1. Introduction
In maize crops, some of the most commonly encountered fungal pathogens are
Aspergillus and
Fusarium spp. These fungi are known to produce mycotoxins, such as aflatoxins and fumonisins which are linked to many different health problems in humans and animals, and are known to be carcinogenic, immunotoxic, mutagenic, nephrotoxic, neurotoxic and teratogenic [
1]. Mycotoxin-producing fungi not only contaminate crops in the field but the toxins they generate have been detected at appreciable levels in harvested products, threatening the safety of food for animals and humans.
Sarocladium zeae (synonym:
Acremonium zeae) [
2] is a protective fungal endophyte, typically producing symptomless kernel association, that acts antagonistically towards the kernel-rotting and mycotoxin-producing fungi
Aspergillus flavus and
Fusarium verticillioides in maize [
3,
4,
5] and
F. graminearum in wheat [
6].
Sarocladium zeae is recognized as a seedborne endophyte of maize [
7] and a systemic endophyte of wheat [
6]. Earlier histopathological studies and fungal isolations from dissections of asymptomatic
Zea mays kernels revealed that
S. zeae was frequently isolated from excised embryos and endosperm [
8], although further evidence is needed to confirm embryo colonisation. There have been no reports that
S. zeae isolates from maize produce any metabolites toxic to animals or plants [
5,
9]. There are no records of naturally occurring
S. zeae having been isolated from plants other than maize or sorghum [
10,
11] which suggests an evolutionary relationship between
S. zeae and these genera of cereal hosts. Synthetic infections can be established in other genera such as
Triticum [
6].
Sarocladium zeae produces secondary metabolites—pyrrocidine A and B, which are polyketide-amino acid-derived antibiotics displaying significant antifungal activity in vitro against
A. flavus and
F. verticillioides as well as inhibiting the growth of
F. verticillioides within
S. zeae colonised kernels [
4]. Pyrrocidine A also exhibits potent activity against major stalk and ear rot pathogens of maize, including
F. graminearum,
Nigrospora oryzae,
Stenocarpella (Diplodia) maydis and
Rhizoctonia zeae in addition to potent activity against
Clavibacter michiganense subsp.
nebraskense, the causal agent of Goss’s bacterial wilt of maize [
3,
4]. Most interestingly, a recent study revealed that pyrrocidines act, likely through interacting with the
FvZBD1 gene, to switch off fumonisin biosynthesis in
F. verticillioides [
12]. This suggests that
S. zeae can manipulate the secondary metabolism of a competitor fungus, thereby repressing the production of this mycotoxin, with the potential to reduce concerns for food safety [
12]. Thus, bioactive
S. zeae strains producing pyrrocidines are potentially a confounding variable in maize variety trials for resistance to pathogenic microbes and their mycotoxins [
3].
Due to genetic variation,
Sarocladium zeae (a haploid) strains differ in their ability to produce antibiotic secondary metabolites and not all
S. zeae strains produce pyrrocidines. An evaluation of 154
S. zeae isolates accessioned by the USDA Agricultural Research Service Culture Collection (NRRL) and CBS Culture Collections from 1969 to 1992 revealed that the proportion of pyrrocidine-producing strains in all
S. zeae strains isolated from various seed populations ranged from <1% to 98%, largely depending on differing climate conditions where the maize populations were grown [
7].
Sarocladium zeae endophytes differing in their ability to produce pyrrocidines were suggested to be naturally selected by climate and distributed with the seeds of maize cultivars grown in commercial plantings. Nevertheless, we speculated that the selection of pyrrocidine-producing
S. zeae strains may also be associated with the evolutionary history of host
Zea species/varieties, particularly within the non-commercialised wild teosintes population. We further investigated this by performing genotyping by sequencing (GBS) [
13] to reveal the inherent genetic variation within
S. zeae strains, isolated from diverse
Zea germplasm including modern
Z. mays and its wild progenitors, and through the development of a rapid polymerase chain reaction (PCR)-based method screened strains of
S. zeae for their ability to produce pyrrocidines.
Detection of pyrrocidine-producing
S. zeae strains by conventional methods is based upon chemical analysis, which is costly, labour intensive and requires significant time to obtain a result [
3,
4]. To meet increasing demands for economic and timely analysis, new approaches for identifying pyrrocidine-producing
S. zeae strains need to be explored. Most recently, biosynthetic gene clusters have been functionally characterised for pyrrocidine B [
14] and identified for pyrrocidine A ([
15], not released), and through whole genome-sequencing, we have also detected a PKS-NRPS (hybrid polyketide synthase-non-ribosomal peptide synthetase) gene cluster linked to pyrrocidine A biosynthesis (this study, see below). The identification of the pyrrocidine biosynthetic pathways [
14,
15] has provided a basis for using a molecular approach to rapidly screen pyrrocidine-producing
S. zeae strains. This paper describes a specific, sensitive and robust PCR detection assay that was developed to preliminarily screen
S. zeae strains for their potential to produce pyrrocidines, before further investigation with chemical analyses.
Whilst our own studies, and others [
6,
7,
8], have documented
S. zeae as being endophytic fungi, research into how host plants are colonised and how the endophyte is transmitted is lacking. To study the
S. zeae transmission modes in more detail, we created a genetically modified strain expressing the green fluorescent protein (GFP) that was inoculated into maize seedlings and examined using fluorescence microscopy.
