Great Abilities of Shinella zoogloeoides Strain from a Landfarming Soil for Crude Oil Degradation and a Synergy Model for Alginate-Bead-Entrapped Consortium Efficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Illumina Sequencing and Processing of Assembled Genomes
2.2. Bacterial Isolation and Enrichment Microcosms
2.3. DNA Extraction and Identification of Bacterial Strains
2.4. Biosurfactant Production Assays
2.4.1. Oil-Coated Agar
2.4.2. Emulsification Test
2.5. Building the Bacterial Consortium
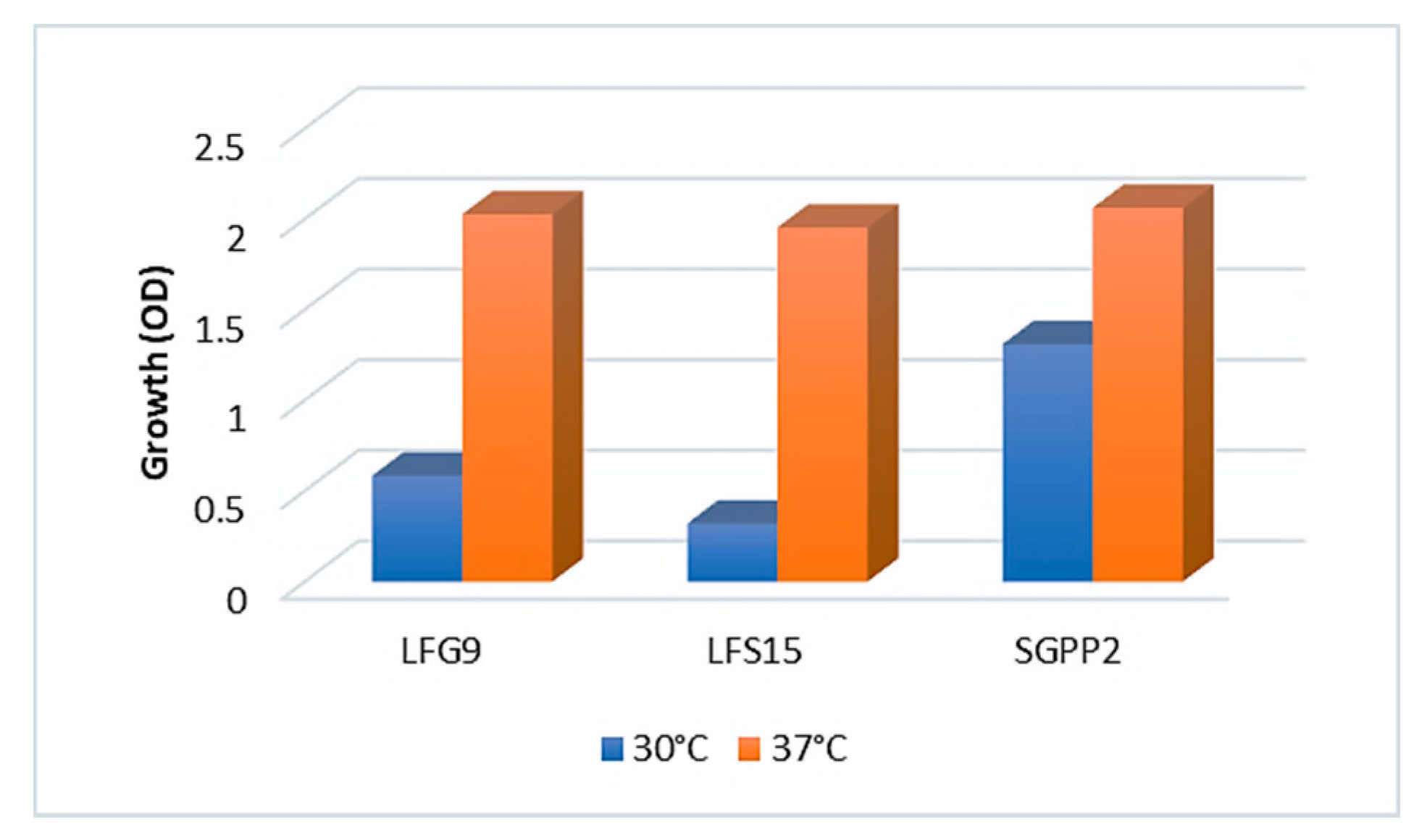
2.5.1. Choice of Temperature for Biodegradation Tests Setup
2.5.2. Biodegradation Tests Setup
2.6. Entrapment of the Bacterial Consortium
2.7. Degradation of Crude Oil by the Free Isolates and the Bacterial Consortium
3. Results
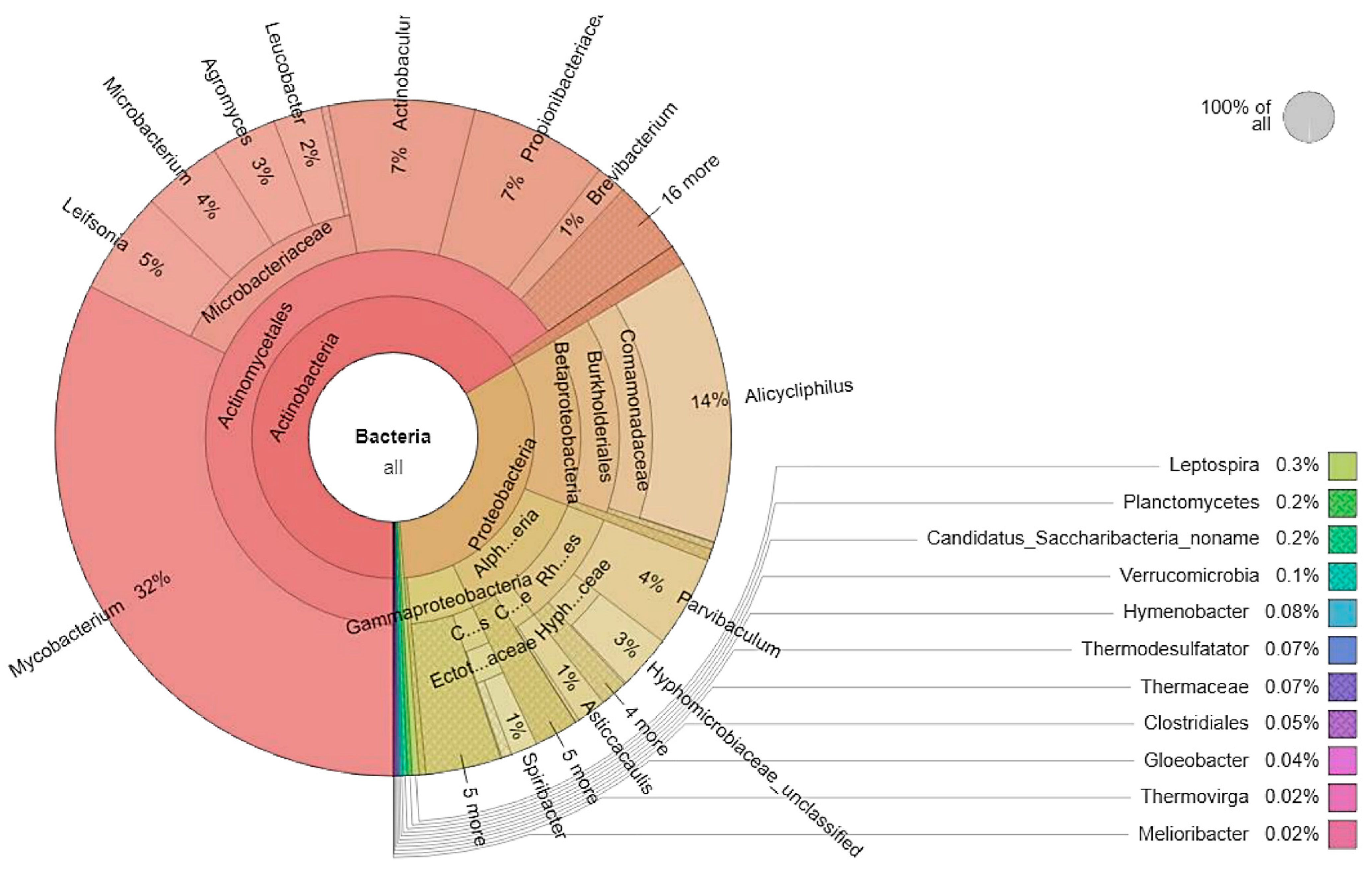
3.1. Microbial Community
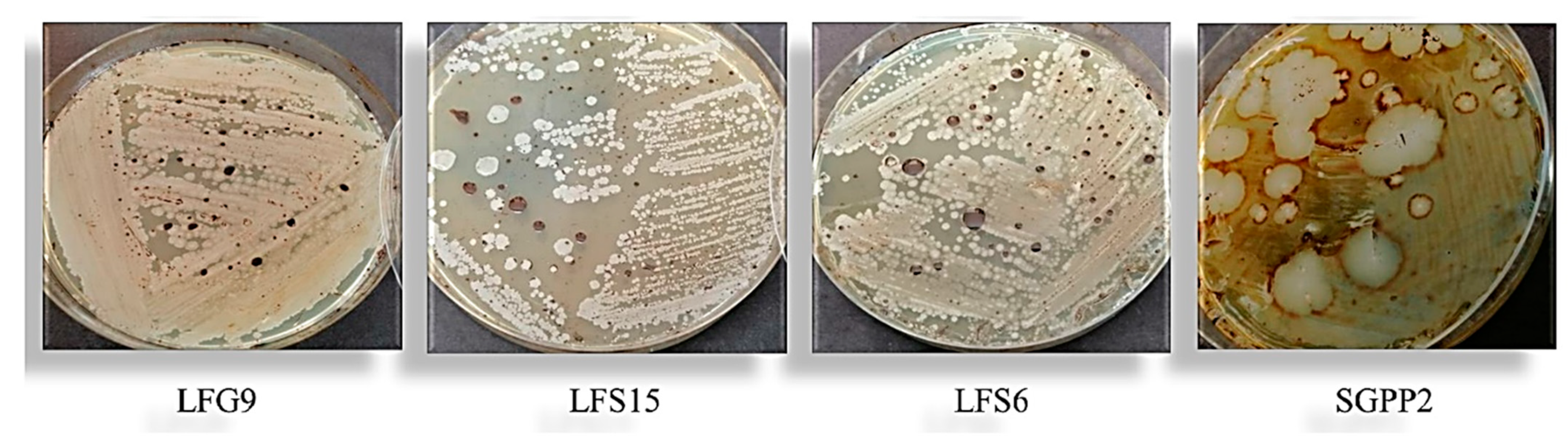
3.2. Isolation and Identification of Bacteria
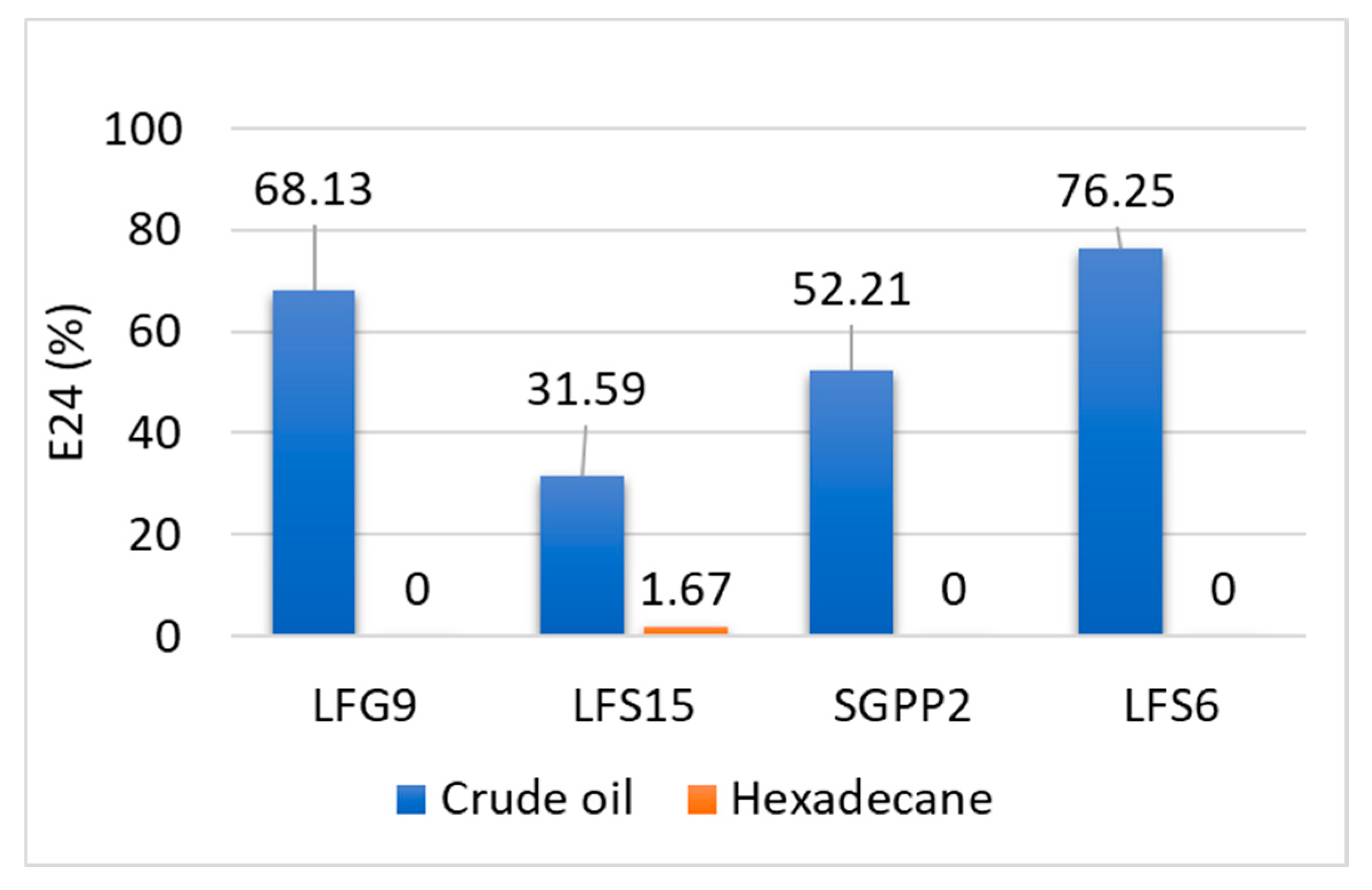
3.3. Biosurfactant Production
3.4. Investigation of Crude Oil Degradation in Microcosms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khandelwal, A.; Sugavanam, R.; Ramakrishnan, B.; Dutta, A.; Varghese, E.; Nain, L.; Banerjee, T.; Singh, N. Free and Immobilized Microbial Culture–Mediated Crude Oil Degradation and Microbial Diversity Changes Through Taxonomic and Functional Markers in a Sandy Loam Soil. Front. Environ. Sci. 2022, 9, 794303. [Google Scholar] [CrossRef]
- Tian, W.; Yao, J.; Liu, R.; Zhu, M.; Wang, F.; Wu, X.; Liu, H. Effect of natural and synthetic surfactants on crude oil biodegradation by indigenous strains. Ecotoxicol. Environ. Saf. 2016, 129, 171–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, N.; Chandran, P. Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef] [Green Version]
