Utilizing the Gastrointestinal Microbiota to Modulate Cattle Health through the Microbiome-Gut-Organ Axes
Abstract
1. Introduction
2. The Gut Microbiota
2.1. Development of the Gut Microbiota
2.2. Dietary Effects on the Gut Microbiota
2.3. Effects on the Immune System
2.4. Therapies to Modulate GIT Health
2.4.1. Probiotics
2.4.2. Prebiotics
2.4.3. Gut Microbial Transplants
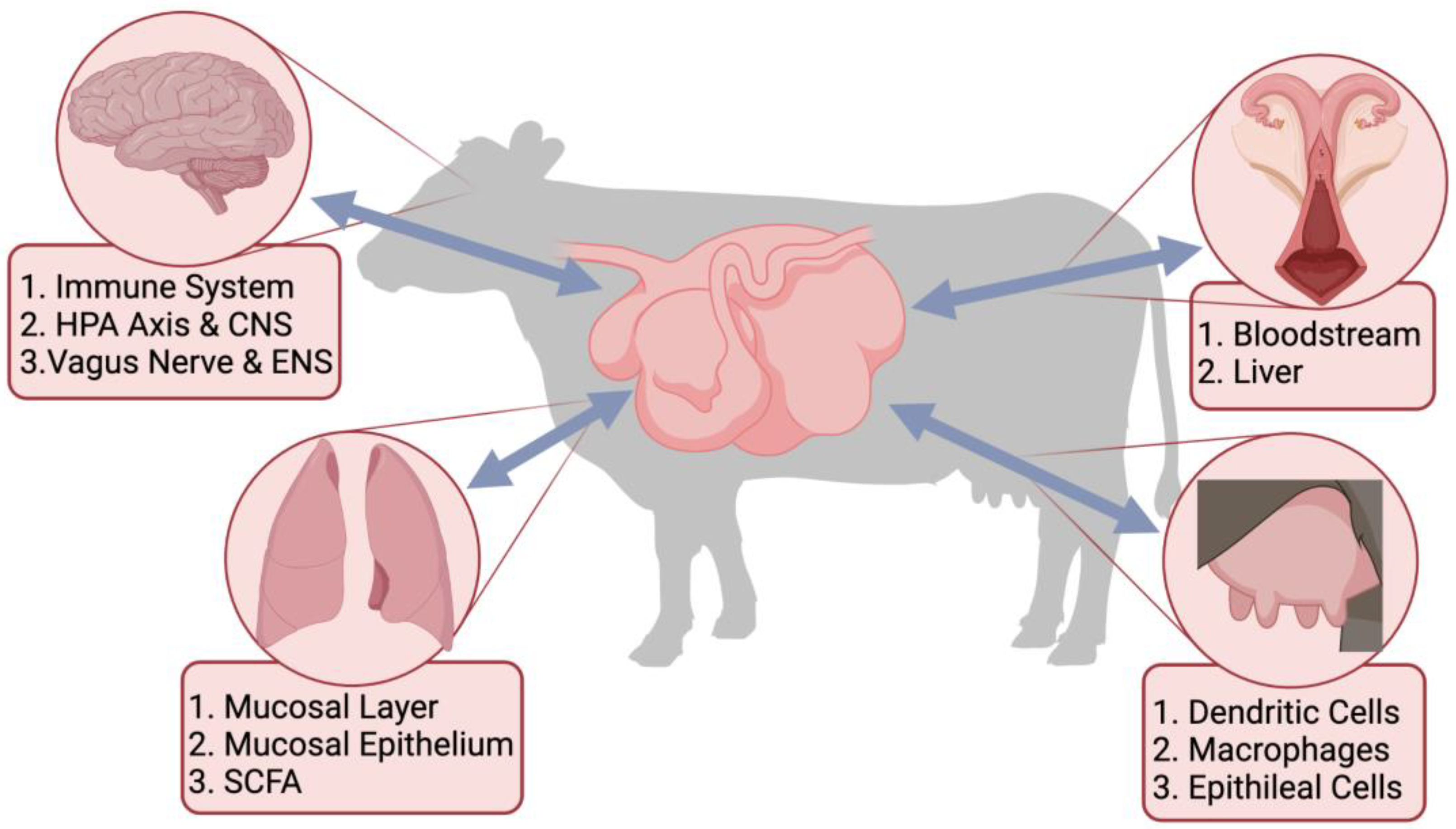
3. Microbiome-Gut-Organ Axes
3.1. Microbiome-Gut-Brain Axis
3.2. Microbiome-Gut-Lung Axis
3.3. Microbiome-Gut-Mammary Axis
3.4. Microbiome-Gut-Reproductive Axis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hungate, R.E. The Rumen and Its Microbes; Elsevier: Amsterdam, The Netherlands, 1966. [Google Scholar]
- Flint, H.J. The rumen microbial ecosystem—some recent developments. Trends Microbiol. 1997, 5, 483–488. [Google Scholar] [CrossRef]
- Mizrahi, I. Rumen Symbioses. In The Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Springer: Berlin/Heidelberg, Germany, 2013; pp. 533–544. [Google Scholar]
- Mackie, R.I. Mutualistic fermentative digestion in the gastrointestinal tract: Diversity and evolution. Integr. Comp. Biol. 2002, 42, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Hume, I.; Torell, D. Development of microbial activity in the alimentary tract of lambs. J. Anim. Sci. 1972, 35, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.B.; Hespell, R.B. Microbial rumen fermentation. J. Dairy Sci. 1981, 64, 1153–1169. [Google Scholar] [CrossRef]
- Yeoman, C.J.; White, B.A. Gastrointestinal tract microbiota and probiotics in production animals. Annu. Rev. Anim. Biosci. 2014, 2, 469–486. [Google Scholar] [CrossRef]
- Hungate, R. Ruminal fermentation. In Handbook of Physiology No. 5; Oxford University Press: New York, NY, USA, 1968; pp. 2725–2745. [Google Scholar]
- Russell, J. Ecology of rumen microorganisms: Energy use. In Aspects of Digestive Physiology in Ruminants; Cornell University Press: Ithaca, NY, USA, 1988; pp. 74–98. [Google Scholar]
- Bryant, M.P.; Wolin, E.A.; Wolin, M.J.; Wolfe, R.S. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Microbiol. 1967, 59, 20–31. [Google Scholar] [CrossRef]
- Iannotti, E.L.; Kafkewitz, D.; Wolin, M.J.; Bryant, M.P. Glucose fermentation products of Ruminococcus albus grown in continuous culture with Vibrio succinogenes: Changes caused by interspecies transfer of H2. J. Bacteriol. 1973, 114, 1231–1240. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Das, P.; Ranjan, R. Role of Gut Microbiome in Improving Animal Health and Productivity. Indian J. Anim. Health 2020, 59, 146–155. [Google Scholar] [CrossRef]
- Fan, P.; Nelson, C.D.; Driver, J.D.; Elzo, A.E.; Penagaricano, F.; Jeong, K.C. Host genetics exerts lifelong effects upon hindgut microbiota and its association with bovine growth and immunity. ISME J. 2021, 15, 2306–2321. [Google Scholar] [CrossRef]
- Fonty, G.; Gouet, P.; Jouany, J.-P.; Senaud, J. Establishment of the microflora and anaerobic fungi in the rumen of lambs. Microbiology 1987, 133, 1835–1843. [Google Scholar] [CrossRef]
- Bicalho, M.L.S.; Santin, T.; Rodrigues, M.X.; Marques, C.E.; Lima, S.F.; Bicalho, R.C. Dynamics of the microbiota found in the vaginas of dairy cows during the transition period: Associations with uterine diseases and reproductive outcome. J. Dairy Sci. 2017, 100, 3043–3058. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.E.; Galvão, K.N.; Rodriguez-Lecompte, J.C.; Costa, M.C. The cattle microbiota and the immune system: An evolving field. Vet. Clin. Food Anim. Pract. 2019, 35, 485–505. [Google Scholar] [CrossRef] [PubMed]