Overall, in this study, we aimed to (i) reveal the genetic variation within the S. zeae isolates by using GBS techniques; (ii) develop a PCR-based array to screen S. zeae strains that produce antibiotic pyrrocidines; and (iii) further investigate the transmission modes of S. zeae endophytes in maize tissues.
2. Materials and Methods
2.1. Seed Material
The maize kernels used for isolation of
S. zeae fungal strains were imported from various germplasm collections, including The U.S. National Plant Germplasm System (NPGS, Ames, IA, USA), The International Maize and Wheat Improvement Centre (CIMMYT, El Batán, Mexico), Forage Genetics International (Davis, CA, USA) and PGG Wrightson Seeds (Christchurch, New Zealand). A total of 384 seed accessions, consisting of 275 landraces/cultivars of
Zea mays and 109 wild teosinte progenitors (51
Z. m. parviglumis, 50
Z. m. mexicana, 5
Z. diploperennis, 2
Z. perennis and 1
Z. m. nicarraguensis), were screened for seed-associated endophytes. Those accessions were originally collected from 51 countries, mostly from North, Central and South America (
Supplementary Table S1). For each accession, at least 5 individual kernels were subjected to endophyte screening.
2.2. Isolation of Fungal Endophytes
Due to the difference in germination features, two protocols were used for isolating endophytes in modern maize (Z. mays) and wild teosinte progenitors, respectively. For Z. mays, kernels (~25) were surface sterilised by soaking in 95% ethanol for 1 min, followed by rinsing in sterile H2O. The kernels were further sterilised in 30 mL 100% commercial Janola bleach containing 42 g/L active sodium hypochlorite (Pental Products Pty Ltd., Melbourne, Australia) for 4 min while gently shaking, followed by thorough rinsing of the kernels at least 5 times using sterile H2O. Surface-disinfected kernels were germinated on sterile 2% water agar plates (12 cm × 12 cm; 5 seeds per plate) in the dark at 23 °C for 4–7 days. Germinated seedlings free of microbial growth were aseptically dissected into ~2–3 mm sections. The sections were placed on PDA (potato dextrose agar, Oxoid Ltd., Basingstoke, UK) plates and incubated at 23 °C in a growth room and assessed for the emergence of fungal endophytes every day. For teosintes, the kernels were first surface sterilised with 12% Janola bleach for 20 min (vortexed frequently), then rinsed in sterile H2O. The bleach-treated kernels were sown in sterile vermiculite (in 50 mL falcon tubes) for germination. Post germination (shoot length > 3 cm), seedlings were harvested and surface sterilised again using 12% Janola bleach for 2–3 min, followed by 70% ethanol for ~30 s and thoroughly rinsed with sterile H2O. The treated seedlings were aseptically dissected into ~2–3 mm sections for endophyte isolation as described above.
Fungi emerging from tissue sections were isolated and purified through hyphal tip sub-culture on PDA plates. Fresh mycelia were harvested after 2 weeks and stored in 20% glycerol solution at −80 °C for long-term storage. Pure cultures on PDA plates were used to produce inoculum for experiments after identification. Notably, due to the concerns of intellectual property, we are not able to share those S. zeae collections. This limitation also applies to our genome sequencing data and GBS dataset as stated below.
2.3. Isolate Identification
Morphological characteristics were examined under 100-400x magnification using a compound microscope (BX53, Olympus, Tokyo, Japan). Genomic DNA of the fungus was extracted using the Quick-DNATM Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA) and the ITS gene was amplified using universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) following the instructions of the SapphireAmp® Fast PCR Master Mix Kit (Takara Bio USA Inc., San Jose, CA, USA). The ITS amplicon was further purified (DNA Clean & ConcentratorTM-5, Zymo Research, Irvine, CA, USA) and sequenced (Massey Genome Service, Palmerston North, New Zealand). The ITS gene sequences of S. zeae strains were blasted (BlastN) against the NCBI GenBank NR database and phylogenetically analysed with the top GenBank hits using Geneious R10 software (Biomatters Ltd., Auckland, New Zealand) with the neighbour-joining method (bootstrap = 1000) to determine their relatedness.