- Zahed, M.A.; Matinvafa, M.A.; Azari, A.; Mohajeri, L. Biosurfactant, a green and effective solution for bioremediation of petroleum hydrocarbons in the aquatic environment. Discov. Water 2022, 2, 5. [Google Scholar] [CrossRef]
- Alnuaimi, M.T.; Taher, T.A.; Aljanabi, Z.Z.; Adel, M.M. High-resolution GC/MS study of biodegradation of crude oil by Bacillus megaterium. Res. Crops 2020, 21, 650–657. [Google Scholar] [CrossRef]
- Okafor, C.P.; Udemang, N.L.; Chikere, C.B.; Akaranta, O.; Ntushelo, K. Indigenous microbial strains as bioresource for remediation of chronically polluted Niger Delta soils. Sci. Afr. 2021, 11, e00682. [Google Scholar] [CrossRef]
- Mandree, P.; Masika, W.; Naicker, J.; Moonsamy, G.; Ramchuran, S.; Lalloo, R. Bioremediation of Polycyclic Aromatic Hydrocarbons from Industry Contaminated Soil Using Indigenous Bacillus spp. Processes 2021, 9, 1606. [Google Scholar] [CrossRef]
- Kumar, A.G.; Rajan, N.N.; Kirubagaran, R.; Dharani, G. Biodegradation of crude oil using self-immobilized hydrocarbonoclastic deep sea bacterial consortium. Mar. Pollut. Bull. 2019, 146, 741–750. [Google Scholar] [CrossRef]
- Lebonguy, A.A.; Goma-Tchimbakala, J.; Miambi, E.; Keleke, S. Isolation and characterization of petroleum product emulsifying Pseudomonas strains from a generating set fuel tank. Afr. J. Microbiol. Res. 2017, 11, 920–926. [Google Scholar] [CrossRef] [Green Version]
- Agarry, S.E.; Salam, K.K.; Arinkoola, A.; Aremu, M.O. Biosurfactant production by indigenous Pseudomonas and Bacillus species isolated from auto-mechanic soil environment towards microbial enhanced oil recovery. Eur. J. Eng. Technol. 2015, 3, 27–39. [Google Scholar]