- Laguardia-Nascimento, M.; Branco, K.M.G.R.; Gasparini, M.R.; Giannattasio-Ferraz, S.; Leite, L.R.; Araujo, F.M.G.; Salim, A.C.D.M.; Nicoli, J.R.; De Oliveira, G.C.; Barbosa-Stancioli, E.F. Vaginal microbiome characterization of nellore cattle using metagenomic analysis. PLoS ONE 2015, 10, e0143294. [Google Scholar] [CrossRef] [PubMed]
- Zambrano-Nava, S.; Bosco’n-Ocando, J.; Nava, J. Normal bacterial flora from vaginas of Criollo Limonero cows. Trop. Anim. Health Prod. 2011, 43, 291–294. [Google Scholar] [CrossRef]
- Mayer, M.; Abenthum, A.; Matthes, J.M.; Kleeberger, D.; Ege, M.J.; Hölzel, C.; Bauer, J.; Schwaiger, K. Development and genetic influence of the rectal bacterial flora of newborn calves. Vet. Microbiol. 2012, 161, 179–185. [Google Scholar] [CrossRef]
- Dehority, B.A. Rumen Microbiology; Nottingham University Press: Nottingham, UK, 2003. [Google Scholar]
- Trinci, A.P.; Lowe, S.E.; Milne, A.; Theodorou, M.K. Growth and survival of rumen fungi. Biosystems 1988, 21, 357–363. [Google Scholar] [CrossRef]
- Fecteau, G.; Baillargeon, P.; Higgins, R.; Paré, J.; Fortin, M. Bacterial contamination of colostrum fed to newborn calves in Que´bec dairy herds. Can. Vet. J. 2002, 43, 523–527. [Google Scholar]
- Minato, H.; Otsuka, M.; Shirasaka, S.; Itabashi, H.; Mitsumori, M. Colonization of microorganisms in the rumen of young calves. J. Gen. Appl. Microbiol. 1992, 38, 447–456. [Google Scholar] [CrossRef]
- Song, Y.; Malmuthuge, N.; Steele, M.A.; Guan, L.L. Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol. Ecol. 2018, 94, fix179. [Google Scholar] [CrossRef]
- Bryant, M.; Small, N.; Bouma, C.; Robinson, I. Studies on the composition of the ruminal flora and fauna of young calves. J. Dairy Sci. 1958, 41, 1747–1767. [Google Scholar] [CrossRef]
- Alipour, M.J.; Jalanka, J.; Pessa-Morikawa, T.; Kokkonen, T.; Satokari, R.; Hynönen, U.; Iivanainen, A.; Niku, M. The composition of the perinatal intestinal microbiota in cattle. Sci. Rep. 2018, 8, 10437. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.; Marcondes, M.I.; Motta de Souza, S.; Cardoso da Mata e Silva, B.; Fontes Noronha, M.; Tassinari Resende, R.; Machado, F.S.; Cuquetto Mantovani, H.; Dill-McFarland, K.A.; Suen, G. Bacterial community dynamics across the gastrointestinal tracts of dairy calves during preweaning development. Appl. Environ. Microbiol. 2018, 84, e02675-17. [Google Scholar] [CrossRef] [PubMed]
- Klein-Jöbstl, D.; Schornsteiner, E.; Mann, E.; Wagner, M.; Drillich, M.; Schmitz-Esser, S. Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front. Microbiol. 2014, 5, 622. [Google Scholar] [CrossRef] [PubMed]
- Oikonomou, G.; Bicalho, M.L.; Meira, E.; Rossi, R.; Foditsch, C.; Machado, V.S.; Teixeira, A.G.V.; Santisteban, C.; Schukken, Y.; Bicalho, R.C. Microbiota of cow’s milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PLoS ONE 2014, 9, e85904. [Google Scholar] [CrossRef] [PubMed]
- Carberry, C.A.; Kenny, D.A.; Han, S.; McCabe, M.S.; Waters, S.M. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 2012, 78, 4949–4958. [Google Scholar] [CrossRef]
- de Menezes, A.B.; Lewis, E.; O’Donovan, M.; O’Neill, B.F.; Clipson, N.; Doyle, E.M. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 2011, 78, 256–265. [Google Scholar] [CrossRef]
- Tajima, K.; Aminov, R.I.; Nagamine, T.; Matsui, H.; Nakamura, M.; Benno, Y. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 2001, 67, 2766–2774. [Google Scholar] [CrossRef]
- Nagaraja, T.G.; Chengappa, M.M. Liver abscesses in feedlot cattle: A review. J. Anim. Sci. 1998, 76, 287–298. [Google Scholar] [CrossRef]
- USDA/APHIS. Feedlot Health Management Report: Part II: Cattle on Feed Evaluation; N172.0195; USDA/APHIS/Veterinary Services, Centers for Epidemiology and Animal Health: Fort Collins, CO, USA, 1995.
