared to non-inoculated control plants. The results are expressed as the means ± SE. Different lowercase letters indicate significant differences according to the least significant difference test at p < 0.05.
3.4. Susceptibility of Two Bacteria to Various Chemical Agents and Antibiotics
The antibacterial activities of the antibiotics and chemical agents in different concentrations against the Pseudomonas ST1 and Pantoea paga strains were examined for the presence or absence of the inhibition zones by the agar disc diffusion method. Sensitivity values of each antibiotic and chemical agent used for antibacterial activity are shown in Table 2.

Table 2.
In vitro antibacterial activity of the antibiotics and chemical agents tested against Pseudomonas ST1 and Pantoea paga strains. Tested concentrations are shown in mg/mL or percentage (%).
Pseudomonas ST1 strain showed the highest susceptibility to all of the used antibiotics, while Pantoea paga strain showed reduced sensitivity to hygromycin and vancomycin. Both strains were resistant to iron (II) citrate, magnesium carbonate, mineral oil, p-hydroxybenzoic acid, polyoxyethylenesorbitan monolaurate, potassium iodide, and potassium sulfate. The sodium molybdate was not effective against Pantoea paga. The high efficiency of sodium dodecyl sulfate on Pseudomonas ST1 was confirmed. Both bacterial strains were very susceptible to copper salts. Hydrogen peroxide showed the highest activity against both bacteria and it is one of the best candidates in the protection of olive plants from the bacteria causing olive knot.
4. Discussion
Pseudomonas savastanoi and Pantoea aglomerans are recognized as causal agents of plant knot disease, causing aerial tumorous growth that evolve to necrosis [1,3,4,34,35,50]. Each bacterium alone or together can induce tumor formation on plant tissue [10,51].
The occurrence and intensity of symptoms in olive cultivars was assessed in the Olive Germplasm Bank of Croatia. In earlier studies, the two most common bacterial species associated with the olive-knot microbiome were isolated from the knots of olive trees grown in Croatia and their genomes have been sequenced [37,38]. Two bacterial strains of Pseudomonas [52] and Pantoea [53] are known as causative agents of knot formation on plants. In silico analysis of Pseudomonas ST1 genome shows presence of all virulence genes necessary for infection such as type T3SS secretion system, protected polymers production, alginate, levan, and production of the phytohormone IAA [54,55]. The strain has two putative operons for production of IAA that could be related to high infectivity of this endemic strain in knot formation. This strain shares a high proportion of genes in common with other Pseudomonas strains. However, strain ST1 has 568 specific genes with unknown function. These genes may contribute to its specific ecological niche. It possesses three gene clusters coding non-ribosomal peptide synthetase that could synthetize bicornutin, rhizomide, and mangotoxin. According to phylogenetic analyses the Pseudomonas ST1 strain belongs to the Pseudomonas amygdali species.
In contrast, Pantoea paga has no panel of virulence genes in its genome. This strain belongs to the Pantoea agglomerans species and probably uses Pseudomonas ST1 to invade plant tissue. It contains antimicrobial genes involved in microcin biosynthesis that is known as antibacterial peptide inhibiting DNA gyrase [56]. Therefore, the presence of Pantoea paga can affect the growth of other bacteria.
The genomes were further analyzed in the present study to determine differences between the two bacteria. These two bacteria were used in the new experiment to determine whether their co-infection intensifies the symptoms of olive knot disease in two olive cultivars Lastovka and Oblica and to determine whether each of them is sufficient to cause the symptoms. Pseudomonas ST1 is the causative agent of knot formation as it was shown in the experiment where the healthy olive plants were inoculated with two bacteria. Inoculation with the Pantoea strain did not induce knot formation when inoculated alone; however, join inoculation with Pseudomonas ST1 strain increased the percentage of knot formation in Oblica cultivar. The differences in occurrence and intensity of symptoms in cultivars Oblica and Lastovka were less pronounced in the greenhouse experiment, where the young plants were artificially infected compared to the evaluation carried out in the collection orchard, where the knots were formed over the longer period before the assessment and without our interference.
