Antimicrobial Susceptibility and Molecular Characterization of Escherichia coli Recovered from Milk and Related Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Isolation and Identification of E. coli
2.3. Antimicrobial Susceptibility of E. coli
2.4. Molecular Identification
2.4.1. DNA Isolation
2.4.2. PCR Amplification of Partial uidA Gene
2.4.3. DNA Sequencing and Analysis
2.5. Statistical Analysis
3. Results
3.1. Occurrence of E. coli in Milk and Related Samples
3.2. Antimicrobial Susceptibility of E. coli Isolated from Raw Cow’s Milk and Related Samples
3.3. Antimicrobial Resistance Profile and Multiple Antibiotic Index of Individual E. coli
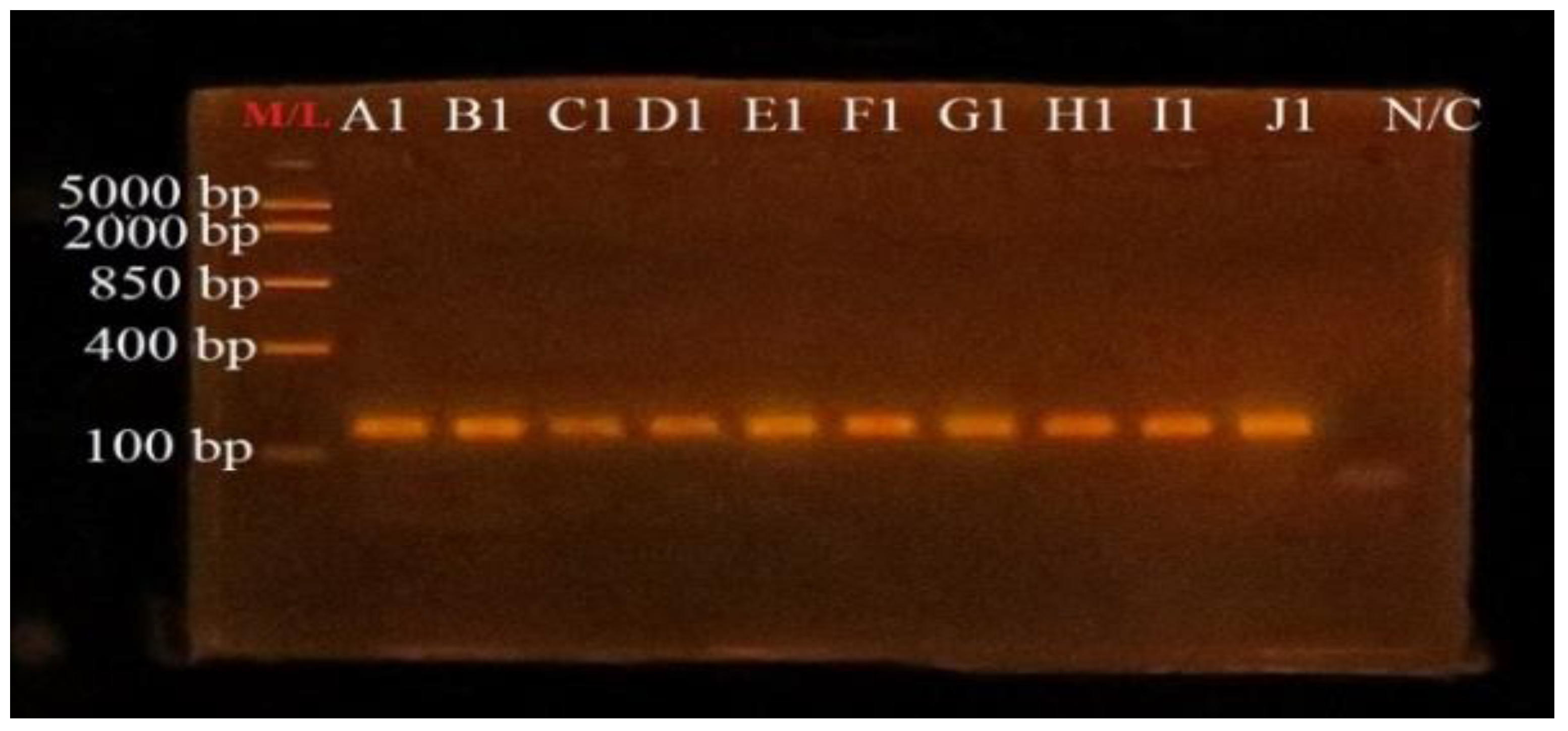
3.4. PCR Amplification of uidA Gene for Confirmation of E. coli
3.5. Sequencing and Species Identification
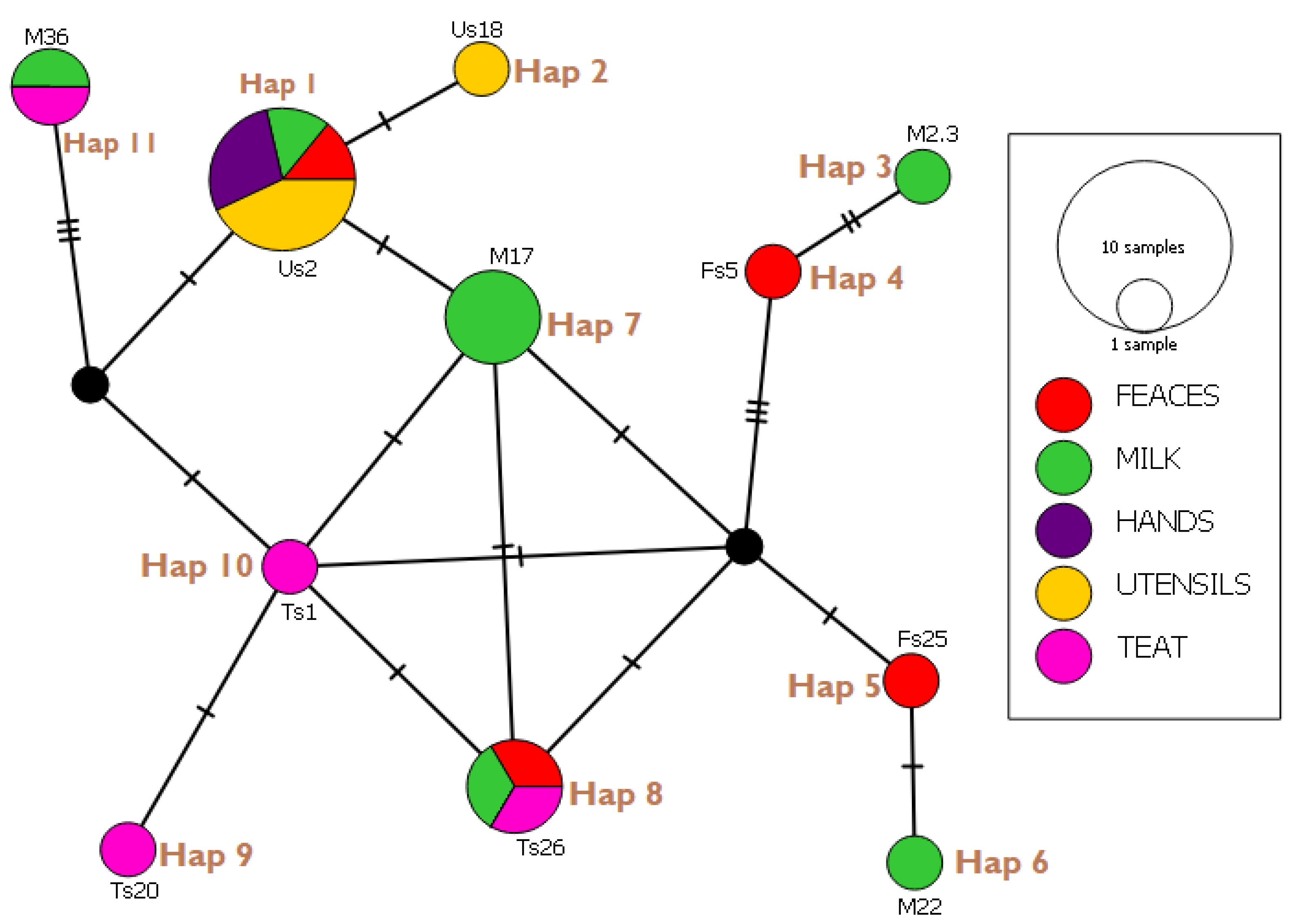
3.6. Haplotype Network Analysis and Indices
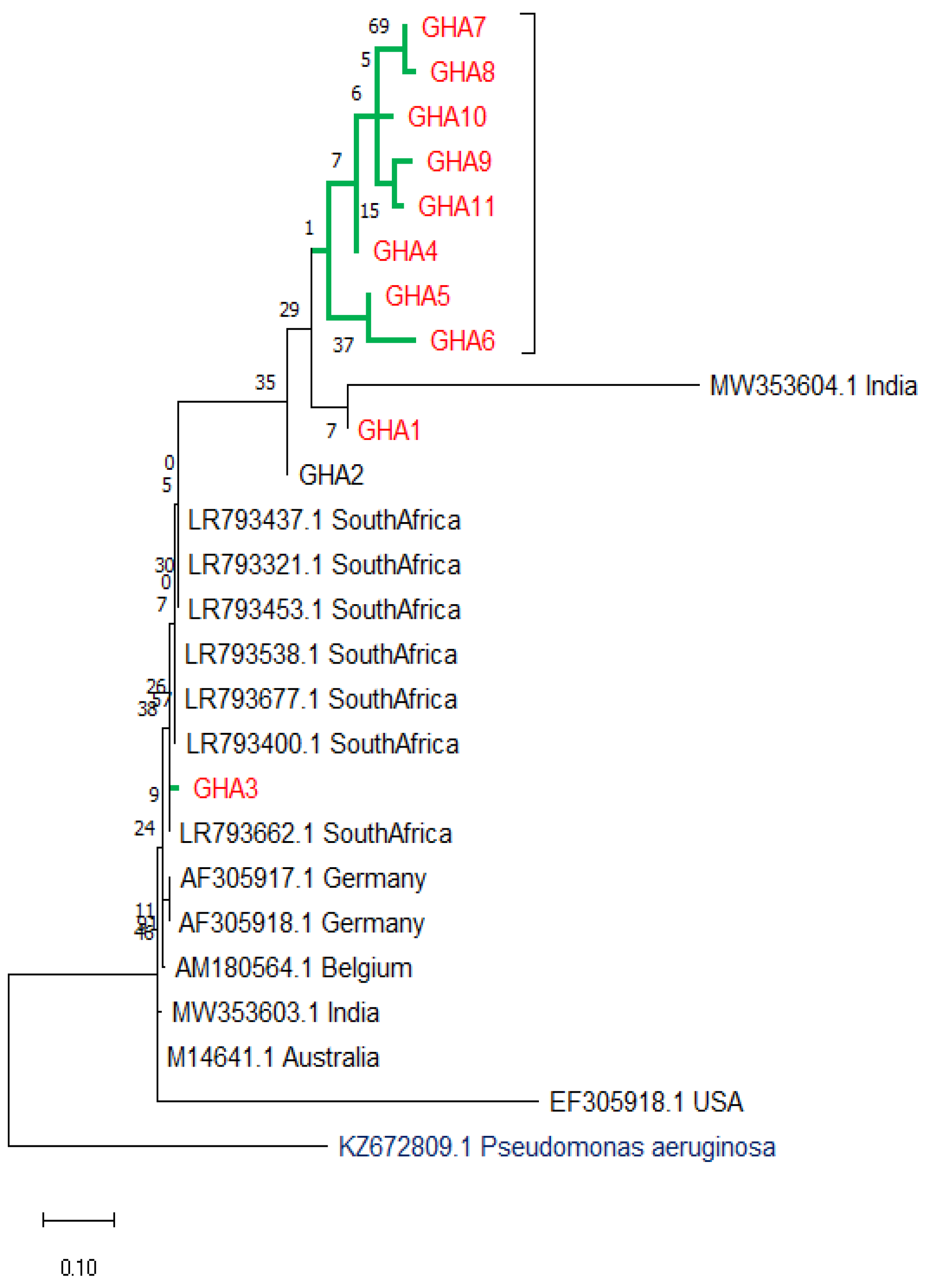
3.7. Evolutionary Relationships (Phylogenetic Tree)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ababu, A.; Endashaw, D.; Fesseha, H. Isolation and antimicrobial susceptibility profile of E. coli O157:H7 from raw milk of dairy cattle in Holeta District, Central Ethiopia. Int. J. Microbiol. 2020, 2020, 6626488. [Google Scholar] [CrossRef]