- Durso, L.M.; Miller, D.N.; Schmidt, T.B.; Callaway, T. Tracking bacteria through the entire gastrointestinal tract of a beef steer. Agric. Environ. Lett. 2017, 2, 170016. [Google Scholar] [CrossRef]
- Welch, C.B.; Lourenco, J.M.; Krause, T.R.; Seidel, D.S.; Fluharty, F.L.; Pringle, T.D.; Callaway, T.R. Evaluation of the Fecal Bacterial Communities of Angus Steers with Divergent Feed Efficiencies Across the Lifespan from Weaning to Slaughter. Front. Vet. Sci. 2021, 8, 597405. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Hernandez-Sanabria, E.; Guan, L.L. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 2009, 75, 6524–6533. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, J.M.; Krause, T.R.; Welch, C.B.; Callaway, T.R.; Pringle, T.D. Longitudinal Changes of the Ruminal Microbiota in Angus Beef Steers. Animals 2022, 12, 1066. [Google Scholar] [CrossRef]
- Weimer, P.J. Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol. 2015, 6, 296. [Google Scholar] [CrossRef]
- Flint, H.J.; Bayer, E.A. Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann. N. Y. Acad. Sci. 2008, 1125, 280–288. [Google Scholar] [CrossRef]
- Flint, H.J.; Bayer, E.A.; Rincon, M.T.; Lamed, R.; White, B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hobson, P.N.; Stewart, C.S. The Rumen Microbial Ecosystem; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Biddle, A.; Stewart, L.; Blanchard, J.L.; Leschine, S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Brulc, J.M.; Antonopoulos, D.A.; Miller, M.E.B.; Wilson, M.K.; Yannarell, A.C.; Dinsdale, E.A.; Edwards, R.E.; Frank, E.D.; Emerson, J.B.; Wacklin, P.; et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Nat. Acad. Sci. USA 2009, 106, 1948–1953. [Google Scholar] [CrossRef]
- Ding, S.-Y.; Rincon, M.T.; Lamed, R.; Martin, J.C.; McCrae, S.I.; Aurilia, V.; Shoham, Y.; Bayer, E.A.; Flint, H.J. Cellulosomal scaffolding-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 2001, 183, 1945–1953. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis, H.T.; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; DeSilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef]
- McAllister, T. Learning more about rumen bugs: Genetic and environmental factors affecting rumen bugs. South. Alta. Beef Rev. 2000, 2. [Google Scholar]
- Krause, D.O.; Denman, S.; Mackie, R.I.; Morrison, M.; Rae, A.L.; Attwood, G.; McSweeney, C. Opportunities to improve fiber degradation in the rumen: Microbiology, ecology, and genomics. FEMS Microbiol. Rev. 2003, 27, 663–693. [Google Scholar] [CrossRef]
- Barrington, G.M.; Parish, S.M. Bovine neonatal immunology. Vet. Clin. N. Am. Food Anim. Pract. 2001, 17, 463–476. [Google Scholar] [CrossRef]
- Felippe, M.J. The immune system. In Equine Clinical Immunology; Felippe, M.J., Ed.; Wiley Blackwell: Ames, IA, USA, 2015; pp. 1–10. [Google Scholar]
- Mulder, I.E.; Schmidt, B.; Lewis, M.; Delday, M.; Stokes, C.R.; Bailey, M.; Aminov, R.; Gill, B.P.; Pluske, J.; Mayer, C.-D.; et al. Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS ONE 2011, 6, e28279. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef]
- O’Hara, E.; Neves, A.L.; Song, Y.; Guan, L.L. The role of the gut microbiome in cattle production and health: Driver or passenger? Annu. Rev. Anim. Biosci. 2020, 8, 199–220. [Google Scholar] [CrossRef]
- Rajasekaran, S.A.; Beyenbach, K.W.; Rajasekaran, A.K. Interactions of tight junctions with membrane channels and transporters. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2008, 1778, 757–769. [Google Scholar] [CrossRef]
- Sawada, N.; Murata, M.; Kikuchi, K.; Osanai, M.; Tobioka, H.; Kojima, T.; Chiba, H. Tight junctions and human diseases. Med. Electron Microsc. 2003, 36, 147–156. [Google Scholar] [CrossRef]
- Bush, L.J.; Staley, T.E. Absorption of colostral immunoglobulins in newborn calves. J. Dairy Sci. 1980, 63, 672–680. [Google Scholar] [CrossRef]
- Besser, T.E.; Gay, C.C. The importance of colostrum to the health of the neonatal calf. Vet. Clin. N. Am. Food Anim. Pract. 1994, 10, 107–117. [Google Scholar] [CrossRef]
- Araujo, G.; Yunta, C.; Terré, M.; Mereu, A.; Ipharraguerre, I.; Bach, A. Intestinal permeability and incidence of diarrhea in newborn calves. J. Dairy Sci. 2015, 98, 7309–7317. [Google Scholar] [CrossRef] [PubMed]
- Ewaschuk, J.B.; Diaz, H.; Meddings, L.; Diederichs, B.; Dmytrash, A.; Backer, J.; Langen, M.L.-V.; Madsen, K.L. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, E.; O’Callaghan, J.; Buttó, L.F.; Hurley, G.; Melgar, S.; Tanabe, S.; Shanahan, F.; Nally, K.; O’Toole, P.W. Mechanism of protection of transepithelial barrier function by Lactobacillus salivarius: Strain dependence and attenuation by bacteriocin production. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; McBain, A.J.; O’Neill, C.A. Strain-dependent augmentation of tight junction barrier function in human primary epidermal keratinocytes by Lactobacillus and Bifidobacterium lysates. Appl. Environ. Microbiol. 2013, 79, 4887–4894. [Google Scholar] [CrossRef]
- Song, Y.; Malmuthuge, N.; Li, F.; Guan, L.L. Colostrum feeding shapes the hindgut microbiota of dairy calves during the first 12 h of life. FEMS Microbiol. Ecol. 2019, 95, fiy203. [Google Scholar] [CrossRef]
- Liang, G.; Malmuthuge, N.; Bao, H.; Stothard, P.; Griebel, P.J.; Guan, L.L. Transcriptome analysis reveals regional and temporal differences in mucosal immune system development in the small intestine of neonatal calves. BMC Genom. 2016, 17, 602. [Google Scholar] [CrossRef]
- Stillie, R.; Farooq, S.M.; Gordon, J.R.; Stadnyk, A.W. The functional significance behind expressing two IL-8 receptor types on PMN. J. Leukoc. Biol. 2009, 86, 529–543. [Google Scholar] [CrossRef]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef]
- Smits, H.H.; Engering, A.; Van Der Kleij, D.; De Jong, E.C.; Schipper, K.; Van Capel, T.M.M.; Zaat, B.A.J.; Yazdanbakhsh, M.; Wierenga, E.A.; Van Kooyk, Y.; et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 2005, 115, 1260–1267. [Google Scholar] [CrossRef]
