Effect of Solar Radiation on Skin Microbiome: Study of Two Populations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sampling
2.3. DNA Extraction and PCR Amplification
2.4. Data Analysis
3. Results
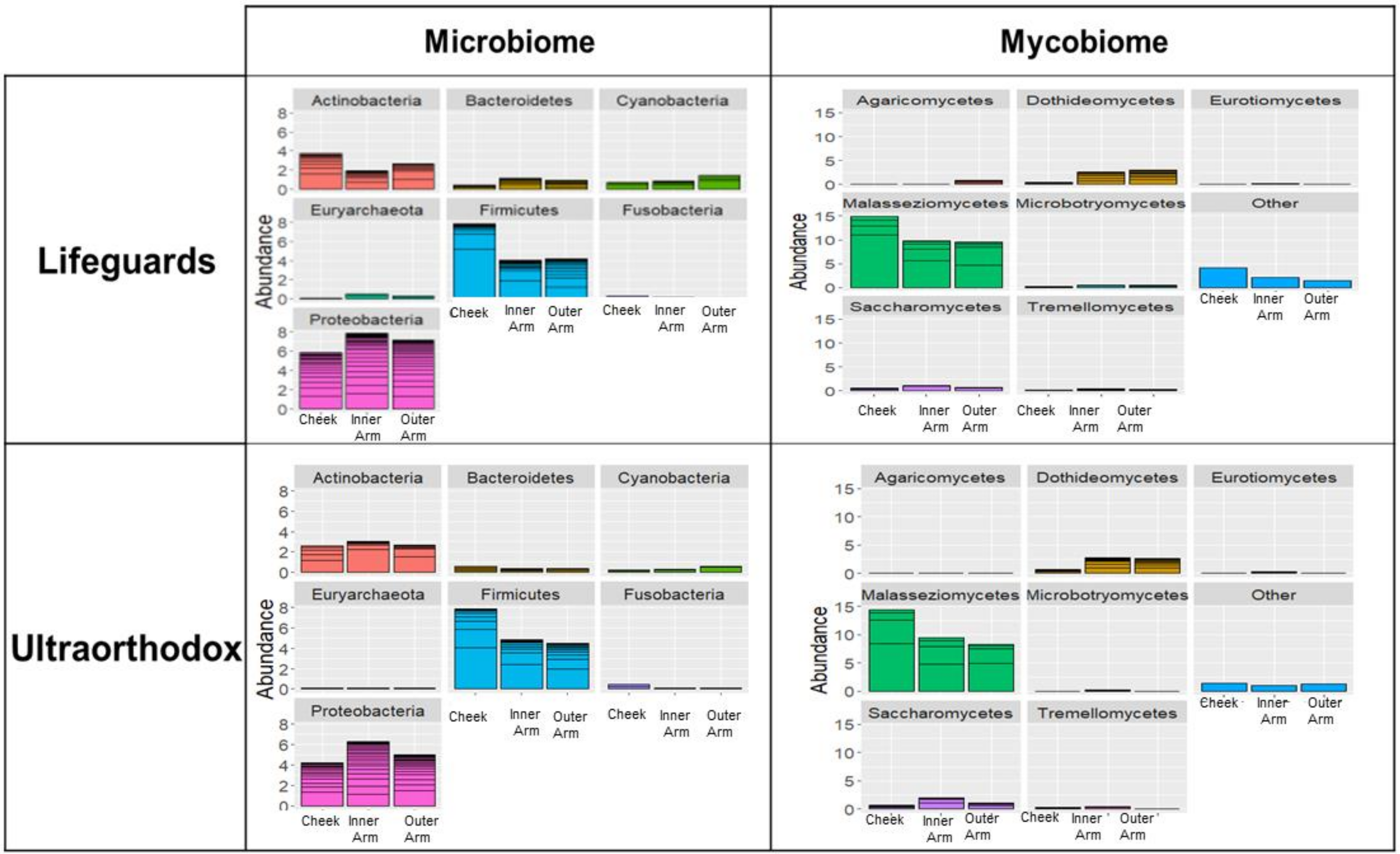
3.1. The Microbiomes of Lifeguards and Ultraorthodox Males Are Broadly Similar
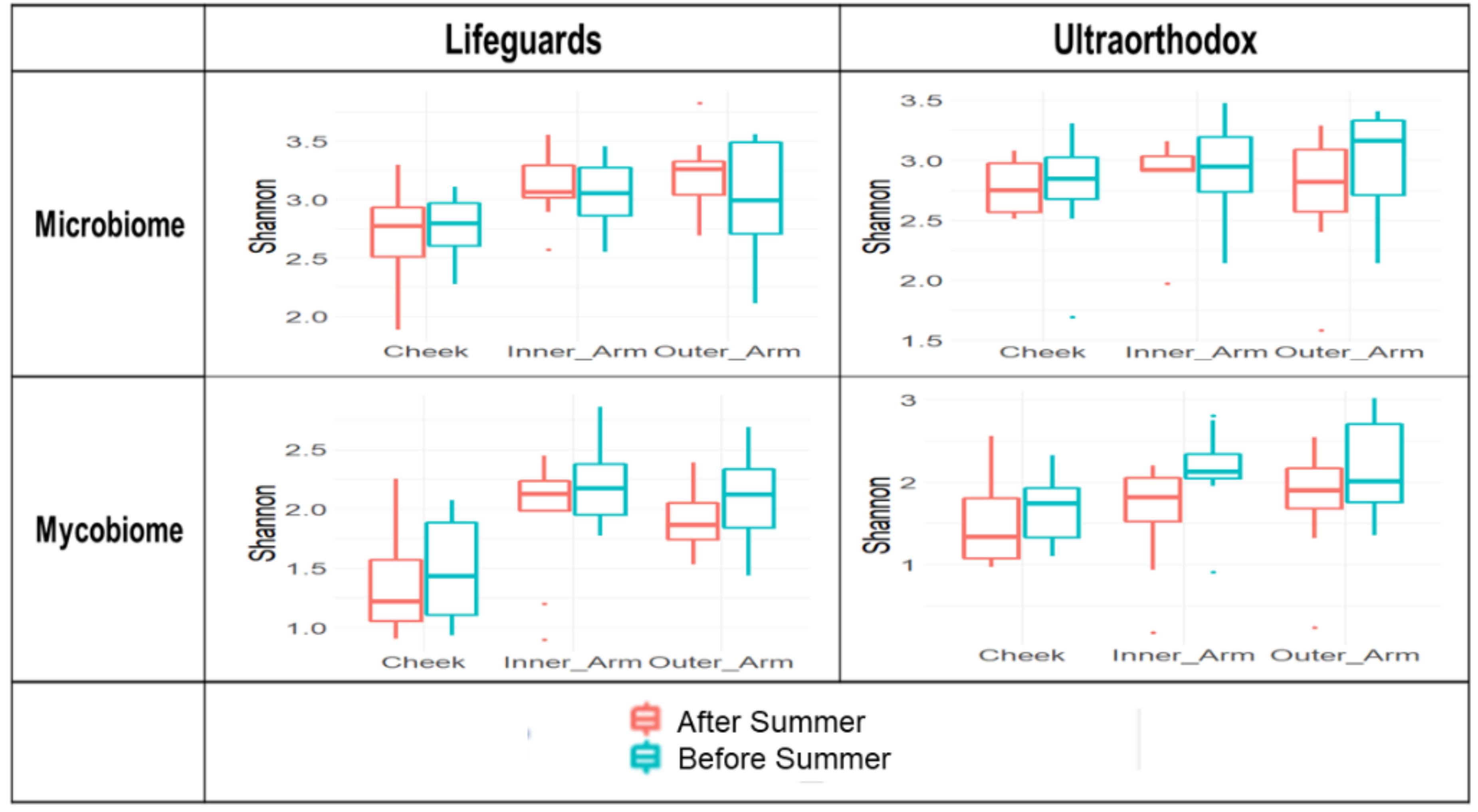
3.2. Microbial Diversity Is Maintained after the Summer Season in Both Lifeguards and Ultraorthodox
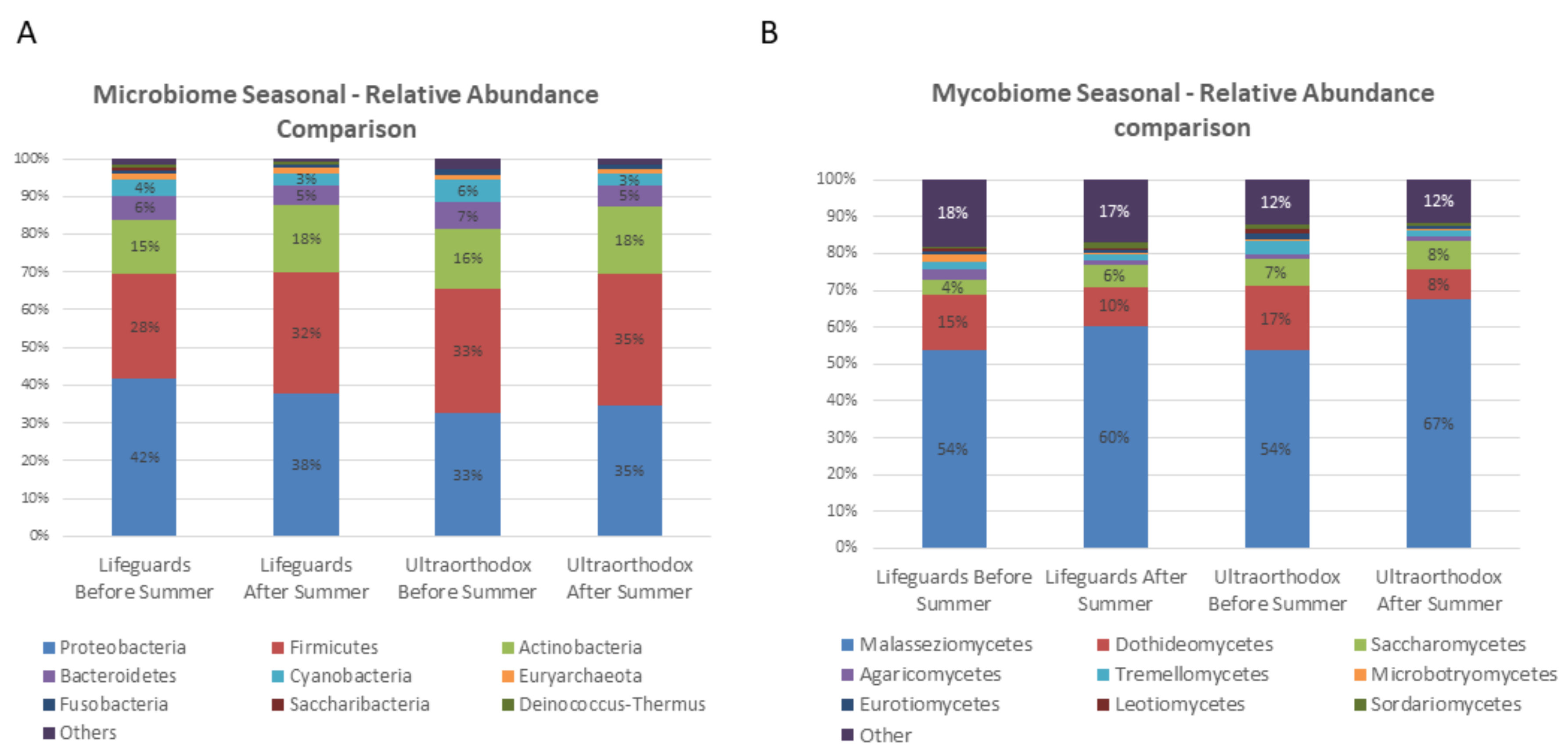
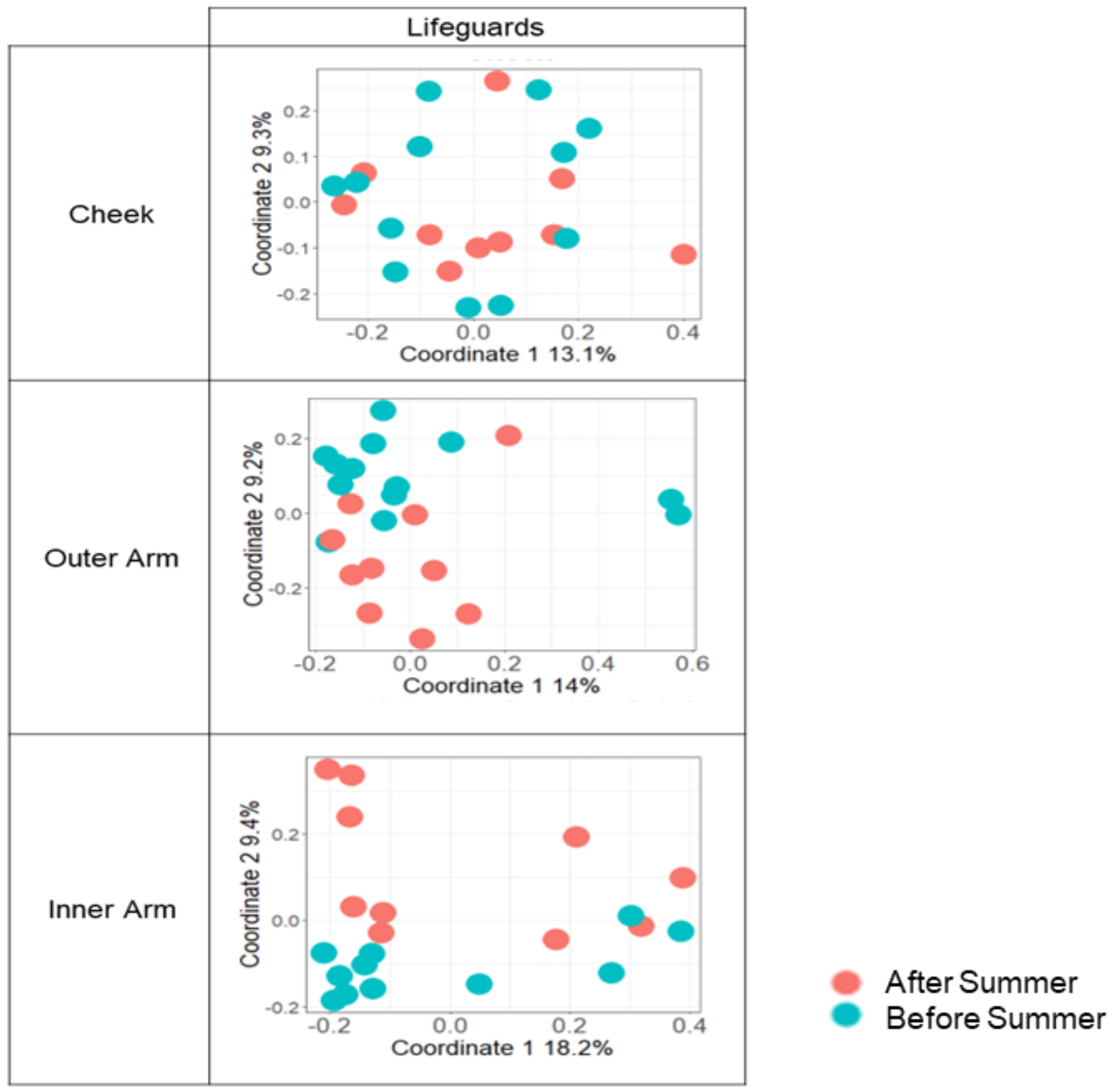
3.3. The Microbial Composition of the Lifeguards’ Changes Following Summer
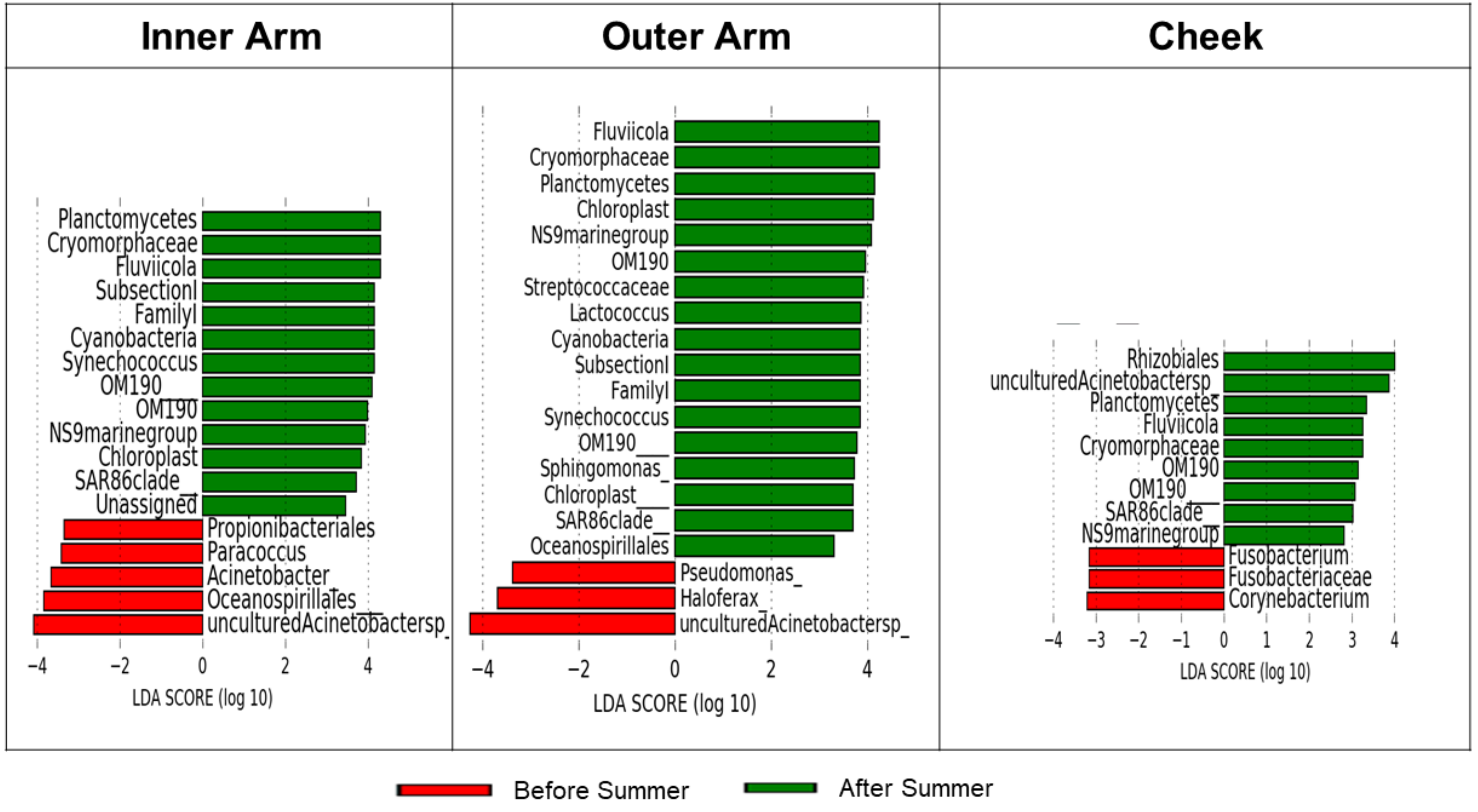
3.4. Specific Taxa Driving Seasonal Differences in the Lifeguards’ Microbiome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Microbiol. 2014, 14, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Meisel, J.S.; Sfyroera, G.; Bartow-McKenney, C.; Gimblet, C.; Bugayev, J.; Horwinski, J.; Kim, B.; Brestoff, J.R.; Tyldsley, A.S.; Zheng, Q.; et al. Commensal microbiota