To avoid the spread of P. savastanoi or other pathogens, it is important to develop strain specific identification. Here we successfully used strain specific PCR oligonucleotide primers for the detection of two bacteria. Furthermore, in order to improve measures to control the spread of this disease in the process of propagation of olive plant material, the efficiency of antibiotics and other chemical agents in control of pathogens has been investigated. Our results provide new insights on the epidemiology of this disease and enable Pseudomonas and Pantoea control.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10081529/s1. Figure S1: Disc diffusion method for antibiotic and chemical agent’s susceptibility testing. Figure S2: Venn diagram showing two genomes (Pseudomonas ST1 and Pseudomonas savastanoi pv. savastanoi PVFi1) with core genome and strain specific genes (Table S3). Table S1: Conserved Pseudomonas ST1 virulence genes. Table S2: Cluster of genes for biosynthesis of secondary metabolites; Table S3: Strain specific and common genes in Pseudomonas ST1.
Author Contributions
Conceptualization and methodology, G.V.S., P.P. and M.R.B.; data analysis, G.V.S. and P.P.; formal analysis and investigation, G.V.S., P.P., M.R.B., D.A. and M.B.; writing—original draft preparation, G.V.S. and P.P.; writing—review and editing, G.V.S., P.P., K.Ž., S.P. and P.N.; supervision, G.V.S. and P.P.; project administration and funding acquisition, G.V.S., K.Ž. and S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the project Biodiversity and Molecular Plant Breeding, Centre of Excellence for Biodiversity and Molecular Plant Breeding (CoE CroP-BioDiv), Zagreb, Croatia, grant number KK.01.1.1.01.0005; by the Split-Dalmatia County grant for the project “Agents, transmitters and bio-control of olive knot disease in the olive groves of the Split Dalmatia County”. M.R.B. is a Ph.D. fellow supported by a French government scholarship in 2019/2020 that was coordinated by the Ministry of Science and Education of Republic of Croatia, Agency for Mobility and EU Programmes and Embassy of the French Republic to the Republic of Croatia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions generated for this study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Acknowledgments
We thank Betty Bigai and Delphine Poncet for their valuable help and support and platform Genomique Environnementale PGE, UMR CNRS 5557 for technical support. The authors would like to thank Rosa Crnov and Stanley Tam for valuable comments and corrections.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Quesada, J.M.; Penyalver, R.; López, M.M. Epidemiology and control of plant diseases caused by phytopathogenic bacteria: The case of olive knot disease caused by Pseudomonas savastanoi pv. savastanoi. In Plant Pathology; Cumagun, C.J., Ed.; Intech: Kayl, Luxembourg, 2012; pp. 299–326. [Google Scholar]
- Vivian, A.; Mansfield, J.A. Proposal for a uniform genetic nomenclature for avirulence genes in phytopathogenic pseudomonads. Mol. Plant-Microbe Interact. 1993, 6, 9–10. [Google Scholar]
- Quesada, J.M.; Penyalver, R.; Pérez-Panades, J.; Salcedo, C.I.; Carbonell, E.A.; López, M.M. Dissemination of Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathol. 2010, 59, 262–269. [Google Scholar] [CrossRef]
- Comai, L.; Kosuge, T. Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J. Bacteriol. 1980, 143, 950–957. [Google Scholar]
- Surico, G.; Iacobellis, N.S.; Sisto, A. Studies on the role of indole-3-acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. savastanoi. Physiol. Plant Pathol. 1985, 26, 309–320. [Google Scholar] [CrossRef]