- Marchiando, A.M.; Graham, W.V.; Turner, J.R. Epithelial barriers in homeostasis and disease. Annu. Rev. Pathol. 2010, 5, 119–144. [Google Scholar] [CrossRef]
- Okumura, R.; Takeda, K. Maintenance of intestinal homeostasis by mucosal barriers. Inflamm. Regen. 2018, 38, 5. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Vogelzang, A.; Maruya, M.; Miyajima, M.; Murata, M.; Son, A.; Kuwahara, T.; Tsuruyama, T.; Yamada, S.; Matsuura, M.; et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J. Exp. Med. 2018, 215, 2019–2034. [Google Scholar] [CrossRef] [PubMed]
- Cunningham-Rundles, C. Physiology of IgA and IgA deficiency. J. Clin. Immunol. 2001, 21, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Shroff, K.E.; Meslin, K.; Cebra, J.J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 1995, 63, 3904–3913. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, K.S.B.; Kissoon-Singh, V.; Gibson, D.L.; Ma, C.; Montero, M.; Sham, H.P.; Ryz, N.; Huang, T.; Velcich, A.; Finlay, B.B.; et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010, 6, e1000902. [Google Scholar] [CrossRef]
- Zarepour, M.; Bhullar, K.; Montero, M.; Ma, C.; Huang, T.; Velcich, A.; Xia, L.; Vallance, B.A. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect. Immun. 2013, 81, 3672–3683. [Google Scholar] [CrossRef]
- Enss, M.L.; Grosse-Siestrup, H.; Schmidt-Wittig, U.; Gärtner, K. Changes in colonic mucins of germfree rats in response to the introduction of a “normal” rat microbial flora. Rat colonic mucin. J. Exp. Anim. Sci. 1992, 35, 110–119. [Google Scholar]
- Szentkuti, L.; Riedesel, H.; Enss, M.L.; Gaertner, K.; Von Engelhardt, W. Pre-epithelial mucus layer in the colon of conventional and germ-free rats. Histochem. J. 1990, 22, 491–497. [Google Scholar] [CrossRef]
- Enss, M.L.; Schmidt-Wittig, U.; Müller, H.; Mai, U.E.; Coenen, M.; Hedrich, H.J. Response of germfree rat colonic mucous cells to peroral endotoxin application. Eur. J. Cell Biol. 1996, 71, 99–104. [Google Scholar]
- Malmuthuge, N.; Li, M.; Goonewardene, L.A.; Oba, M.; Guan, L.L. Effect of calf starter feeding on gut microbial diversity and expression of genes involved in host immune responses and tight junctions in dairy calves during weaning transition. J. Dairy Sci. 2013, 96, 3189–3200. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation associated diseases in organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef]
- Ogunade, I.; Pech-Cervantes, A.; Schweickart, H. Metatranscriptomic Analysis of Sub-Acute Ruminal Acidosis in Beef Cattle. Animals 2019, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.C.; Srivastava, A.; Lall, R. (Eds.) Nutraceuticals in Veterinary Medicine; Springer: Cham, Switzerland, 2019; Volume 2109. [Google Scholar]
- Anadón, A.; Martínez-Larrañaga, M.R.; Aresi, M.M.A. Chapter 54. Prebiotics: Safety and toxicity considerations. In Nutraceuticals: Efficacy, Safety and Toxicity; Gupta, R.C., Ed.; Academic: Amsterdam, The Netherlands, 2016; pp. 757–775. [Google Scholar]
- Fuller, R. The effect of probiotics on the gut micro-ecology of farm animals. In The Lactic Acid Bacteria; Springer: Boston, MA, USA, 1992; Volume 1, pp. 171–192. [Google Scholar]
- Hamilton, M.J.; Weingarden, A.R.; Unno, T.; Khoruts, A.; Sadowsky, M.J. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 2013, 4, 125–135. [Google Scholar] [CrossRef]
- Liou, A.P.; Paziuk, M.; Luevano, J.M.J.; Machineni, S.; Turnbaugh, P.J.; Kaplan, L.M. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci. Transl. Med. 2013, 5, 178ra41. [Google Scholar] [CrossRef]
- Manichanh, C.; Reeder, J.; Gibert, P.; Varela, E.; Llopis, M.; Antolin, M.; Guigo, R.; Knight, R.; Guarner, F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010, 20, 1411–1419. [Google Scholar] [CrossRef]
- Weingarden, A.; González, A.; Vázquez-Baeza, Y.; Weiss, S.; Humphry, G.; Berg-Lyons, D.; Knights, D.; Unno, T.; Bobr, A.; Kang, J.; et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 2015, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Vacharaksa, A.; Croxen, M.; Thanachayanont, T.; Finlay, B.B. Altering host resistance to infections through microbial transplantation. PLoS ONE 2011, 6, e26988. [Google Scholar] [CrossRef] [PubMed]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef] [PubMed]
- Anadón, A.; Martínez-Larrañaga, M.R.; Arés, I.; Martínez, M.A. Chapter 1. Prebiotics and probiotics: An assessment of their safety and health benefits. In Probiotics, Prebiotics, and Synbiotics: Bioactive Foods in Health Promotion; Ross Watson, R., Preedy, V.R., Eds.; Academic: San Diego, CA, USA, 2016; pp. 3–23. [Google Scholar]
- Kraimi, N.; Calandreau, L.; Zemb, O.; Germain, K.; Dupont, C.; Velge, P.; Guitton, E.; Lavillatte, S.; Parias, C.; Leterrier, C. Leterrier. Effects of a gut microbiota transfer on emotional reactivity in Japanese quails (Coturnix japonica). J. Exp. Biol. 2019, 222, jeb.202879. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef]
- Signorini, M.; Soto, L.; Zbrun, M.; Sequeira, G.; Rosmini, M.; Frizzo, L. Impact of probiotic administration on the health and fecal microbiota of young calves: A meta-analysis of randomized controlled trials of lactic acid bacteria. Res. Vet. Sci. 2012, 93, 250–258. [Google Scholar] [CrossRef]
- Faubladier, C.; Chaucheyras-Durand, F.; da Veiga, L.; Julliand, V. Julliand. Effect of transportation on fecal bacterial communities and fermentative activities in horses: Impact of Saccharomyces cerevisiae CNCM I-1077 supplementation. J. Anim. Sci. 2013, 91, 1736–1744. [Google Scholar] [CrossRef] [PubMed]
- Julien, C.; Marden, J.P.; Auclair, E.; Moncoulon, R.; Cauquil, L.; Peyraud, J.L.; Bayourthe, C. Interaction between live yeast and dietary rumen degradable protein level: Effects on diet utilization in early-lactating dairy cows. Agric. Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Adeyemi, J.A.; Peters, S.O.; De Donato, M.; Cervantes, A.P.; Ogunade, I.M. Effects of a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products on plasma carbonyl-metabolome and fecal bacterial community of beef steers. J. Anim. Sci. Biotechnol. 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Al-Saiady, M.Y. Effect of probiotic bacteria on immunoglobulin G concentration and other blood components of Newborn calves. J. Anim. Vet. Adv. 2010, 9, 604–609. [Google Scholar] [CrossRef]