modulate gene expression in the skin. Microbiome 2018, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Segre, J.A. Dialogue between skin microbiota and immunity. Science 2014, 346, 954–959. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [Green Version]
- Chiller, K.; Selkin, B.A.; Murakawa, G.J. Skin microflora and bacterial infections of the skin. J. Investig. Dermatol. Symp. Proc. 2001, 6, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.C.H.; McMillan, N.A.; Antonsson, A. Human papillomavirus type spectrum in normal skin of individuals with or without a history of frequent sun exposure. J. Gen. Virol. 2008, 89, 2891–2897. [Google Scholar] [CrossRef]
- Garcia-Pichel, F. A model for internal self-shading in planktonic organisms and its implications for the usefulness of ultraviolet sunscreens. Limnol. Oceanogr. 1994, 39, 1704–1717. [Google Scholar] [CrossRef]
- Jeffrey, W.H.; Pledger, R.J.; Aas, P.; Hager, S.; Coffin, R.B.; Von Haven, R.; Mitchell, D.L. Diel and depth profiles of DNA photodamage in bacterioplankton exposed to ambient solar ultraviolet radiation. Mar. Ecol. Prog. Ser. 1996, 137, 283–291. [Google Scholar] [CrossRef]
- Bailey, C.A.; Neihof, R.A.; Tabor, P.S. Inhibitory effect of solar radiation on amino acid uptake in Chesapeake Bay bacteria. Appl. Environ. Microbiol. 1983, 46, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Müller-Niklas, G.; Heissenberger, A.; Puskaric, S.; Herndl, G.J. Ultraviolet-B radiation and bacterial metabolism in coastal waters. Aquat. Microb. Ecol. 1995, 9, 111–116. [Google Scholar] [CrossRef]
- Pakulski, J.D.; Aas, P.; Jeffrey, W.; Lyons, M.; van Waasbergen, L.G.; Mitchell, D.; Coffin, R. Influence of light on bacterioplankton production and respiration in a subtropical coral reef. Aquat. Microb. Ecol. 1998, 14, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.; Morris, A.; Ghedin, E. The human mycobiome in health and disease. Genome Med. 2013, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High resolution sample inference from amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [Green Version]