- Silva, R.D.C.F.S.; Almeida, D.G.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Applications of Biosurfactants in the Petroleum Industry and the Remediation of Oil Spills. Int. J. Mol. Sci. 2014, 15, 12523–12542. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.E.; Brant, J.L.; Campo, P.; Clark, D.R.; Coulon, F.; Gregson, B.H.; McGenity, T.J.; McKew, B.A. Effects of Dispersants and Biosurfactants on Crude-Oil Biodegradation and Bacterial Community Succession. Microorganisms 2021, 9, 1200. [Google Scholar] [CrossRef]
- Zakaria, N.N.; Gomez-Fuentes, C.; Abdul Khalil, K.; Convey, P.; Roslee, A.F.A.; Zulkharnain, A.; Sabri, S.; Shaharuddin, N.A.; Cárdenas, L.; Ahmad, S.A. Statistical optimisation of diesel biodegradation at low temperatures by an Antarctic marine bacterial consortium isolated from non-contaminated seawater. Microorganisms 2021, 9, 1213. [Google Scholar] [CrossRef] [PubMed]
- Goma-Tchimbakala, J.; Lebonguy, A.A. Assessment of Bacterial Diversity of Sandy-Loam Soil Polluted by Hydrocarbons using 454 Pyrosequencing. Am. J. Microbiol. Res. 2020, 8, 34–41. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Patowary, K.; Patowary, R.; Kalita, M.C.; Deka, S. Development of an Efficient Bacterial Consortium for the Potential Remediation of Hydrocarbons from Contaminated Sites. Front. Microbiol. 2016, 7, 1092. [Google Scholar] [CrossRef]
- Sohail, R.; Jamil, N. Isolation of biosurfactant producing bacteria from Potwar oil fields: Effect of non-fossil fuel based carbon sources. Green Process. Synth. 2020, 9, 77–86. [Google Scholar] [CrossRef]
- Chen, Q.; Li, J.; Liu, M.; Sun, H.; Bao, M. Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PLoS ONE 2017, 12, e0174445. [Google Scholar] [CrossRef] [PubMed]
- Kottek, M.; Grieser, J.; Beck, C.; Rudolf, B.; Rubel, F. World map of the Köppen-Geiger climate classification updated. Meteorol. Z. 2006, 15, 259–263. [Google Scholar] [CrossRef]