- Rehman, M.U.; Rashid, M.; Sheikh, J.A.; Bhat, M.A. Molecular epidemiology and antibiotic resistance pattern of enteropathogenic E. coli isolated from bovines and their handlers in Jammu, India. J. Adv. Vet. Anim. Res. 2014, 1, 177–181. [Google Scholar] [CrossRef]
- Soomro, A.H.; Arain, M.A.; Khaskheli, M.; Bhutto, B. Isolation of E. coli from raw milk and milk products in relation to public health sold under market conditions at Tandojam, Pakistan. Pak. J. Nutr. 2002, 1, 151–152. [Google Scholar] [CrossRef][Green Version]
- Costard, S.; Espejo, L.; Groenendaal, H.; Zagmutt, F.J. Outbreak-related disease burden associated with consumption of unpasteurized cow’s milk and cheese, United States, 2009–2014. Emerg. Infect. Dis. 2017, 23, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, H.A.; Gidey, N.B.; Workelule, K.; Hailu, A.B.; Gidey, S.; Bsrat, A.; Taddele, H. Antimicrobial resistance profile of E. coli isolated from raw cow milk and fresh fruit juice in Mekelle, Tigray, Ethiopia. Vet. Med. Int. 2018, 2018, 8903142. [Google Scholar] [CrossRef]
- Gajdács, M.; Albericio, F. Antibiotic resistance: From the bench to patients. Antibiotics 2019, 8, 129. [Google Scholar] [CrossRef]