- Chen, L.; Shen, Y.; Wang, C.; Ding, L.; Zhao, F.; Wang, M.; Fu, J.; Wang, H. Megasphaera elsdenii Lactate Degradation Pattern Shifts in Rumen Acidosis Models. Front. Microbiol. 2019, 10, 162. [Google Scholar] [CrossRef]
- Roberfroid, M. Prebiotics: The concept revisited. J. Nutr. 2007, 137, 830–837. [Google Scholar] [CrossRef]
- Macfarlane, S.M.G.T.; Macfarlane, G.T.; Cummings, J.T. Prebiotics in the gastrointestinal tract. Aliment Pharmacol. Ther. 2006, 24, 701–714. [Google Scholar] [CrossRef]
- De Vrese, M.; Schrezenmeir, J. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 2008, 111, 1–66. [Google Scholar] [CrossRef]
- Quigley, J.D., III; Drewry, J.J.; Murray, L.M.; Ivey, S.J. Body weight gain, feed efficiency, and fecal scores of dairy calves in response to galactosyllactose or antibiotics in milk replacers. J. Dairy Sci. 1997, 80, 1751–1754. [Google Scholar] [CrossRef]
- Ghosh, S.; Mehla, R.K. Influence of dietary supplementation of prebiotics (mannooligosaccharide) on the performance of crossbred calves. Trop. Anim. Health Prod. 2012, 44, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Roodposhti, P.M.; Dabiri, N. Effects of probiotic and prebiotic on average daily gain, fecal shedding of Escherichia coli, and immune system status in newborn female calves. Asian-Australas. J Anim. Sci. 2012, 25, 1255–1261. [Google Scholar] [CrossRef]
- Quigley, J.D., III; Kost, C.J.; Wolfe, T.A. Effects of spray-dried animal plasma in milk replacers or additives containing serum and oligosaccharides on growth and health of calves. J. Dairy Sci. 2002, 85, 413–421. [Google Scholar] [CrossRef]
- Hartemink, R.; Van Laere, K.M.J.; Rombouts, F.M. Growth of enterobacteria on fructo-oligosaccharides. J. Appl. Microbiol. 1997, 83, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, J.M.; Welch, C.B. Using microbiome information to understand and improve animal performance. Ital. J. Anim. Sci. 2022, 21, 899–913. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.J.S. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Zhu, W.; Mao, S. Dynamic changes in rumen fermentation and bacterial community following rumen fluid transplantation in a sheep model of rumen acidosis: Implications for rumen health in ruminants. FASEB J. 2019, 33, 8453–8467. [Google Scholar] [CrossRef]
- Yu, S.; Shi, W.; Yang, B.; Gao, G.; Chen, H.; Cao, L.; Yu, Z.; Wang, J. Effects of repeated oral inoculation of artificially fed lambs with lyophilized rumen fluid on growth performance, rumen fermentation, microbial population and organ development. Anim. Feed Sci. Technol. 2020, 264, 114465. [Google Scholar] [CrossRef]
- Kim, H.S.; Whon, T.W.; Sung, H.; Jeong, Y.-S.; Jung, E.S.; Shin, N.-R.; Hyun, D.-W.; Kim, P.S.; Lee, J.-Y.; Lee, C.H.; et al. Longitudinal evaluation of fecal microbiota transplantation for ameliorating calf diarrhea and improving growth performance. Nat. Commun. 2021, 12, 161. [Google Scholar] [CrossRef]
- Brag, S.; Hansen, H. Treatment of ruminal indigestion according to popular belief in Sweden. Rev. Sci. Tech. 1994, 13, 529–535. [Google Scholar] [CrossRef]
- Forkosh, E.; Ilan, Y. The heart-gut axis: New target for atherosclerosis and congestive heart failure therapy. Open Heart 2019, 6, e000993. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Ahlawat, S.; Sharma, K.K. Gut–organ axis: A microbial outreach and networking. Lett. Appl. Microbiol. 2021, 72, 636–668. [Google Scholar] [CrossRef] [PubMed]
- El Aidy, S.; Dinan, T.G.; Cryan, J.F. Gut microbiota: The conductor in the orchestra of immune-neuroendocrine communication. Clin. Ther. 2015, 37, 954–967. [Google Scholar] [CrossRef]
- Wells, J.M.; Rossi, O.; Meijerink, M.; Van Baarlen, P. Epithelial crosstalk at the microbiota-mucosal interface. Proc. Natl. Acad. Sci. USA 2011, 108, 4607–4614. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Conlon, M.; Bird, A. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef]
- Bienenstock, J.; Kunze, W.; Forsythe, P. Microbiota and the gut-brain axis. Nutr. Rev. 2015, 73, 28–31. [Google Scholar] [CrossRef]
- Bercik, P. The microbiota-gut-brain axis: Learning from intestinal bacteria? Gut 2011, 60, 288–290. [Google Scholar] [CrossRef]
- Feng, Q.; Chen, W.D.; Wang, Y.D. Gut microbiota: An integral moderator in health and disease. Front. Microbiol. 2018, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Raybould, H.E. Gut chemosensing: Interactions between gut endocrine cells and visceral afferents. Auton. Neurosci. 2010, 153, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Powley, T.L.; Wang, X.Y.; Fox, E.A.; Phillips, R.J.; Liu, L.W.C.; Huizinga, J.D. Ultrastructural evidence for communication between intramuscular vagal mechanoreceptors and interstitial cells of Cajal in the rat fundus. Neurogastroenterol. Motil. 2008, 20, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kasper, L.H. The role of microbiome in central nervous system disorders. Brain Behav. Immun. 2014, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kraimi, N.; Dawkins, M.; Gebhardt-Henrich, S.G.; Velge, P.; Rychlik, I.; Volf, J.; Creach, P.; Smith, A.; Colles, F.; Leterrier, C. Influence of the microbiota-gut-brain axis on behavior and welfare in farm animals: A review. Physiol. Behav. 2019, 210, 112658. [Google Scholar] [CrossRef]
- Li, F.; Shah, A.M.; Wang, Z.; Peng, Q.; Hu, R.; Zou, H.; Tan, C.; Zhang, X.; Liao, Y.; Zeng, L.; et al. Effects of Land Transport Stress on Variations in Ruminal Microbe Diversity and Immune Functions in Different Breeds of Cattle. Animals 2019, 9, 599. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, L.; Luo, R.; Chen, H.; Nie, C.; Niu, J.; Chen, C.; Xu, Y.; Li, X.; Zhang, W. Effect of a Multispecies Probiotic Mixture on the Growth and Incidence of Diarrhea, Immune Function, and Fecal Microbiota of Pre-weaning Dairy Calves. Front. Microbiol. 2021, 12, 681014. [Google Scholar] [CrossRef]