- Pessoa, T.B.A.; De Souza, S.S.; Cerqueira, A.F.; Rezende, R.P.; Pirovani, C.P.; Dias, J.C.T. Construction and validation of metagenomic DNA libraries from landfarm soil microorganisms. Genet. Mol. Res. 2013, 12, 2148–2155. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kadam, V.; Patil, Y. Isolation and development of a microbial consortium for the treatment of automobile service station wastewater. J. Appl. Microbiol. 2021, 132, 1048–1061. [Google Scholar] [CrossRef]
- Gagandeep, S.; Malik, D.K. Utilization of 2T engine oil by Pseudomonas sp. isolated from automobile workshop contaminated soil. Int. J. Chem. Anal. Sci. 2013, 4, 80–84. [Google Scholar]
- Ebakota, O.D.; Osarueme, J.O.; Gift, O.N.; Odoligie, I.; Osazee, J.O. Isolation and Characterization of Hydrocarbon-Degrading Bacteria in Top and Subsoil of selected Mechanic Workshops in Benin City Metropolis, Nigeria. J. Appl. Sci. Environ. Manag. 2017, 21, 641–645. [Google Scholar] [CrossRef] [Green Version]
- Chikere, C.B.; Tekere, M.; Adeleke, R. Microbial communities in field-scale oil-polluted soil remediation using 16S rRNA amplicon sequencing. Int. J. Environ. Stud. 2021, 78, 410–426. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.H.; Jung, W.C.; Park, M.; Kim, M.S.; Lee, S.J.; Park, H. Changes in Soil Health with Remediation of Petroleum Hydrocarbon Contaminated Soils Using Two Different Remediation Technologies. Sustainability 2020, 12, 10078. [Google Scholar] [CrossRef]
- Bergsveinson, J.; Perry, B.J.; Simpson, G.L.; Yost, C.; Schutzman, R.J.; Hall, B.D.; Cameron, A.D.S. Spatial analysis of a hydrocarbon waste-remediating landfarm demonstrates influence of management practices on bacterial and fungal community structure. Microb. Biotechnol. 2019, 12, 1199–1209. [Google Scholar] [CrossRef]
- Pourbabaee, A.A.; Shahriari, M.H.; Garousin, H. Biodegradation of phenanthrene as a model hydrocarbon: Power display of a super-hydrophobic halotolerant enriched culture derived from a saline-sodic soil. Biotechnol. Rep. 2019, 24, e00388. [Google Scholar] [CrossRef]