- Boor, K.J.; Wiedmann, M.; Murphy, S.; Alcaine, S. A 100-year review: Microbiology and safety of milk handling. J. Dairy Sci. 2017, 100, 9933–9951. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.P.; Petti, C.A. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar] [CrossRef]
- Yadav, B.; Ronda, V.; Vashista, D.P.; Sharma, B. Sequencing and computational approaches to identification and characterization of microbial organisms. Biomed. Eng. Comput. Biol. 2013, 5, 43–49. [Google Scholar] [CrossRef]
- Gebeyehu, A.; Taye, M.; Abebe, R. Isolation, molecular detection and antimicrobial susceptibility profile of Salmonella from raw cow milk collected from dairy farms and households in southern Ethiopia. BMC Microbiol. 2022, 22, 84. [Google Scholar] [CrossRef]
- Hassani, S.; Moosavy, M.-H.; Gharajalar, S.N.; Khatibi, S.A.; Hajibemani, A.; Barabadi, Z. High prevalence of antibiotic resistance in pathogenic foodborne bacteria isolated from bovine milk. Sci. Rep. 2022, 12, 3878. [Google Scholar] [CrossRef] [PubMed]
- Manishimwe, R.; Moncada, P.M.; Bugarel, M.; Scott, H.M.; Loneragan, G.H. Antibiotic resistance among E. coli and Salmonella isolated from dairy cattle feces in Texas. PLoS ONE 2021, 16, e0242390. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Krikó, E.; Tőzsér, D.; Maróti, G.; Patai, V.; Makrai, L.; Szita, G.; Solymosi, N. Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 2020, 10, 7464. [Google Scholar] [CrossRef]
- Feng, P.; Weagant, S.D.; Jinneman, K.; Bacteriological Analytical Manuel, Chapter 4A: Diarrheagenic E. coli. 2020. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-4a-diarrheagenic-escherichia-coli (accessed on 23 August 2021).
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turk, M. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standard Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2022. [Google Scholar]
- Krumperman, P.H. Multiple antibiotic resistance indexing of E. coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Tamerat, N.; Muktar, Y. Application of molecular diagnostic techniques for the detection of E. coli O157:H7: A review. J. Vet. Sci. Technol. 2016, 7, 5. [Google Scholar] [CrossRef]
- Bej, A.K.; DiCesare, J.L.; Haff, L.; Atlas, R.M. Detection of E. coli and Shigella spp. in water by using the polymerase chain Reaction and gene probes for uid. Appl. Environ. Microbiol. 1991, 57, 2445. [Google Scholar] [CrossRef]
- Manske, M. GENtle, a Free Multi-Purpose Molecular Biology Tool. Ph.D. Thesis, der Universität zu Köln, Köln, Germany, 2006. Available online: https://core.ac.uk/download/pdf/12009749.pdf (accessed on 23 August 2021).