- Han, Z.; Willer, T.; Li, L.; Pielsticker, C.; Rychlik, I.; Velge, P.; Kaspers, B.; Rautenschlein, S. Rautenschlein. Influence of the gut microbiota composition on Campylobacter jejuni colonization in chickens. Infect. Immun. 2017, 85, e00380-17. [Google Scholar] [CrossRef]
- Nagata, R.; Kim, Y.-H.; Ohkubo, A.; Kushibiki, S.; Ichijo, T.; Sato, S. Effects of repeated subacute ruminal acidosis challenges on the adaptation of the rumen bacterial community in Holstein bulls. J. Dairy Sci. 2018, 101, 4424–4436. [Google Scholar] [CrossRef]
- Thomson, R.G. Rumenitis in cattle. Can. Vet. J. 1967, 8, 189–192. [Google Scholar]
- Desnoyers, M.; Giger-Reverdin, S.; Sauvant, D.; Bertin, G.; Duvaux-Ponter, C. The influence of acidosis and live yeast (Saccharomyces cerevisiae) supplementation on time-budget and feeding behaviour of dairy goats receiving two diets of differing concentrate proportion. Appl. Anim. Behav. Sci. 2009, 121, 108–119. [Google Scholar] [CrossRef]
- Devries, T.; Chevaux, E. Modification of the feeding behavior of dairy cows through live yeast supplementation. J. Dairy Sci. 2014, 97, 6499–6510. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the respiratory microbiome in healthy non-smokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Schuijt, T.J.; Lankelma, J.M.; Scicluna, B.P.; e Melo, F.D.S.; Roelofs, J.J.; de Boer, J.D.; Hoogendijk, A.J.; de Beer, R.; de Vos, A.; Belzer, C.; et al. The gut microbiota plays a protective role in the host defense against pneumococcal pneumonia. Gut 2016, 65, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.W.; Chen, P.H.; Hsu, C.M. Commensal microflora contribute to host defense against Escherichia coli pneumonia through Toll-like receptors. Shock 2011, 36, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef]
- Beers, M.H. The Merck Manual of Diagnosis and Therapy; Merck Research Laboratories: Whitehouse Station, NJ, USA, 2001. [Google Scholar]
- Zeineldin, M.; Lowe, J.; Aldridge, B. Contribution of the mucosal microbiota to bovine respiratory health. Trends Microbiol. 2019, 27, 753–770. [Google Scholar] [CrossRef]
- Goddard, A.F.; Staudinger, B.J.; Dowd, S.E.; Joshi-Datar, A.; Wolcott, R.D.; Aitken, M.L.; Fligner, C.L.; Singh, P.K. Direct sampling of cystic fibrosis lungs indicates that DNA- based analyses of upper-airway specimens can misrepresent lung microbiota. Proc. Natl. Acad. Sci. USA 2012, 109, 13769–13774. [Google Scholar] [CrossRef]
- Nicola, I.; Cerutti, F.; Grego, E.; Bertone, I.; Gianella, P.; D’Angelo, A.; Peletto, S.; Bellino, C. Characterization of the upper and lower respiratory tract microbiota in Piedmontese calves. Microbiome 2017, 5, 152. [Google Scholar] [CrossRef]
- Zeineldin, M.M.; Lowe, J.F.; Grimmer, E.D.; De Godoy, M.R.C.; Ghanem, M.M.; El-Raof, Y.M.A.; Aldridge, B.M. Relationship between nasopharyngeal and bronchoalveolar microbial communities in clinically healthy feedlot cattle. BMC Microbiol. 2017, 17, 138. [Google Scholar] [CrossRef] [PubMed]
- Woldehiwet, Z.; Mamache, B.; Rowan, T.G. Effects of age, environmental temperature and relative humidity on the colonization of the nose and trachea of calves by Mycoplasma spp. Br. Vet. J. 1990, 146, 419–424. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The microbiome and the respiratory tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Lemon, K.P.; Klepac-Ceraj, V.; Schiffer, H.K.; Brodie, E.L.; Lynch, S.V.; Kolter, R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 2010, 1, e00129-10. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.T.; Kirkeby, L.P.; Poulsen, K.; Reinholdt, J.; Kilian, M. Resident aerobic microbiota of the adult human nasal cavity. APMIS 2000, 108, 663–675. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Hall, J.A.; Isaiah, A.; Estill, C.T.; Pirelli, G.J.; Suchodolski, J.S. Weaned beef calves fed selenium-biofortified alfalfa hay have an enriched nasal microbiota compared with healthy controls. PLoS ONE 2017, 12, e0179215. [Google Scholar] [CrossRef]
- Timsit, E.; Holman, D.B.; Hallewell, J.; Alexander, T.W. The nasopharyngeal microbiota in feedlot cattle and its role in respiratory health. Anim. Front. 2016, 6, 44–50. [Google Scholar] [CrossRef]
- McMullen, C.; Orsel, K.; Alexander, T.W.; van der Meer, F.; Plastow, G.; Timsit, E. Evolution of the nasopharyngeal bacterial microbiota of beef calves from spring processing to 40 days after feedlot arrival. Vet. Microbiol. 2018, 225, 139–148. [Google Scholar] [CrossRef]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037-15. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wen, Q.; Yao, F.; Xu, D.; Huang, Y.; Wang, J. Gut- lung axis: The microbial contributions and clinical implications. Crit. Rev. Microbiol. 2017, 43, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, N.; Khare, A.; Oriss, T.B.; Raundhal, M.; Morse, C.; Yarlagadda, M.; Wenzel, S.E.; Moore, M.L.; Peebles, R.S.; Ray, A.; et al. Early infection with respiratory syncytial virus impairs regulatory T cell function and increases susceptibility to allergic asthma. Nat. Med. 2012, 18, 1525–1530. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.G.; Livraghi-Butrico, A.; Fletcher, A.A.; McElwee, M.M.; Evans, S.E.; Boerner, R.M.; Alexander, S.N.; Bellinghausen, L.K.; Song, A.S.; Petrova, Y.M.; et al. Muc5b is required for airway defense. Nature 2014, 505, 412–416. [Google Scholar] [CrossRef]
- Eisele, N.A.; Anderson, D.M. Host defense and the airway epithelium: Frontline responses that protect against bacterial invasion and pneumonia. J. Pathog. 2011, 2011, 249802. [Google Scholar] [CrossRef]
- Vareille, M.; Kieninger, E.; Edwards, M.R.; Regamey, N. The airway epithelium: Soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 2011, 24, 210–229. [Google Scholar] [CrossRef]
- Osman, R.; Malmuthuge, N.; Gonzalez-Cano, P.; Griebel, P. Development and function of the mucosal immune system in the upper respiratory tract of neonatal calves. Annu. Rev. Anim. Biosci. 2018, 6, 141–155. [Google Scholar] [CrossRef]
- Uehara, A.; Fujimoto, Y.; Fukase, K.; Takada, H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol. Immunol. 2007, 44, 3100–3111. [Google Scholar] [CrossRef]