2.4. Whole Genome Sequencing and AntiSMASH Analysis of S. zeae Strain PI159-1.1
Sarocladium zeae strain PI159-1.1, a strain producing pyrrocidine A, was isolated in this study from a landrace of Z. mays, collected in Thailand by USA-NPGS. Fungal mycelia of the strain, grown on sterile cellophane film overlaid on PDA medium for ~2 weeks, were collected and ground in liquid nitrogen. High-quality genomic DNA was extracted according to the manual of the Quick-DNATM Fungal/Bacterial Midiprep Kit (Zymo Research, Irvine, CA, USA) and sent to BGI Tech Solution (Hong Kong) Co. Limited for de novo sequencing using the platform of PacBio Sequel with 20k library. Genome assembly was performed using the program Canu version 1.6, with an estimated genome size parameter of 40 Mb. All other parameters were left with default values, and the command was modified to skip the Gnuplot test (gnuplotTested = true) and to run on a single computer (useGrid = false). The taxonomy affiliation of the assembly was verified by blasting it against the NCBI NR database.
The gene clusters associated with secondary metabolites were predicted by using antiSMASH (Fungi version 5.1 [
16]), with the criterion of 80% shared gene content to identify similar gene clusters. The sequences of gene clusters that are responsible for producing pyrrocidine metabolites in PI159-1.1 were compared with the biosynthetic gene cluster MW690134, a gene cluster of
S. zeae strain NRRL13540 that produces pyrrocidine B and has recently been functionally characterised by Ohashi et al. [
14].
2.5. Profiling Genetic Variation of S. zeae Isolates with GBS
Eighty-seven DNA samples of S. zeae, isolated from diverse hosts including various wild teosintes, landraces and cultivated varieties, were sequenced using GBS. Fungal growth and extraction of high-quality gDNA were performed as described above for whole-genome sequencing. DNA quality was checked via visualisation on ethidium bromide stained 0.8% agarose/TAE gels and then quantified using an Invitrogen Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA concentrations were normalised to 20 ng per μL and subsequently used for GBS library preparation.
A GBS library was generated following the methodology of Faville et al. [
17], with 100 ng of DNA digested using restriction enzyme PstI (New England Biolabs, Ipswich, MA, USA) and ligated to a unique barcoded adapter and a common adapter (20 ng, pre-determined by a titration) [
17]. The library was developed in 96-plex which includes a blank and samples, with samples AM56-4.F1N and PI147-2.1 in quadruplicate and sample AM13-2.1 in triplicate. The library was sequenced on one lane of an Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) at AgResearch Invermay, New Zealand. This generated a library containing 1.2 M single reads of 100 nt. Initial quality control and trimming was performed using Trimmomatic [
18]. The GBS reads were analysed using Tassel 5, version 5.2.40 [
19] and the assembled genome as a reference with standard parameters, except that the minimum minor allele frequency was set to 0.001. The vcf table produced by this pipeline was used for further downstream analysis. The vcf table was then subject to filtering using a custom Ruby script, where SNPs (single nucleotide polymorphism) with scores less than 100, multi-allelic SNPs, SNPs with less than 10× coverage in total and indels were removed. Diagnostic plots, particularly minimum allele frequencies and SNP depth frequencies (
supplementary Figures S1 and S2, respectively) were generated using the KGD package [
20]. To classify isolates based on SNP similarity, the SNPs were converted into genotype calls where 0 represented homozygous reference calls and 1 represented homozygous alternative calls, and SNPs with heterozygosity were removed. A Euclidean distance matrix was generated from the genotype calls and a dendrogram of the isolates from different locations and different hosts was created using a hierarchical clustering method with complete linkage.