- Clement, M.J.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K.A. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, Lauderdale, FL, USA, 15–19 April 2002. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ribeiro, L.F.; Barbosa, M.M.C.; Pinto, F.R.; Lavezzo, L.F.; Rossi, G.A.M.; Almeida, H.M.S.; Amaral, L.A. Diarrheagenic E. coli in raw milk, water, and cattle feces in non-technified dairy farms. Braz. Anim. Sci. 2019, 20, 1–9. [Google Scholar] [CrossRef]
- Disassa, N.; Sibhat, B.; Mengistu, S.; Muktar, Y.; Belina, D. Prevalence and antimicrobial susceptibility pattern of E. coli O157:H7 isolated from traditionally marketed raw cow milk in and around Asosa Town, Western Ethiopia. Vet. Med. Int. 2017, 2017, 7581531. [Google Scholar] [CrossRef] [PubMed]
- Yohannes, G. Isolation, identification and antimicrobial susceptibility testing of Escherichia coli isolated from selected dairy farms in and around mekelle, Ethiopia. J. Dairy Vet. Anim. Res. 2018, 7, 287–291. [Google Scholar] [CrossRef]
- Caine, L.-A.; Nwodo, U.U.; Okoh, A.I.; Ndip, R.N.; Green, E. Occurrence of virulence genes associated with diarrheagenic Escherichia coli isolated from raw cow’s milk from two commercial dairy farms in the Eastern Cape Province, South Africa. Int. J. Environ. Res. Public Health 2014, 11, 11950–11963. [Google Scholar] [CrossRef] [PubMed]
- Bali, O.S.; Lajnef, R.; Felfoul, I.; Attia, H.; Ayadi, M.A. Detection of E. coli in unpasteurized raw milk. Int. J. Agric. Food Sci. 2013, 2013, 53–55. [Google Scholar]
- Chye, F.Y.; Abdullah, A.; Ayob, M.K. Bacteriological quality and safety of raw milk in Malaysia. Food Microbiol. 2004, 21, 535–541. [Google Scholar] [CrossRef]
- Salman, A.M.A.; Hamad, I.M. Enumeration and identification of coliform bacteria from raw milk in Khartoum State, Sudan. J. Cell Anim. Biol. 2011, 5, 121–128. [Google Scholar]
- Fadaei, A. Bacteriological quality of raw cow milk in Shahrekord, Iran. Vet. World 2014, 7, 240–243. [Google Scholar] [CrossRef]
- Beauvais, W.; Gart, E.V.; Bean, M.; Blanco, A.; Wilsey, J.; McWhinney, K.; Bryan, L.K.; Krath, M.; Yang, C.-Y.; Alvarez, D.M.; et al. The prevalence of Escherichia coli O157:H7 fecal shedding in feedlot pens is affected by the water-to-cattle ratio: A randomized controlled trial. PLoS ONE 2018, 13, e0192149. [Google Scholar] [CrossRef]
- Adzitey, F.; Sumaila, N.; Saba, C.K.S. Isolation of E. coli from drinking water sources for humans and farm animals in Nyankpala Community of Ghana. Res. J. Microbiol. 2015, 10, 126–131. [Google Scholar] [CrossRef]
- Akansale, R.; Adzitey, F.; Teye, G.A. Knowledge of farmers in antibiotic usage and investigation of antibiotic residues in meats in Sunyani Municipality, Ghana. J. Food Saf. Hyg. 2019, 5, 155–164. [Google Scholar] [CrossRef]
- Ekli, R.; Adzitey, F.; Agbolosu, A.A. Farmers’ knowledge in antibiotic usage, antibiotic residues, and susceptibility of Salmonella enterica in beef samples from the Wa Municipality, Ghana. Bull. Anim. Health Prod. Afr. 2020, 68, 89–101. [Google Scholar]
- Tuem, K.B.; Gebre, A.K.; Atey, T.M.; Bitew, H.; Yimer, E.M.; Berhe, D.F. Drug resistance patterns of Escherichia coli in Ethiopia: A meta-analysis. BioMed Res. Int. 2018, 2018, 4536905. [Google Scholar] [CrossRef] [PubMed]