- Ackermann, M.R.; Derscheid, R.; Roth, J.A. Innate immunology of bovine respiratory disease. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 215–228. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Gaudier, E.; Jarry, A.; Blottière, H.M.; de Coppet, P.; Buisine, M.P.; Aubert, J.P.; Laboisse, C.; Cherbut, C.; Hoebler, C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G1168–G1174. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, L.E.M.; Koetsier, M.A.; Van Deventer, S.J.H.; Van Tol, E.A.F. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Ohata, A.; Usami, M.; Miyoshi, M. Short-chain fatty acids alter tight junction permeability in intestinal monolayer cells via lipoxygenase activation. Nutrition 2005, 21, 838–847. [Google Scholar] [CrossRef]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut Microbial metabolites fuel host antibody responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y.; et al. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef]
- Guilloteau, P.; Martin, L.; Eeckhaut, V.; Ducatelle, R.; Zabielski, R.; Van Immerseel, F. From the gut to the peripheral tissues: The multiple effects of butyrate. Nutr. Res. Rev. 2010, 23, 366–384. [Google Scholar] [CrossRef]
- Meale, S.J.; Li, S.; Azevedo, P.; Derakhshani, H.; Plaizier, J.C.; Steele, M.; Khafipour, E. Does weaning age affect the development of ruminal and fecal microbiomes in dairy calves? J. Anim. Sci. 2016, 94, 785. [Google Scholar] [CrossRef]
- Zeineldin, M.; Lowe, J.; de Godoy, M.; Maradiaga, N.; Ramirez, C.; Ghanem, M.; Abd El-Raof, Y.; Al-dridge, B. Disparity in the nasopharyngeal microbiota between healthy cattle on feed, at entry processing and with respiratory disease. Vet. Microbiol. 2017, 208, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Timsit, E.; Workentine, M.; Schryvers, A.B.; Holman, D.B.; van der Meer, F.; Alexander, T.W. Evolution of the nasopharyngeal microbiota of beef cattle from weaning to 40 days after arrival at a feedlot. Vet. Microbiol. 2016, 187, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Caswell, J.L. Failure of respiratory defenses in the pathogenesis of bacterial pneumonia of cattle. Vet. Pathol. 2014, 51, 393–409. [Google Scholar] [CrossRef]
- Lyte, M. The effect of stress on microbial growth. Anim. Health Res. Rev. 2014, 15, 172–174. [Google Scholar] [CrossRef]
- Lima, S.F.; Teixeira, A.G.V.; Higgins, C.H.; Lima, F.S.; Bicalho, R.C. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci. Rep. 2016, 6, 29050. [Google Scholar] [CrossRef]
- Buckham Sporer, K.R.; Weber, P.S.D.; Burton, J.L.; Earley, B.; Crowe, M.A. Transportation of young beef bulls alters circulating physiological parameters that may be effective biomarkers of stress. J. Anim. Sci. 2008, 86, 1325–1334. [Google Scholar] [CrossRef]
- Apley, M. The clinical syndrome of BRD: What it is and what it is not. Anim. Health Res. Rev. 2014, 15, 135–137. [Google Scholar] [CrossRef]
- Zeineldin, M.M.; El-Raof, Y.M.A.; El-attar, H.A.; Ghanem, M.M. Lung ultrasonography and computer-aided scoring system as a diagnostic aid for bovine respiratory disease in feedlot cattle. Glob. Vet. 2016, 17, 588–594. [Google Scholar] [CrossRef]
- Alexander, T.W.; Plaizier, J.C. The importance of microbiota in ruminant production. Anim. Front. 2016, 6, 4. [Google Scholar] [CrossRef][Green Version]
- Rice, J.A.; Carrasco-Medina, L.; Hodgins, D.C.; Shewen, P.E. Mannheimia haemolytica and bovine respiratory disease. Anim. Health Res. Rev. 2007, 8, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Timsit, E.; Workentine, M.; van der Meer, F.; Alexander, T. Distinct bacterial metacommunities inhabit the upper and lower respiratory tracts of healthy feedlot cattle and those diagnosed with bronchopneumonia. Vet. Microbiol. 2018, 221, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Capik, S.F.; Kegley, B.; Richeson, J.; Powell, J.; Zhao, J. Bovine respiratory microbiota of feedlot cattle and its association with disease. Vet. Res. 2022, 53, 4. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Bicalho, R.C.; Moroni, P. The bovine milk microbiota: Insights and perspectives from -omics studies. Mol. BioSyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef]
- Cremonesi, P.; Ceccarani, C.; Curone, G.; Severgnini, M.; Pollera, C.; Bronzo, V.; Riva, F.; Addis, M.F.; Filipe, J.; Amadori, M.; et al. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS ONE 2018, 13, e0205054. [Google Scholar] [CrossRef]
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 2011, 150, 81–94. [Google Scholar] [CrossRef]
- Bhatt, V.; Ahir, V.; Koringa, P.; Jakhesara, S.; Rank, D.; Nauriyal, D.; Kunjadia, A.; Joshi, C. Milk microbiome signatures of subclinical mastitis-affected cattle analysed by shotgun sequencing. J. Appl. Microbiol. 2012, 112, 639–650. [Google Scholar] [CrossRef]
- Young, W.; Hine, B.C.; Wallace, O.A.; Callaghan, M.; Bibiloni, R. Transfer of intestinal bacterial components to mammary secretions in the cow. PeerJ 2015, 3, e888. [Google Scholar] [CrossRef]
- Rodriguez, J.M. The origin of human milk bacteria: Is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef]
- Roux, M.E.; Mcwilliams, M.; Phillips-Quaglita, J.M.; Weisz-Carrington, P.; Lamm, M.E. Origin of IgA-secreting plasma cells in the mammary gland. J. Exp. Med. 1977, 146, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004, 303, 1662–1665. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Guo, J.; Zhao, C.; Jiang, P.; Maimai, T.; Yanyi, L.; Cao, Y.; Fu, Y.; Zhang, N. The gut microbiota contributes to the development of Staphylococcus aureus-induced mastitis in mice. ISME J. 2020, 14, 1897–1910. [Google Scholar] [CrossRef] [PubMed]
- Melchior, M.; Vaarkamp, H.; Fink-Gremmels, J. Biofilms: A role in recurrent mastitis infections? Vet. J. 2006, 171, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Andrews, T.; Neher, D.A.; Weicht, T.R.; Barlow, J.W. Mammary microbiome of lactating organic dairy cows varies by time, tissue site, and infection status. PLoS ONE 2019, 14, e0225001. [Google Scholar] [CrossRef] [PubMed]