- Calvo, C.; Manzanera, M.; Silva-Castro, G.; Uad, I.; González-López, J. Application of bioemulsifiers in soil oil bioremediation processes. Future prospects. Sci. Total Environ. 2009, 407, 3634–3640. [Google Scholar] [CrossRef] [PubMed]
- An, D.-S.; Im, W.-T.; Yang, H.-C.; Lee, S.-T. Shinella granuli gen. nov., sp. nov., and proposal of the reclassification of Zoogloea ramigera ATCC 19623 as Shinella zoogloeoides sp. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Yang, Y.; Zhang, J.; Wang, H.; Ma, Y.; He, J.; Lu, Z. The Complete Genome Sequence of the Nicotine-Degrading Bacterium Shinella sp. HZN7. Front. Microbiol. 2016, 7, 1348. [Google Scholar] [CrossRef] [PubMed]
- Biala, S.; Chadha, P.; Saini, H.S. Biodegradation of 4-aminobenzenesulfonate by Indigenous Isolate Shinella yambaruensis SA1 and its Validation by Genotoxic Analysis. Biotechnol. Bioprocess Eng. 2014, 19, 1034–1041. [Google Scholar] [CrossRef]
- Wu, H.; Shen, J.; Wu, R.; Sun, X.; Li, J.; Han, W.; Wang, L. Biodegradation mechanism of 1H-1,2,4-triazole by a newly isolated strain Shinella sp. NJUST26. Sci. Rep. 2016, 6, 29675. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Sun, Q.; Zhao, C.; Wen, D.; Tang, X. Aerobic degradation of pyridine by a new bacterial strain, Shinella zoogloeoides BC026. J. Ind. Microbiol. Biotechnol. 2009, 36, 1391–1400. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, Y. The Synthesis of PbS NPs and Biosorption of Pb(II) by Shinella Zoogloeoides PQ7 in Aqueous Conditions. Water 2020, 12, 2065. [Google Scholar] [CrossRef]
- Ntougias, S.; Melidis, P.; Navrozidou, E.; Tzegkas, F. Diversity and efficiency of anthracene-degrading bacteria isolated from a denitrifying activated sludge system treating municipal wastewater. Int. Biodeterior. Biodegradation 2015, 97, 151–158. [Google Scholar] [CrossRef]