- Saba, C.K.S.; Yankey, E.; Adzitey, F. Prevalence of E. coli and shiga toxin producing E. coli in cattle faeces and raw cow milk sold in the Tamale Metropolis, Ghana. J. Dairy Vet. Anim. Res. 2015, 2, 191–193. [Google Scholar]
- Adzitey, F.; Saba, C.K.S.; Teye, G.A. Antibiotic susceptibility of E. coli isolated from milk and hands of milkers in Nyankpala community of Ghana. Curr. Res. Dairy Sci. 2016, 8, 6–11. [Google Scholar] [CrossRef]
- Adzitey, F.; Amposah, C.; Teye, G. Prevalence and antimicrobial resistance patterns of E. coli isolates from cow milk, milk products and handlers in the tamale metropolis of Ghana. Niger. Vet. J. 2019, 39, 338–345. [Google Scholar] [CrossRef][Green Version]
- Atnafie, B.; Paulos, D.; Abera, M.; Tefera, G.; Hailu, D.; Kasaye, S.; Amenu, K. Occurrence of Escherichia coli O157:H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa, Ethiopia. BMC Microbiol. 2017, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.A.; Katz, D.E. Escherichia coli, cattle and the propagation of disease. FEMS Microbiol. Lett. 2017, 364, fnx050. [Google Scholar] [CrossRef]
- Adzitey, F. Antibiotic classes and antibiotic susceptibility of bacterial isolates from selected poultry: A mini review. Worlds Vet. J. 2015, 5, 36–41. [Google Scholar] [CrossRef]
- Uddin, A.; Motazzim-Ul-Haque, H.M.; Noor, R. Isolation and identification of pathogenic Escherichia coli, Klebsiella spp. and Staphylococcus spp. in raw milk samples collected from different areas of Dhaka City, Bangladesh. Stamford J. Microbiol. 1970, 1, 19–23. [Google Scholar] [CrossRef]
- Hossain, M.F.; Rahman, T.; Kabir, S.L. Microbial assessment of milk collected from different markets of Mymensingh, Gazipur and Sherpur districts of Bangladesh and determination of antimicrobial resistance patterns of the isolated bacteria. Asian-Australas. J. Food Saf. Secur. 2017, 1, 7–16. [Google Scholar] [CrossRef]
- Lorian, V. Antibiotics in Laboratory Medicine, 5th ed.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2005; ISBN 0781749832. [Google Scholar]
- Adzitey, F.; Rusul, G.; Huda, N. Prevalence and antibiotic resistance of Salmonella serovars in ducks, duck rearing and processing environments in Penang, Malaysia. Food Res. Int. 2012, 45, 947–952. [Google Scholar] [CrossRef]
- Davis, R.; Brown, P.D. Multiple antibiotic resistance index, fitness and virulence potential in respiratory Pseudomonas aeruginosa from Jamaica. J. Med. Microbiol. 2016, 65, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Adzitey, F.; Asiamah, P.; Boateng, E. Prevalence and antibiotic susceptibility of Salmonella enterica isolated from cow milk, milk products and hands of sellers in the Tamale Metropolis of Ghana. J. Appl. Sci. Environ. Manag. 2020, 24, 59–64. [Google Scholar] [CrossRef]
- Maheux, A.F.; Picard, F.J.; Boissinot, M.; Bissonnette, L.; Paradis, S.; Bergeron, M.G. Analytical comparison of nine PCR primer sets designed to detect the presence of Escherichia coli/Shigella in water samples. Water Res. 2009, 43, 3019–3028. [Google Scholar] [CrossRef]