- Hillerton, J.E. Whatever happened to mastitis pathogenesis? J. Dairy Res. 2020, 87, 273–276. [Google Scholar] [CrossRef]
- Rainard, P. Mammary microbiota of dairy ruminants: Fact or fiction? Vet. Res. 2017, 48, 25. [Google Scholar] [CrossRef]
- Hanage, W.P. Microbiology: Microbiome science needs a healthy dose of scepticism. Nature 2014, 512, 247–248. [Google Scholar] [CrossRef]
- Taponen, S.; McGuinness, D.; Hiitiö, H.; Simojoki, H.; Zadoks, R.; Pyorala, S. Bovine milk microbiome: A more complex issue than expected. Vet. Res. 2019, 50, 44. [Google Scholar] [CrossRef]
- Schoenmakers, S.; Steegers-Theunissen, R.; Faas, M. The matter of the reproductive microbiome. Obstet. Med. 2019, 12, 107–115. [Google Scholar] [CrossRef]
- Chen, C.; Song, X.; Wei, W.; Zhong, H.; Dai, J.; Lan, Z.; Li, F.; Yu, X.; Feng, Q.; Wang, Z.; et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017, 8, 875. [Google Scholar] [CrossRef] [PubMed]
- Scolari, F.; Attardo, G.M.; Aksoy, E.; Weiss, B.; Savini, G.; Takac, P.; Abd-Alla, A.; Parker, A.G.; Aksoy, S.; Malacrida, A.R. Symbiotic microbes affect the expression of male reproductive genes in Glossina m. morsitans. BMC Microbiol. 2018, 18, 169. [Google Scholar] [CrossRef] [PubMed]
- Appiah, M.; Wang, J.; Lu, W. Microflora in the reproductive tract of cattle: A review. Agriculture 2020, 10, 232. [Google Scholar] [CrossRef]
- Piersanti, R.L.; Bromfield, J.J. The Consequence of Postpartum Uterine Disease on Dairy Cow Fertility. EDIS 2019, 2019, 107174. [Google Scholar] [CrossRef]
- Sheldon, I.M.; Dobson, H. Postpartum uterine health in cattle. Anim. Reprod. Sci. 2004, 82–83, 295–306. [Google Scholar] [CrossRef]
- Nesengani, L.T.; Wang, J.; Yang, Y.; Yang, L.; Lu, W. Unravelling vaginal microbial genetic diversity and abundance between Holstein and Fleckvieh cattle. RSC Adv. 2017, 7, 56137–56143. [Google Scholar] [CrossRef]
- Giannattasio-Ferraz, S.; Laguardia-Nascimento, M.; Gasparini, M.R.; Leite, L.R.; Araujo, F.M.G.; de Matos Salim, A.C.; de Oliveira, A.P.; Nicoli, J.R.; de Oliveira, G.C.; da Fonseca, F.G.; et al. A common vaginal microbiota composition among breeds of Bos taurus indicus (Gyr and Nellore). Braz. J. Microbiol. 2019, 50, 1115–1124. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, H.; Fu, K.; Pang, B.; Yang, Y.; Liu, Y.; Tian, W.; Cao, R. Characterization of the cervical bacterial community in dairy cows with metritis and during different physiological phases. Theriogenology 2018, 108, 306–313. [Google Scholar] [CrossRef]
- Santos, T.M.; Bicalho, R.C. Diversity and succession of bacterial communities in the uterine fluid of postpartum metritic, endometritic and healthy dairy cows. PLoS ONE 2012, 7, e53048. [Google Scholar] [CrossRef]
- Jeon, S.J.; Cunha, F.; Vieira-Neto, A.; Bicalho, R.C.; Lima, S.; Bicalho, M.L.; Galvão, K.N. Blood as a route of transmission of uterine pathogens from the gut to the uterus in cows. Microbiome 2017, 5, 109. [Google Scholar] [CrossRef]
- Nagaraja, T.; Lechtenberg, K.F. Liver abscesses in feedlot cattle. Vet. Clin. N. Am. Food Anim. Pract. 2007, 23, 351–369. [Google Scholar] [CrossRef]
- Jones, K.; Cunha, F.; Jeon, S.J.; Pérez-Báez, J.; Casaro, S.; Fan, P.; Liu, T.; Lee, S.; Jeong, K.C.; Yang, Y.; et al. Tracing the source and route of uterine colonization by exploring the genetic relationship of Escherichia coli isolated from the reproductive and gastrointestinal tract of dairy cows. Vet. Microbiol. 2020, 266, 109355. [Google Scholar] [CrossRef] [PubMed]
- Ulfina, G.G.; Kimothi, S.P.; Oberoi, P.S.; Baithalu, R.K.; Kumaresan, A.; Mohanty, T.K.; Imtiwati, P.; Dang, A.K. Modulation of post-partum reproductive performance in dairy cows through supplementation of long- or short-chain fatty acids during transition period. J. Anim. Physiol. Anim. Nutr. 2015, 99, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Sha, L.; Li, Y.; Zhu, L.; Wang, Z.; Li, K.; Lu, H.; Bao, T.; Guo, L.; Zhang, X.; et al. Dietary alinolenic acid-rich flaxseed oil exerts beneficial effects on polycystic ovary syndrome through sex steroid hormones—microbiota—inflammation Axis in rats. Front. Endocrinol. 2020, 11, 284. [Google Scholar] [CrossRef] [PubMed]
- Boukhliq, R.; Martin, G.B. Administration of fatty acids and gonadotrophin secretion in the mature ram. Anim. Reprod. Sci. 1997, 49, 143–159. [Google Scholar] [CrossRef]
- Miranda-CasoLuengo, R.; Lu, J.; Williams, E.J.; Miranda-CasoLuengo, A.A.; Carrington, S.D.; Evans, A.C.; Meijer, W.G. Delayed differentiation of vaginal and uterine microbiomes in dairy cows developing postpartum endometritis. PLoS ONE 2019, 14, e0200974. [Google Scholar] [CrossRef]
- Galvão, K.N.; Bicalho, R.C.; Jeon, S.J. Symposium review: The uterine microbiome associated with the development of uterine disease in dairy cows. J. Dairy Sci. 2019, 102, 11786–11797. [Google Scholar] [CrossRef]
- Moreno, C.G.; Luque, A.T.; Galvão, K.N.; Otero, M.C. Bacterial communities from vagina of dairy healthy heifers and cows with impaired reproductive performance. Res. Vet. Sci. 2022, 142, 15–23. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Welch, C.B.; Ryman, V.E.; Pringle, T.D.; Lourenco, J.M. Utilizing the Gastrointestinal Microbiota to Modulate Cattle Health through the Microbiome-Gut-Organ Axes. Microorganisms 2022, 10, 1391. https://doi.org/10.3390/microorganisms10071391
Welch CB, Ryman VE, Pringle TD, Lourenco JM. Utilizing the Gastrointestinal Microbiota to Modulate Cattle Health through the Microbiome-Gut-Organ Axes. Microorganisms. 2022; 10(7):1391. https://doi.org/10.3390/microorganisms10071391
Chicago/Turabian StyleWelch, Christina B., Valerie E. Ryman, T. Dean Pringle, and Jeferson M. Lourenco. 2022. "Utilizing the Gastrointestinal Microbiota to Modulate Cattle Health through the Microbiome-Gut-Organ Axes" Microorganisms 10, no. 7: 1391. https://doi.org/10.3390/microorganisms10071391
APA StyleWelch, C. B., Ryman, V. E., Pringle, T. D., & Lourenco, J. M. (2022). Utilizing the Gastrointestinal Microbiota to Modulate Cattle Health through the Microbiome-Gut-Organ Axes. Microorganisms, 10(7), 1391. https://doi.org/10.3390/microorganisms10071391