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2020. Available online: http://www.rstudio.com/ (accessed on 24 June 2022).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Conlan, S.; Polley, E.C.; Segre, J.A.; Kong, H.H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012, 4, 77. [Google Scholar] [CrossRef] [Green Version]
- Burns, E.M.; Ahmed, H.; Isedeh, P.N.; Kohli, I.; Van Der Pol, W.; Shaheen, A.; Muzaffar, A.F.; Al-Sadek, C.; Foy, T.M.; Abdelgawwad, M.S.; et al. Ultraviolet radiation, both UVA and UVB, influences the composition of the skin microbiome. Exp. Dermatol. 2019, 28, 136–141. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Q.; Zhou, R.; Feng, T.; Hilal, M.G.; Li, H. Nationality and body location alter human skin microbiome. Appl. Microbiol. Biotechnol. 2021, 105, 5241–5256. [Google Scholar] [CrossRef]
- Leung, M.H.; Tong, X.; Wilkins, D.; Cheung, H.H.; Lee, P.K. Individual and household attributes influence the dynamics of the personal skin microbiota and its association network. Microbiome 2018, 6, 26. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. NISC Comparative Sequencing Program. Temporal stability of the human skin microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, M.C.; Jiang, S.C. Alterations of the human skin microbiome after ocean water exposure. Mar. Pollut. Bull. 2019, 145, 595–603. [Google Scholar] [CrossRef]
- Patra, V.; Byrne, S.N.; Wolf, P. The skin microbiome: Is it affected by UV-induced immune suppression? Front. Microbiol. 2016, 7, 1235. [Google Scholar] [CrossRef] [Green Version]
- Peleg, A.; Hogan, D.; Mylonakis, E. Medically important bacterial–fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef] [PubMed]

| Microbiome | ||||
| Outer Arm | Inner Arm | Cheek | ||
| Lifeguards | 10 | 10 | 10 | After Summer |
| 13 | 11 | 12 | Before Summer | |
| Ultraorthodox | 8 | 9 | 9 | After Summer |
| 11 | 11 | 10 | Before Summer | |
| Mycobiome | ||||
| Outer Arm | Inner Arm | Cheek | ||
| Lifeguards | 9 | 9 | 10 | After Summer |
| 11 | 11 | 12 | Before Summer | |