- Liu, H.; Geagea, H.; Rousseau, G.M.; Labrie, S.J.; Tremblay, D.M.; Liu, X.; Moineau, S. Characterization of the Escherichia coli Virulent Myophage ST32. Viruses 2018, 10, 616. [Google Scholar] [CrossRef]
- Vejborg, R.M.; Hancock, V.; Schembri, M.A.; Klemm, P. Comparative genomics of Escherichia coli strains causing urinary tract Infections. Appl. Environ. Microbiol. 2011, 77, 3268–3278. [Google Scholar] [CrossRef]
- Mare, A.D.; Ciurea, C.N.; Man, A.; Tudor, B.; Moldovan, V.; Decean, L.; Toma, F. Enteropathogenic Escherichia coli—A summary of the literature. Gastroenterol. Insights 2021, 12, 4. [Google Scholar] [CrossRef]
- Godambe, L.P.; Bandekar, J.; Shashidhar, R. Species specific PCR based detection of Escherichia coli from Indian foods. 3 Biotech 2017, 7, 130. [Google Scholar] [CrossRef]
- Zhang, J.-J. A control program of Bovine viral diarrhoea virus (BVDV)-infection on large dairy farms in Beijing, China in 2009 and 2010. Afr. J. Microbiol. Res. 2012, 6, 3821–3823. [Google Scholar] [CrossRef]
| Sample Type | Number of Samples | Number of Positives | % Occurrence |
|---|---|---|---|
| Milk | 50 | 34 | 68.0 |
| Feces | 50 | 33 | 66.0 |
| Utensils | 50 | 21 | 42.0 |
| Teat | 50 | 7 | 14.0 |
| Hands | 50 | 6 | 12.0 |
| Overall | 250 | 101 | 40.4 |
| Antibiotics | % Resistance | % Intermediate Resistance | % Susceptibility |
|---|---|---|---|
| Amoxycillin (A) 30 µg | 50.0 | 47.6 | 2.4 |
| Azithromycin (Azm) 15 µg | 0.0 | 7.1 | 92.9 |
| Ceftriaxone (Cro) 30 µg | 9.5 | 31.0 | 59.5 |
| Chloramphenicol (C) 30 µg | 2.4 | 2.4 | 95.2 |
| Ciprofloxacin (Cip) 5 µg | 19.0 | 26.2 | 54.8 |
| Gentamicin (Gm) 10 µg | 2.4 | 14.3 | 83.3 |
| Imipenem (Imi) 10 µg | 9.5 | 16.7 | 73.8 |
| Teicoplanin (Tec) 30 µg | 100.0 | 0.0 | 0.0 |
| Tetracycline (Te) 30 µg | 35.7 | 2.4 | 61.9 |
| Sulphamethoxazole/trimethoprim (Sxt) 22 µg | 26.2 | 2.4 | 71.4 |
| Overall | 25.5 | 15.0 | 59.5 |
| Isolate Code | Sources | No. of Antibiotics | Antibiotics Resistance | MAR Index |
|---|---|---|---|---|
| FS17 | Fecal | 2 | Tec-Te | 0.2 |
| FS22 | Fecal | 5 | A-Tec-T-Cro-Sxt | 0.5 |
| FS25 | Fecal | 1 | Tec | 0.1 |
| FS34 | Fecal | 4 | Cip-Tec-Te-Sxt | 0.4 |
| FS48 | Fecal | 4 | Cip-Tec-Te-Sxt | 0.4 |
| FS5 | Fecal | 2 | Tec-Imi | 0.2 |
| FS50 | Fecal | 1 | Tec | 0.1 |
| FS6 | Fecal | 1 | Tec | 0.1 |
| FS8 | Fecal | 1 | Tec | 0.1 |
| HS1 | Hand | 2 | A-Tec | 0.2 |
| HS11 | Hand | 4 | Cip-A-Tec-Te | 0.4 |
| HS12 | Hand | 3 | A-Tec-Te | 0.3 |
| HS18 | Hand | 1 | Tec | 0.1 |
| HS3 | Hand | 3 | A-Tec-Te | 0.3 |
| HS9 | Hand | 1 | Tec | 0.1 |
| M15 | Milk | 1 | Tec | 0.1 |
| M17 | Milk | 1 | Tec | 0.1 |
| M2 | Milk | 1 | Tec | 0.1 |
| M25 | Milk | 2 | A-Tec | 0.2 |
| M39 | Milk | 4 | A-Tec-Cro-Imi | 0.4 |
| M45 | Milk | 2 | A-Tec | 0.2 |
| M50 | Milk | 4 | A-Tec-Te-Sxt | 0.4 |
| M51 | Milk | 1 | Tec | 0.1 |
| M6 | Milk | 5 | A-Tec-C-Cro-Sxt | 0.5 |
| M9 | Milk | 5 | A-Tec-Te-Gm-Sxt | 0.5 |
| TS1 | Teat | 1 | Tec | 0.1 |
| TS10 | Teat | 4 | A-Tec-Te-Imi | 0.4 |
| TS20 | Teat | 2 | A-Tec | 0.2 |
| TS26 | Teat | 5 | Cip-A-Tec-Te-Sxt | 0.5 |
| TS27 | Teat | 1 | Tec | 0.1 |
| TS36 | Teat | 2 | Cip-Tec | 0.2 |
| TS45 | Teat | 3 | Tec-Cro-Sxt | 0.3 |