- Ramos, C.; Matas, I.M.; Bardaji, L.; Aragon, I.M.; Murillo, J. Pseudomonas savastanoi pv. savastanoi: Some like it knot. Mol. Plant Pathol. 2012, 13, 998–1009. [Google Scholar] [CrossRef]
- Schroth, M.N.; Osgood, J.W.; Miller, T.D. Quantitative assessment of the effect of the olive knot disease on olive yield and quality. Phytopathology 1973, 63, 1064–1065. [Google Scholar]
- Wilson, E.E. The olive knot disease: Its inception, development and control. Hilgardia 1935, 9, 231–264. [Google Scholar]
- Passos da Silva, D.; Castaneda-Ojeda, P.M.; Moretti, C.; Buonaurio, R.; Ramos, C.; Venturi, V. Bacterial multispecies studies and microbiome analysis of a plant disease. Microbiology 2014, 160, 556–566. [Google Scholar] [CrossRef]
- Marchi, G.; Sisto, A.; Cimmino, A.; Andolfi, A.; Cipriani, M.G.; Evidente, A.; Surico, G. Interaction between Pseudomonas savastanoi pv. savastanoi and Pantoea agglomerans in olive knots. Plant Pathol. 2006, 55, 614–624. [Google Scholar] [CrossRef]
- Penyalver, R.; García, A.; Ferrer, A.; Bertolini, E.; Quesada, J.M.; Salcedo, C.I.; Piquer, J.; Pérez-Panadés, J.; Carbonell, E.A.; del Río, C.; et al. Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculations and their use for evaluation of olive cultivar susceptibility. Phytopathology 2006, 96, 313–319. [Google Scholar] [CrossRef]
- Hosni, T.; Moretti, C.; Devescovi, G.; Suarez-Moreno, Z.R.; Fatmi, M.B.; Guarnaccia, C.; Pongor, S.; Onofri, A.; Buonaurio, R.; Venturi, V.; et al. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 2011, 5, 1857–1870. [Google Scholar] [CrossRef]
- Buonaurio, R.; Moretti, C.; Cortese, C.; da Silva, D.P.; Ramos, C.; Venturi, V. The olive knot disease as a model to study the role of interspecies bacterial communities in plant disease. Front. Plant Sci. 2015, 6, 434. [Google Scholar] [CrossRef] [Green Version]
- Bodenhausen, N.; Bortfeld-Miller, M.; Ackermann, M.; Vorholt, J.A. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLOS Genet. 2014, 10, e1004283. [Google Scholar] [CrossRef] [Green Version]
- Gomes, T.; Pereira, J.A.; Lino-Neto, T.; Bennett, A.E.; Baptista, P. Bacterial disease induced changes in fungal communities of olive tree twigs depend on host genotype. Sci. Rep. 2019, 9, 5882. [Google Scholar] [CrossRef] [Green Version]
- Mina, D.; Pereira, J.A.; Lino-Neto, T.; Baptista, P. Impact of plant genotype and plant habitat in shaping bacterial pathobiome: A comparative study in olive tree. Sci. Rep. 2020, 10, 3475. [Google Scholar] [CrossRef] [Green Version]
- Sisto, A.; Cipriani, M.G.; Morea, M. Knot formation caused by Pseudomonas syringae subsp. savastanoi on olive plants is hrp-dependent. Phytopathology 2004, 94, 484–489. [Google Scholar] [CrossRef] [Green Version]
- Cunnac, S.; Lindeberg, M.; Collmer, A. Pseudomonas syringae type III secretion system effectors: Repertoires in search of functions. Curr. Opin. Microbiol. 2009, 12, 53–60. [Google Scholar] [CrossRef]
- Mansfield, J.W. From bacterial avirulence genes to effector functions via the hrp delivery system: An overview of 25 years of progress in our understanding of plant innate immunity. Mol. Plant Pathol. 2009, 10, 721–734. [Google Scholar] [CrossRef]
- Pérez-Martínez, I.; Rodríguez-Moreno, L.; Lambertsen, L.; Matas, I.M.; Murillo, J.; Tegli, S.; Jiménez, A.J.; Ramos, C. Fate of a Pseudomonas savastanoi pv. savastanoi type III secretion system mutant in olive plants (Olea europaea L.). Appl. Environ. Microbiol. 2010, 76, 3611–3619. [Google Scholar] [CrossRef] [Green Version]