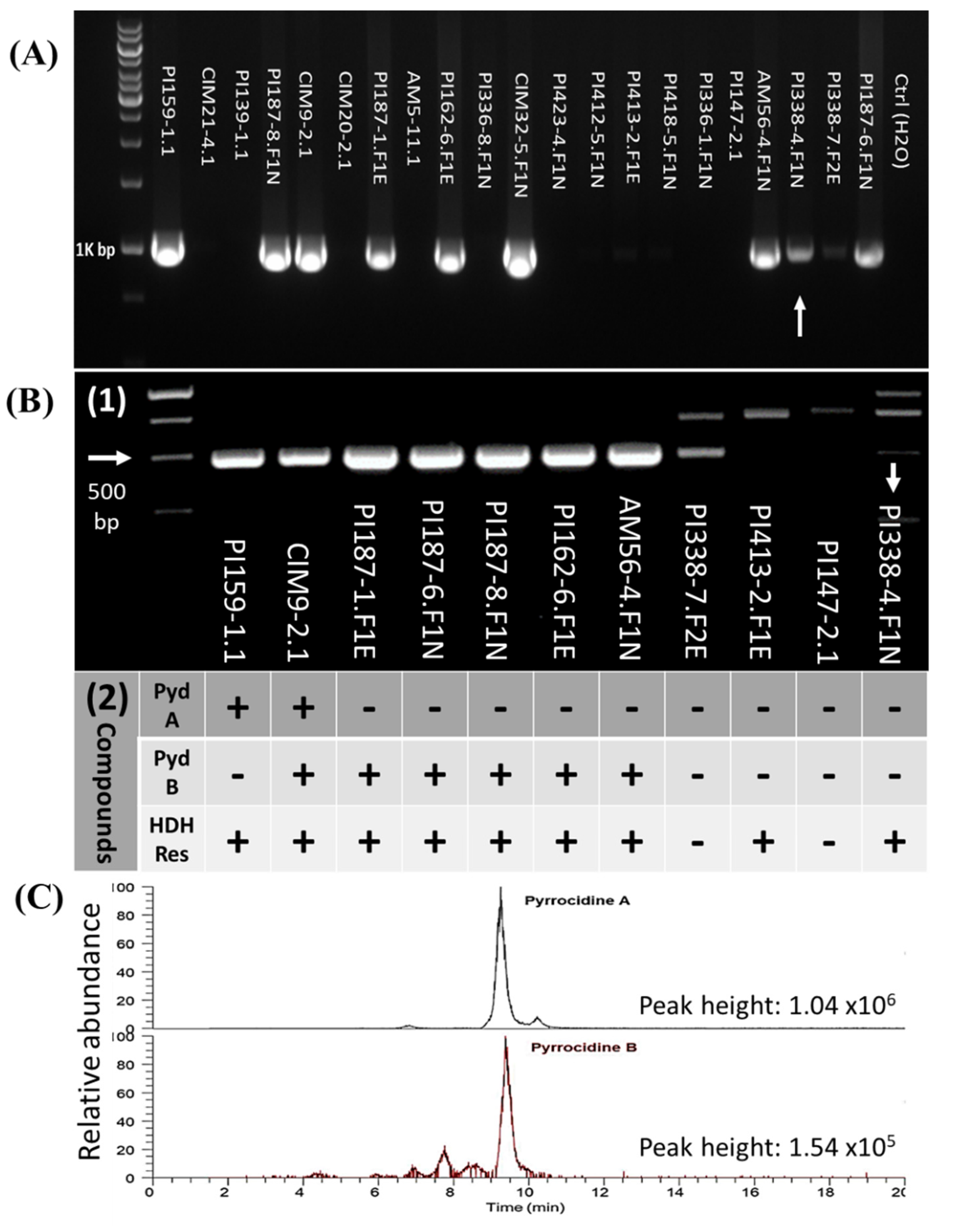
2.6. Detection of Pyrrocidine-Producing S. zeae Strains by PCR Assay
Purified S. zeae isolates were grown on PDA plates for 2 weeks. Mycelia were harvested and genomic DNA of 160 S. zeae isolates was extracted using the Quick-DNATM Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA) according to the kit instructions. PCR detection assays for pyrrocidine-producing S. zeae strains were carried out using two sets of primers: PYD-1 (forward: 5′-CTCTCTATGTCGGCTCTATCAA-3′; reverse: 5′- GGTGAAGTAGCCTCTGTAGTAG -3′; amplicon = 992 bp) and PYD-2 (forward: 5′-AACCTGCGCGTTGAGTTCTA-3′; reverse: 5′-CGATGGAGTATGGTCCTGCC-3′; amplicon = 481 bp). These two sets of primers were both designed from a shared open reading frame (ORF) region of the key pydA gene (i.e., the PKS-NRPS gene) in both pyrrocidine A and B biosynthetic gene clusters. PYD-1 primers were employed to preliminarily distinguish pyrrocidine-producing strains from non-producing S. zeae isolates. The PYD-2 primers were used to further clarify these PCR results. PCR detection arrays were conducted in a T100TM Thermal Cycler (BIO-RAD Laboratories, Singapore) following the instructions of SapphireAmp® Fast PCR Master Mix kit (Takara Bio USA Inc., San Jose, CA, USA); and were programmed for 1 cycle of 3 min at 94 °C, 35 cycles of 5 s 98 °C (denaturalization), 5 s at 60 °C for PYD-1 or 58 °C for PYD-2 (annealing), 15 s at 72 °C (extension), and finally 1 cycle of 5 min at 72 °C. Amplification reaction was carried out in volumes of 25 µL containing ~15–30 ng of template gDNA in 1 µL, 0.5 µL of each primer (10 µM), 12.5 µL of SapphireAmp® Fast PCR Master Mix. PCR products were detected in 1.5% agarose ethidium bromide gels (Life Technologies Inc., Gaithersburg, MD, USA). The GeneRuler 1kb DNA ladder (Thermo Fisher Scientific, Vilnius, Lithuania) was used as a molecular size marker.