- Meliani, A.; Bensoltane, A. The Ability of Some Pseudomonas Strains to Produce Biosurfactant. Poult. Fish. Wildl. Sci. 2014, 2, 112. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-C.; Young, L.-S.; Lin, S.-Y.; Hameed, A.; Hsu, Y.-H.; Lai, W.-A.; Shen, F.-T.; Young, C.-C. Pseudomonas guguanensis sp. nov., a gammaproteobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 4591–4598. [Google Scholar] [CrossRef] [Green Version]
- Ajona, M.; Vasanthi, P. Bio-remediation of crude oil contaminated soil using recombinant native microbial strain. Environ. Technol. Innov. 2021, 23, 101635. [Google Scholar] [CrossRef]
- Asha, D.; Vasudevan, V.; Krishnan, M.E.G. Demonstration of bioprocess factors optimization for enhanced mono-rhamnolipid production by a marine Pseudomonas guguanensis. Int. J. Biol. Macromol. 2018, 108, 531–540. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Schisler, D.A.; Perry, E.B.; Connor, N.; Cohan, F.M.; Rooney, A.P. Bacillus swezeyi sp. nov. and Bacillus haynesii sp. nov., isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 2720–2725. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Xu, H.; Shah, K.J.; Tao, J.; Cai, D. Enhance Pyrene Degradation of Sodium Alginate Embedding Immobilization by Adding PAC. Chem. Eng. Trans. 2020, 81, 19–24. [Google Scholar] [CrossRef]
- Deschênes, L.; Lafrance, P.; Villeneuve, J.-P.; Samson, R. Adding sodium dodecyl sulfate and Pseudomonas aeruginosa UG2 biosurfactants inhibits polycyclic aromatic hydrocarbon biodegradation in a weathered creosote-contaminated soil. Appl. Microbiol. Biotechnol. 1996, 46, 638–646. [Google Scholar] [CrossRef]
- Tiehm1, A.; Schulze, S. Intrinsic Aromatic Hydrocarbon Biodegradation for Groundwater Remediation. Oil Gas Sci. Technol. 2003, 58, 449–462. [Google Scholar] [CrossRef]