- Grant, S.R.; Fisher, E.J.; Chang, J.H.; Mole, B.M.; Dangl, J.L. Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 2006, 60, 425–449. [Google Scholar] [CrossRef] [Green Version]
- Heath, M.C. Hypersensitive response-related death. Plant Mol. Biol. 2000, 44, 312–334. [Google Scholar] [CrossRef]
- Glickmann, E.; Gardan, L.; Jacquet, S.; Hussain, S.; Elasri, M.; Petit, A.; Dessaux, Y. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 1998, 11, 156–162. [Google Scholar]
- Bender, C.L.; Alarcón-Chaidez, F.; Gross, D.C. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar] [CrossRef] [Green Version]
- Caballo-Ponce, E.; Murillo, J.; Martínez-Gil, M.; Moreno-Pérez, A.; Pintado, A.; Ramos, C. Knots untie: Molecular determinants involved in knot formation induced by Pseudomonas savastanoi in woody hosts. Front. Plant Sci. 2017, 8, 1089. [Google Scholar] [CrossRef] [Green Version]
- Dillon, M.M.; Almeida, R.N.D.; Laflamme, B.; Martel, A.; Weir, B.S.; Desveaux, D.; Guttman, D.S. Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front. Plant Sci. 2019, 10, 418. [Google Scholar] [CrossRef] [Green Version]
- Glass, N.L.; Kosuge, T. Role of indoleacetic acid-lysine synthetase in regulation of indoleacetic acid pool size and virulence of Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 1988, 170, 2367–2373. [Google Scholar] [CrossRef] [Green Version]
- Quesada, J.M.; Penyalver, R.; Pérez-Panadés, J.; Salcedo, C.I.; Carbonell, E.A.; López, M.M. Comparison of chemical treatments for reducing epiphytic Pseudomonas savastanoi pv. savastanoi populations and for improving subsequent control of olive knot disease. Crop Prot. 2010, 29, 1413–1420. [Google Scholar] [CrossRef]
- Marcelo, A.; Fernandes, M.; Fatima Potes, M.; Serrano, J.F. Reactions of some cultivars of Olea europaea L. to experimental inoculation with Pseudomonas syringae pv. savastanoi. Acta Hortic. 1999, 474, 581–584. [Google Scholar]
- Hassani, D.; Buonaurio, R.; Tombesi, A. Response of some olive cultivars, hybrid and open pollinated seedling to Pseudomonas savastanoi pv. Savastanoi. In Pseudomonas syringae and Related Pathogens; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003; pp. 489–494. [Google Scholar]
- Valverde, P.; Zucchini, M.; Polverigiani, S.; Lodolini, E.M.; López-Escudero, F.J.; Neri, D. Olive knot damages in ten olive cultivars after late-winter frost in central Italy. Sci. Hortic. 2020, 266, 109274. [Google Scholar] [CrossRef]
- EPPO. Pathogen-tested olive trees and rootstocks. EPPO Bull. 2006, 36, 77–83. [Google Scholar]
- Gavini, F.; Mergaert, J.; Beji, A.; Mielcarek, C.; Izard, D.; Kersters, K.; De Ley, J. Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int. J. Syst. Bacteriol. 1989, 39, 337–345. [Google Scholar]
- Manulis, S.; Gafni, Y.; Clark, E.; Zutra, D.; Ophir, Y.; Barash, I. Identification of a plasmid DNA probe for detection of Erwinia herbicola pathogenic on Gypsophila paniculata. Phytopathology 1991, 81, 54–57. [Google Scholar]
- Manulis, M.; Barash, I. The molecular basis for transformation of an epiphyte into a gall-forming pathogen as exemplified by Erwinia herbicola pv. gypsophilae. In Plant-Microbe Interactions; Stacey, G., Keen, N., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2003; pp. 9–52. [Google Scholar]
- Volcani, Z. Bacterial Diseases of Plants in Israel; Agricultural Research Organization, the Volcani Center: Bet Dagan, Israel, 1985; p. 388. [Google Scholar]