2.7. Validation of Pyrrocidine-Producing S. zeae Strains by Chemical Analysis
Eleven
S. zeae strains with different results based on the above PCR assays were selected for chemical analysis of pyrrocidines. Fungal inoculation/fermentation procedures were carried out as described by Wicklow et al. [
5]. Generally, maize kernels (50 g each) were weighed into 500 mL flasks and soaked in 50 mL H
2O overnight. Kernels were then autoclaved for 60 min and cooled down to room temperature. The autoclaved kernels were inoculated with 3 mL
S. zeae conidial spore suspension (containing 2 × 10
4 conidia per mL in sterile H
2O) and fermented at 23 °C for 20 days.
Post-fermentation, infected kernels were freeze-dried, and subsamples (~1000 mg) were ground with a bead ruptor (6.4 mm zirconium bead, 4 m/s for 30 s) (FastPrep-24, M.P. Biomedicals, Irvine, CA, USA) before being extracted with 10 mL ethyl acetate (Sigma-Aldrich, St. Louis, MO, USA) by end-over-end rotation in the dark (1 h at 30 rpm). After centrifuging (4000× g, 2 min) (Heraeus Megafuge 16, Thermo Scientific, Osterode, Germany), the supernatant was transferred into glass screwcap Kimax tubes (13 mm × 100 mm) and dried under a stream of nitrogen. The residue was resuspended in 2 mL of hexane (Sigma-Aldrich, St. Louis, MO, USA) and transferred to a second glass screwcap Kimax tube (13 mm × 100 mm). Partition with 3 × 2 mL of acetonitrile (Sigma-Aldrich, St. Louis, MO, USA) (rinsing the original Kimax tube), transferring the acetonitrile portion into a fresh screwcap Kimax tube (13 mm × 100 mm). The combined acetonitrile portions were dried under a stream of nitrogen, then resuspended in 1 mL ethyl acetate and transferred to a 2 mL glass HPLC vial. The ethyl acetate was removed under a stream of nitrogen, then the residue dissolved in 200 µL of 50% acetonitrile, and then analysed by LC-HRMS. Compounds were separated using a Polar-RP column (100 × 2.0 mm, 2.5 µm) (Phenomenex, Torrance, CA, USA) using a water/methanol gradient (0.1% acetic acid buffered). Peaks were detected using a Q-Exactive (Thermo Fisher Scientific, Waltham, MA USA) collecting MS1 spectra from 200–500 m/z and selected PRMs for predicted metabolites.
2.8. Investigation of S. zeae Fungal Transmission Modes
The endophytic characterisations of
S. zeae fungi that were inoculated into maize seedlings were investigated through genetically modified
S. zeae strain PI159-1.1-
eGFP
#13, which produces green fluorescence and is helpful for microscopic observation. This strain was generated from PI159-1.1 by randomly cloning a gene coding elected green fluorescence protein (
eGFP) into the PI159-1.1 fungal genome, via an
eGFP-constructed vector (pTefp119-
eGFP-HygR). Protoplast preparation and
eGFP transformation followed the modified protocols described by Vollmer and Yanofsky [
21] and Oliver et al. [
22] as used by Johnson et al. [
23], respectively.
Seeds of the modern Z. mays cultivar N101 (from PGG Wrightson Seeds, NZ)) and fast-flowering mini-maize (FFMM), which was kindly provided by Professor James Birchler via Maize Genetics Coop Stock Center (Urbana, IL, USA), were artificially inoculated with S. zeae strain PI159-1.1-eGFP#13, by immersing the surface-sterilised kernels into fungal conidia suspension (2 × 106 conidia per mL in 0.05% Tween-20 solution) for 1 h. In order to avoid the genetically modified S. zeae releasing into the environment, S. zeae-inoculated N101 or FFMM seeds were sowed and grown either in sealed transparent pots (8.5 cm diameter and 24 cm height) for 3 weeks in a growth room or in sealed Vented Large Propagator (58 cm length × 38 cm width × 84 cm height, Egmont Seed Company, New Plymouth, New Zealand) for 3 months in a growth chamber, respectively. For both trials, pre-autoclaved soil plus potting mix (2:1 v/v) were supplied as growth media; and the growth regime was set up at 25 °C/14 h light and 20 °C/10 h dark. Colonisation and endophytic characterisation of the eGFP-labelled S. zeae strain were microscopically investigated by using an Olympus Fluoview FV10i confocal laser scanning microscope (Tokyo, Japan) and an Olympus BX63 automated fluorescence microscope (Tokyo, Japan). Notably, the mini maize plants growing in the sealed propagator did not complete their full lifecycle, even though the trial has been extended to 3 months, and the plants died at the flowering stage.