| TS9 | Teat | 3 | A-Tec-Imi | 0.3 |
| US18 | Utensils | 2 | Tec-Te | 0.2 |
| US2 | Utensils | 2 | A-Tec | 0.2 |
| US24 | Utensils | 5 | Cip-A-Tec-Te-Sxt | 0.5 |
| US3 | Utensils | 5 | Cip-A-Tec-Te-Sxt | 0.5 |
| US30 | Utensils | 5 | Cip-A-Tec-Te-Sxt | 0.5 |
| US31 | Utensils | 2 | A-Tec | 0.2 |
| US34 | Utensils | 2 | A-Tec | 0.2 |
| US49 | Utensils | 1 | Tec | 0.1 |
| US5 | Utensils | 1 | Tec | 0.1 |
| Haplotypes (Isolates) | E. coli Strain Identified | Gene Bank Reference | Country | Percentage Identity (%) |
|---|---|---|---|---|
| Hap 01 (Us2, Us30, Us34, Hs1, Hs18, Fs6, M15) | STEC2017-197 RHB07-C16 ECS C054 O100:H21 strain Res 13-lact | CP075663.1 CP055973.1 AP024112.1 CP062889.1 | Switzerland USA Japan Canada | 98.97 100 100 98.97 |
| Hap 02 (Us18) | KCJ3K291 L3Cip3 | CP054407.1 CP062211.1 | USA New Zealand | 98.95 98.95 |
| Hap 03 (M23) | STW0522-31 H20 MING6 | AP022409.1 CP069677.1 | Japan Poland | 98.92 98.92 |
| Hap 04 (Fs5) | O176:H45 strain MIN9 chromosome | CP069682.1 | Poland | 97.92 |
| Hap 05 (Fs25) | 19-5 chromosome V14 beta-D-glucuronidase gene | CP047010.1 MW353604.1 | China India | 98.91 98.91 |
| Hap 06 (M22) | EH10-18-47 0126:H45 MING 10 | CP063499.1 CP069677.1 | Laos Poland | 100 100 |
| Hap 07 (M17, M45, M46) | STEC2018-553 WS0115A 65ECOLEC | CP075665.5 CP035882.1 CP070914.1 | Switzerland Egypt Singapore | 100 100 100 |
| Hap 08 (Ts26, Fs50, M51) | STEC- 183 chromosome 039:H21 strain Res13-lact-PEB08-01tcmA_3 | CP0756971.1 CP062865.1 CP059835.1 | Switzerland Canada China | 98.95 100 98.2 |
| Hap 09 (Ts 20) | SH9PTE6 EF7-18-51 | CP073768.1 CP063487.1 | China Laos | 100 100 |
| Hap 10 (Ts1) | EcPF20 CP070920.1 | CP071441.1 CP070920.1 | USA Singapore | 98.25 100 |
| Hap 11 (M36, Ts 36) | TW10722 RH-048-MS 179 chromosomes | CP035841.1 CP050206.1 CP062924.1 | Guinea Bissau Bangladesh Turkey | 96.84 95.1 98.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adzitey, F.; Yussif, S.; Ayamga, R.; Zuberu, S.; Addy, F.; Adu-Bonsu, G.; Huda, N.; Kobun, R. Antimicrobial Susceptibility and Molecular Characterization of Escherichia coli Recovered from Milk and Related Samples. Microorganisms 2022, 10, 1335. https://doi.org/10.3390/microorganisms10071335
Adzitey F, Yussif S, Ayamga R, Zuberu S, Addy F, Adu-Bonsu G, Huda N, Kobun R. Antimicrobial Susceptibility and Molecular Characterization of Escherichia coli Recovered from Milk and Related Samples. Microorganisms. 2022; 10(7):1335. https://doi.org/10.3390/microorganisms10071335
Chicago/Turabian StyleAdzitey, Frederick, Saniyatu Yussif, Roland Ayamga, Sumaila Zuberu, Francis Addy, Gideon Adu-Bonsu, Nurul Huda, and Rovina Kobun. 2022. "Antimicrobial Susceptibility and Molecular Characterization of Escherichia coli Recovered from Milk and Related Samples" Microorganisms 10, no. 7: 1335. https://doi.org/10.3390/microorganisms10071335
APA StyleAdzitey, F., Yussif, S., Ayamga, R., Zuberu, S., Addy, F., Adu-Bonsu, G., Huda, N., & Kobun, R. (2022). Antimicrobial Susceptibility and Molecular Characterization of Escherichia coli Recovered from Milk and Related Samples. Microorganisms, 10(7), 1335. https://doi.org/10.3390/microorganisms10071335