| Ultraorthodox | 8 | 9 | 9 | After Summer |
| 8 | 10 | 10 | Before Summer | |
| Beta Diversity—Lifeguards vs. Ultraorthodox | ||||||||
|---|---|---|---|---|---|---|---|---|
| Before Summer | After Summer | |||||||
| Bray–Curtis | Jaccard | Bray–Curtis | Jaccard | |||||
| p Value | R | p Value | R | p Value | R | p Value | R | |
| Cheek | 0.119 | 0.07 | 0.021 | 0.17 | 0.133 | 0.08 | 0.005 | 0.23 |
| Outer Arm | 0.669 | 0.04 | 0.738 | 0.05 | 0.089 | 0.11 | 0.039 | 0.14 |
| Inner Arm | 0.4 | 0 | 0.32 | 0.02 | 0.085 | 0.11 | 0.039 | 0.14 |
| Microbiome | Mycobiome | |||||||
|---|---|---|---|---|---|---|---|---|
| Bray–Curtis | Jaccard | Bray–Curtis | Jaccard | |||||
| p Value | R | p Value | R | p Value | R | p Value | R | |
| Lifeguards | 0.07 | 0.03 | 0.001 | 0.13 | 0.341 | 0.01 | 0.323 | 0.01 |
| Ultraorthodox | 0.107 | 0.03 | 0.399 | 0 | 0.397 | 0 | 0.061 | 0.03 |
| Bray–Curtis | Jaccard | |||
|---|---|---|---|---|
| p Value | R | p Value | R | |
| Cheek | 0.595 | 0.08 | 0.215 | 0.05 |
| Outer Arm | 0.495 | 0.01 | 0.044 | 0.12 |
| Inner Arm | 0.209 | 0.04 | 0.058 | 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harel, N.; Reshef, L.; Biran, D.; Brenner, S.; Ron, E.Z.; Gophna, U. Effect of Solar Radiation on Skin Microbiome: Study of Two Populations. Microorganisms 2022, 10, 1523. https://doi.org/10.3390/microorganisms10081523
Harel N, Reshef L, Biran D, Brenner S, Ron EZ, Gophna U. Effect of Solar Radiation on Skin Microbiome: Study of Two Populations. Microorganisms. 2022; 10(8):1523. https://doi.org/10.3390/microorganisms10081523
Chicago/Turabian StyleHarel, Nurit, Leah Reshef, Dvora Biran, Sarah Brenner, Eliora Z. Ron, and Uri Gophna. 2022. "Effect of Solar Radiation on Skin Microbiome: Study of Two Populations" Microorganisms 10, no. 8: 1523. https://doi.org/10.3390/microorganisms10081523
APA StyleHarel, N., Reshef, L., Biran, D., Brenner, S., Ron, E. Z., & Gophna, U. (2022). Effect of Solar Radiation on Skin Microbiome: Study of Two Populations. Microorganisms, 10(8), 1523. https://doi.org/10.3390/microorganisms10081523