- Lv, J.-X.; Dong, W.; Zhou, J.-T. Interaction Mechanisms between Anionic Surfactant Micelles and Different Metal Ions in Aqueous Solutions. J. Dispers. Sci. Technol. 2006, 27, 1073–1077. [Google Scholar] [CrossRef]
- Yang, Z.; Li, M.; Yu, M.; Huang, J.; Xu, H.; Zhou, Y.; Song, P.; Xu, R. A novel approach for methylene blue removal by calcium dodecyl sulfate enhanced precipitation and microbial flocculant GA1 flocculation. Chem. Eng. J. 2016, 303, 1–13. [Google Scholar] [CrossRef]

| Parameters | Values |
|---|---|
| Clay (%) | 13.70 ± 2 |
| Slit (%) | 20.92 ± 3.93 |
| Sand (%) | 65.35 ± 2.21 |
| C (g/kg) | 60.93 |
| N (g/kg) | 2.01 |
| P (g/kg) | 0.1 |
| Fe (mg/kg) | 9500 |
| K (mg/kg) | 982 |
| Ca (mg/kg) | 2020 |
| Mg (mg/kg) | 5580 |
| Mn (mg/kg) | 145 |
| pH | 6.11 |
| TPH (g/kg) | 355 |
| Compounds | Rt | Compounds | Rt |
|---|---|---|---|
| Nonanoic acid | 12.00 | Tetradecanoic acid | 18.16 |
| Phthalic anhydride | 12.68 | Pentadecanoic acid | 19.50 |
| n-Decanoic acid | 13.31 | 7.9-Di-tert-butyl-1-oxaspiro (4.5) deca-6.9-diene-2.8-dione | 20.30 |
| 2.4.7.9-Tetramethyl-5-decyn-4.7-diol | 13.91 | n-Hexadecanoic acid | 21.06 |
| Coelution of benzofuran and phthalate | 14.86 | Octadecanoic acid | 23.85 |
| 2.4-bis(1.1-dimethylethyl)-phenol | 15.20 | Hexadecanoic acid. 2-hydroxy-1-(hydroxymethyl)ethyl ester | 27.82 |
| Unknown aromatic compound with lateral unsaturated chain | 15.61 | 13-Docosenamide. (Z)- | 30.43 |
| Dodecanoic acid | 15.81 | Unknown unsaturated long chain alkene | 30.77 |
| Unknown phenolic compound with amino group | 16.75 |
| Strain | Identification | GenBank Accession N |
|---|---|---|
| LFS6 | Bacillus safensis | MT672419.1 |
| LFS15 | Bacillus swezeyi | MT672420.1 |
| LFS26 | Bacillus wiedmannii | MT672421.1 |
| LFS28 | Bacillus paramycoides | MT672336.1 |
| LFG2 | Ochrobactrum haematophilum | MT672422.1 |
| LFG3 | Staphylococcus saprophyticus | MT672338.1 |