- Vuletin Selak, G.; Raboteg, M.; Fournier, P.; Dubost, A.; Abrouk, D.; Žanić, K.; Perica, S.; Normand, P.; Pujić, P. Genome sequence of Pseudomonas sp. Strain ST1, Isolated from Olive (Olea europaea L.) Knot Galls in Croatia. Microbiol. Resour. Announc. 2019, 8, e00986-19. [Google Scholar] [CrossRef] [Green Version]
- Vuletin Selak, G.; Raboteg, M.; Dubost, A.; Abrouk, D.; Žanić, K.; Normand, P.; Pujić, P. Whole-genome sequence of a Pantoea sp. strain isolated from an olive (Olea europaea L.) Knot. Microbiol. Resour. Announc. 2019, 8, e00978-19. [Google Scholar] [CrossRef] [Green Version]
- Vallenet, D.; Calteau, A.; Cruveiller, S.; Gachet, M.; Lajus, A.; Josso, A.; Mercier, J.; Renaux, A.; Rolli, J.; Rouy, Z.; et al. MicroScope in 2017: An expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017, 45, D517–D528. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Natale, D.A.; Garkavtsev, I.V.; Tatusova, T.A.; Shankavaram, U.T.; Rao, B.S.; Kiryutin, B.; Galperin, M.Y.; Fedorova, N.D.; Koonin, E.V. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar]
- Liu, B.; Zheng, D.D.; Zhou, S.Y.; Chen, L.H.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Olofsson, T.C.; Ahrné, S.; Molin, G. Composition of the bacterial population of refrigerated beef, identified with direct 16S rRNA gene analysis and pure culture technique. Int. J. Food Microbiol. 2007, 18, 233–240. [Google Scholar]
- Edwards, U.; Rogall, T.; Böcker, H.; Emde, M.; Böttger, E. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal DNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar]
- Lane, D.J.; Pace, B.; Olson, G.J.; Stahl, D.A.; Sagin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar]
- Rodríguez-Palenzuela, P.; Matas, I.M.; Murillo, J.; López-Solanilla, E.; Bardaji, L.; Pérez-Martínez, I.; Rodríguez-Moskera, M.E.; Penyalver, R.; López, M.M.; Quesada, J.M.; et al. Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of atumour-inducing pathogen of woody hosts. Environ. Microbiol. 2010, 12, 1604–1620. [Google Scholar]
- Bartoli, C.; Carrere, S.; Lamichhane, J.R.; Varvaro, L.; Morris, C.E. Whole-genome sequencing of 10 Pseudomonas syringae strains representing different host range spectra. Genome Announc. 2015, 3, e00379-15. [Google Scholar]
- Turco, S.; Drais, M.I.; Rossini, L.; Chaboteaux, E.; Rahi, Y.J.; Balestra, G.M.; Iacobellis, N.S.; Mazzaglia, A. Complete genome assembly of the levan-positive strain PVFi1 of Pseudomonas savastanoi pv. savastanoi isolated from olive knots in Central Italy. Environ. Microbiol. Rep. 2022, 14, 274–285. [Google Scholar] [CrossRef]
- Barash, I.; Manulis-Sasson, S. Virulence mechanisms and host specificity of gall-forming Pantoea agglomerans. Trends Microbiol. 2007, 15, 538–545. [Google Scholar] [CrossRef]
- Matas, I.M.; Lambertsen, L.; Rodríguez-Moreno, L.; Ramos, C. Identification of novel virulence genes and metabolic pathways required for full fitness of Pseudomonas savastanoi pv. savastanoi in olive (Olea europaea) knots. New Phytol. 2012, 196, 1182–1196. [Google Scholar] [CrossRef] [Green Version]
- Matas, I.M.; Castañeda-Ojeda, M.P.; Aragón, I.M.; Antúnez-Lamas, M.; Murillo, J.; Rodríguez-Palenzuela, P.; López-Solanilla, E.; Ramos, C. Translocation and functional analysis of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III secretion system effectors reveals two novel effector families of the Pseudomonas syringae complex. Mol. Plant-Microbe Interact. 2014, 27, 424–436. [Google Scholar] [CrossRef] [Green Version]
- Sinha Roy, R.; Gehring, A.M.; Milne, J.C.; Belshaw, P.J.; Walsh, C.T. Thiazole and oxazole peptides: Biosynthesis and molecular machinery. Nat. Prod. Rep. 1999, 16, 249–263. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).