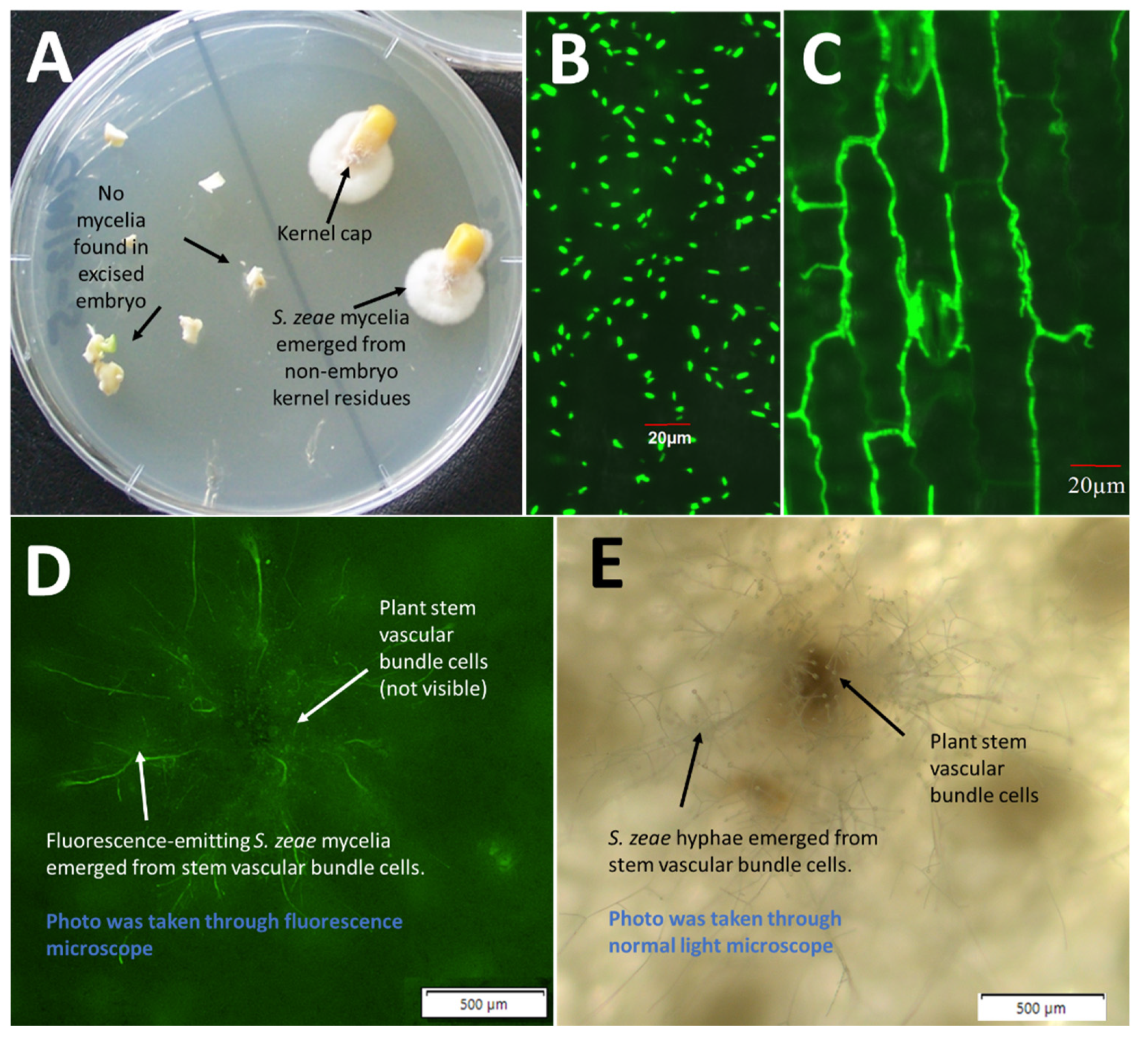
Due to the failure to produce seed from the mini maize plants that have been inoculated with eGFP-labelled S. zeae, multiple kernels (>10) of imported Z. mays that have naturally colonised with S. zeae fungi were selected for investigating colonisation sites of S. zeae fungi in seeds. Seed embryos were dissected out from symptomless and surface-disinfected kernels, and these embryos and the remaining seed residues were plated on PDA to check for the emergence sites of S. zeae.
4. Discussion
Sarocladium zeae is characterized as a “protective fungal endophyte” of maize. The potent antifungal and antibacterial properties of pyrrocidines produced by
S. zeae were first described by Wicklow and Poling [
4] and He et al. [
24], respectively. Recently, pyrrocidine has been demonstrated to switch off fumonisin biosynthesis in
F. verticillioides [
12], a major mycotoxigenic food safety threat. All these studies suggest that
S. zeae is a potential confounding variable in maize variety trials for resistance to pathogenic microbes and their mycotoxins [
7]. In the USA,
Sarocladium zeae has been used to effectively eliminate mycotoxin accumulation in the kernels caused by
F. verticillioides and
A. flavus that may produce fumonisin and aflatoxin [
1].