| LFG4 | Bacillus cereus | MT672423.1 |
| LFG6 | Ochrobactrum sp. | MT672337.1 |
| LFG9 | Shinella zoogloeoides | MT672424.1 |
| SGPP1 | Pseudomonas guguanensis | MT672417.1 |
| SGPP2 | Pseudomonas guguanensis | MT672418.1 |
| SGPP3 | Pseudomonas aeruginosa | MT672425.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goma-Tchimbakala, E.J.C.D.; Pietrini, I.; Dal Bello, F.; Goma-Tchimbakala, J.; Lo Russo, S.; Corgnati, S.P. Great Abilities of Shinella zoogloeoides Strain from a Landfarming Soil for Crude Oil Degradation and a Synergy Model for Alginate-Bead-Entrapped Consortium Efficiency. Microorganisms 2022, 10, 1361. https://doi.org/10.3390/microorganisms10071361
Goma-Tchimbakala EJCD, Pietrini I, Dal Bello F, Goma-Tchimbakala J, Lo Russo S, Corgnati SP. Great Abilities of Shinella zoogloeoides Strain from a Landfarming Soil for Crude Oil Degradation and a Synergy Model for Alginate-Bead-Entrapped Consortium Efficiency. Microorganisms. 2022; 10(7):1361. https://doi.org/10.3390/microorganisms10071361
Chicago/Turabian StyleGoma-Tchimbakala, Emerance Jessica Claire D’Assise, Ilaria Pietrini, Federica Dal Bello, Joseph Goma-Tchimbakala, Stefano Lo Russo, and Stefano Paolo Corgnati. 2022. "Great Abilities of Shinella zoogloeoides Strain from a Landfarming Soil for Crude Oil Degradation and a Synergy Model for Alginate-Bead-Entrapped Consortium Efficiency" Microorganisms 10, no. 7: 1361. https://doi.org/10.3390/microorganisms10071361
APA StyleGoma-Tchimbakala, E. J. C. D., Pietrini, I., Dal Bello, F., Goma-Tchimbakala, J., Lo Russo, S., & Corgnati, S. P. (2022). Great Abilities of Shinella zoogloeoides Strain from a Landfarming Soil for Crude Oil Degradation and a Synergy Model for Alginate-Bead-Entrapped Consortium Efficiency. Microorganisms, 10(7), 1361. https://doi.org/10.3390/microorganisms10071361