Sarocladium zeae is not reported to synthesize secondary metabolites harmful to plants, nor does it cause any ear or stem rot symptoms [
5,
12].
Bioactive
S. zeae strains produce pyrrocidine A and B, with pyrrocidine A being more active than B [
5]. Pyrrocidine A differs from pyrrocidine B by the presence of a double bond in its lactam ring resulting, by an unknown mechanism, in a higher toxicity of pyrrocidine A against a number of fungal and bacterial species, compared to pyrrocidine B [
4,
5,
12]. As demonstrated in strain CIM9-2.1 in this study, pyrrocidine A and B can both be detected in
S. zeae-infected kernels. Biosynthesis of pyrrocidine A and pyrrocidine B appeared to share the same biosynthetic pathway. When sequences of pyrrocidine biosynthetic gene clusters (BGC) were compared between the
S. zeae strains producing various pyrrocidines, the nucleotide sequences of strain PI159-1.1 (pyrrocidine A producing; this study) showed a 94.4% similarity with the sequences in strain NRRL13540 (pyrrocidine B producing [
14]). The translated amino acid sequences of a key PKS-NRPS gene (
pydA) in the pyrrocidine BGC shared a similarity of 98.8% between the two pyrricidine A- and B-producing strains. The
pydA gene has been suggested to be responsible for the biosynthesis of the lactam ring (the tricarbocyclic core) in most pyrrocidine-like compounds [
25]. Since the pyrrocidine biosynthetic gene cluster in strain NRRL13540 has recently been functionally characterised [
14], we believe that this improved knowledge on the biosynthetic pathways of pyrrocidines, particularly the
S. zeae specific
pydA gene, may provide a foundation for, and facilitate the adaptation of, molecular genetic techniques for screening pyrrocidine-producing
S. zeae strains.
In
Zea crops, there is strong selection for pyrrocidine production among
S. zeae fungal populations [
7]. It is therefore important to discriminate pyrrocidine-producing strains from non-pyrrocidine-producing strains for studying the biosynthesis of pyrrocidines, as well as gaining a detailed knowledge of the potential bioactivity of various fungal strains. Current methods for identifying pyrrocidine-producing
S. zeae strains are costly and often involve labour- and time-consuming tasks that require expertise in chemical analysis. To meet increasing demands for accurate, economic, and timely analysis, new tools for identifying pyrrocidine-producing
S. zeae strains must be explored. Nucleic acid-based identification systems have the advantage of differentiating strains based on their genetic information. In this work, PCR primers were designed from
pydA gene with shared sequences for both pyrrocidine A- and B-producing
S. zeae strains; and PCR detection assays were developed for the first time to identify both pyrrocidine A- and B-producing
S. zeae strains. The PCR detection assays provide a simple and rapid method for preliminarily screening pyrrocidine-producing
S. zeae strains. As mentioned above, the PYD-2 primer set was not intended to be used alone, but to filter out the small number of
pydA negative isolates that falsely test positive with PYD-1 primers.
In this work, the PCR-positive
S. zeae strains were mostly associated with well-clustered
Z. mays host Group-A, while PCR negative
S. zeae strains were commonly associated with teosintes Group-B1 (dominated by
Z. m. mexicana), although within-species variation also existed in
Z. mays (between Group-A and Group-B2). The results clearly reflected fungal genetic diversity, as analysed by the GBS. The mechanisms involved in the gene loss or gain in the pyrrocidine biosynthetic gene cluster is unclear. Wicklow et al. [
7] suggested that natural environment, such as different climate conditions, may impact the selection of
S. zeae strains producing pyrrocidines. In regions where maize is particularly vulnerable to pathogen attack following the damaging effects of drought and temperature stress, selection may favour
S. zeae endophytes that produce pyrrocidines as acquired chemical defences [
7]. This kind of stress-induced selection is highly possible and has been observed in other fungal species, e.g., heat stress-induced different gene expression in mycorrhizal
Tuber borchii [
26]. Nevertheless, our results, in terms of the complemented profiles between PCR assay and GBS analysis, strongly suggest that host species/varieties may play a more important role in influencing
S. zeae genetic diversity and selection of pyrrocidine-producing strains. We therefore conclude that, in addition to the natural selection as described above, pyrrocidine biosynthesis in
S. zeae may have different evolutionary histories, possibly associated with host domestication from wild progenitors of modern maize. The observation that a large majority of
S. zeae isolates from wild teosintes do not produce pyrrocidines invites further consideration of endophyte contributions to maize defence from pathogen attack. The findings from this study are expected to help develop effective strategies for the management of biocontrol endophytes in maize.
In this work, a total of 206 S. zeae fungi have been isolated from diverse maize germplasm, including 275 landraces/cultivars and 109 wild teosinte progenitors of Zea crops collected from 51 countries geographically. Those S. zeae fungi were more commonly discovered from accessions collected from North, Central and South America than other regions/countries. There were no S. zeae fungi isolated from New Zealand-produced Z. mays seeds although more than 46 accessions have been screened. In some seed accessions, multiple S. zeae isolates were recovered from a single accession, but usually isolated from different kernels. Those multiple strains mostly shared a similar genetic background in terms of the pydA gene.
Studies via traditional morphological description and ITS amplicon sequencing often revealed no obvious variation in
S. zeae isolates. This study identified genetic diversity within
S. zeae for the first time using GBS technology where SNP detection and genotyping are performed simultaneously [
13,
17]. GBS analysis clustered the
S. zeae fungal isolates into clearly host-related groups, demonstrating significant host lineages of
S. zeae genetic diversity. Host lineages of genetic variation have been reported in other microorganisms such as in
Pseudoperonospora cubensis [
27]. Our observation of the host-linked genetic variation of
S. zeae provided the first view of the
S. zeae genetic diversity at the strain level, in accordance with broad host relationship parameters. We speculated that the genetic diversity of
S. zeae was likely to be linked to host evolution from ancestor teosintes to modern maize.
S. zeae is the most prevalent seedborne fungus associated with maize seed, reportedly as high as 39.5% of the seeds [
7,
28]. This is consistent with our observation that
S. zeae isolates make up ~40% of our whole fungal collection in the present study. Seedborne
S. zeae endophytes in maize were often thought to inhabit the seed embryo and transmit through vascular tissues. This observation of
S. zeae residing in the embryo can be dated back to early histopathological studies on “sound-appearing” maize kernels, which revealed that
S. zeae fungi were often detected in the embryo and endosperm [
5,
8]. It was speculated that
S. zeae fungi are able to vertically transmit via seed and propagate in the next generation of its host, such as the well-studied
Epichloë endophytes [
29]. Since then, however, there has been no further experimental evidence to support the finding of
S. zeae being embryo associated. In the present study, no
S. zeae fungi were recovered from the dissected seed embryos but mycelia were recovered from the pedicel and abscission layer of the seed, suggesting that
S. zeae more likely inhabits the pedicel and abscission layer of asymptomatic seeds, rather than the seed embryo [
5,
8].
S. zeae was also reported to be the predominant fungus recovered from stem core tissues of naturally infected healthy maize plants [
8,
30]. An earlier study by Reddy and Holbert [
31] proposed that
S. zeae is a systemic seedborne fungus of maize that infects the plant through the vascular system, eventually invading the ears and seeds. These observations are consistent with our observation of
eGFP-labelled
S. zeae, clearly showing that
S. zeae can be endophytically transmitted within stem vascular cells. By using scanning electron microscopy, Kemp et al. [
6] demonstrated that
S. zeae spores can be transmitted vertically to the progeny of infected wheat plants, suggesting the possibility that conidia, potentially including the inoculum itself, may be transported within vascular tissue even in the absence of active growth. The authors considered that this type of passive dispersal may explain why recovery of
S. zeae from wheat tissue pieces was sometimes discontinuous; conidia may be carried variable distances before becoming lodged or otherwise being obstructed from continuing with the flow of the transpiration stream [
6].
Regarding the study of
S. zeae transmission in maize, earlier investigation mostly focused on the transmitting modes in non-foliar tissues, such as in seeds and stems. Comparably, less attention was paid to
S. zeae colonisation on leaves via conidia.
Sarocladium zeae are a conidia-rich species. As illustrated in
Figure 3B, conidia repropagated by
S. zeae were often observed on intact leaves of maize seedlings. By using an
eGFP-labelled
S. zeae strain, the confocal laser scanning microscopy clearly revealed that conidia-germinated mycelia of the fungus can transmit and spread within intercellular spaces of the leaf blade, suggesting that
S. zeae colonisation via propagated conidia may be another important transmission strategy of the endophyte, particularly in the tissue of leaf blades. Conidia of
S. zeae are very small compared to most other fungi, at approximately 3.5 to 6 µm in length and 1.2 to 1.8 µm in width [
6,
32]. This suggests the possibility that conidia can penetrate through leaf stomata and grow into leaf intercellular tissues.
In summary, the present study provides further evidence that the effectiveness of biological control of S. zeae species varies among strains, which motivates the continued screening of additional strains. The findings of the present study provide preliminary information on the adaptation of molecular techniques for screening S. zeae strains producing pyrrocidines and significantly expands our knowledge of the genetic variation within this fungal species at the strain level. Our study also demonstrated that S. zeae endophytes can be vertically transmitted through stem tissues into next-generation seeds where they may inhabit seed caps rather than embryos. Endophytic colonisation of conidia-germinated mycelia of S. zeae in maize leaf blades may provide another important transmission strategy of the endophyte fungi.