Effect of Biltong Dried Beef Processing on the Reduction of Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus, and the Contribution of the Major Marinade Components
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions, and Antibiotic Resistance
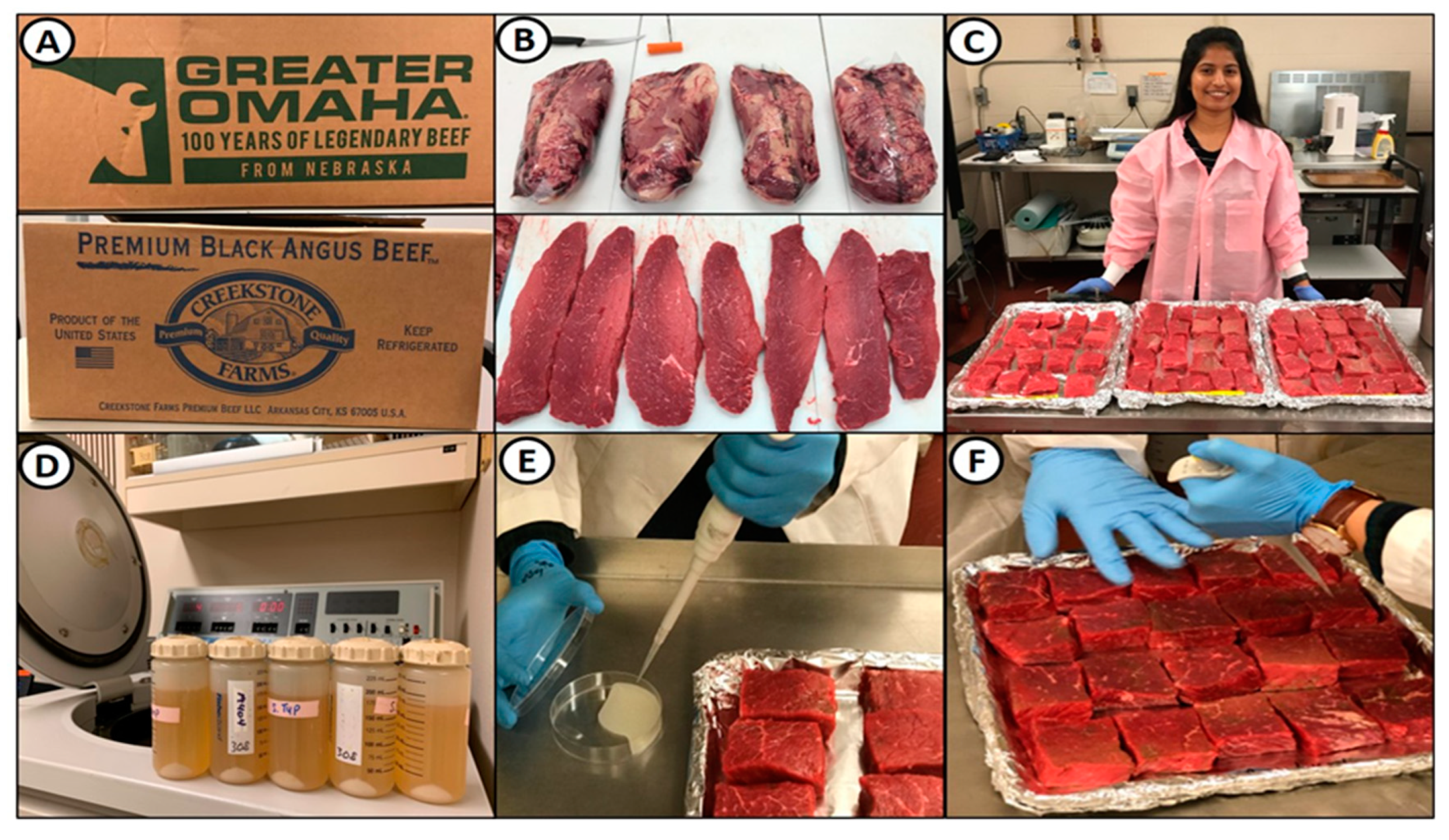
2.2. Beef Handling, Fabrication, and Inoculation
2.3. Marination of Inoculated Beef Pieces
2.4. Drying of Marinaded Beef Pieces
2.5. Beef Sampling, Microbial Enumeration, Water Activity Testing, and Enterotoxin Detection
2.6. Statistical Analysis
3. Results and Discussion
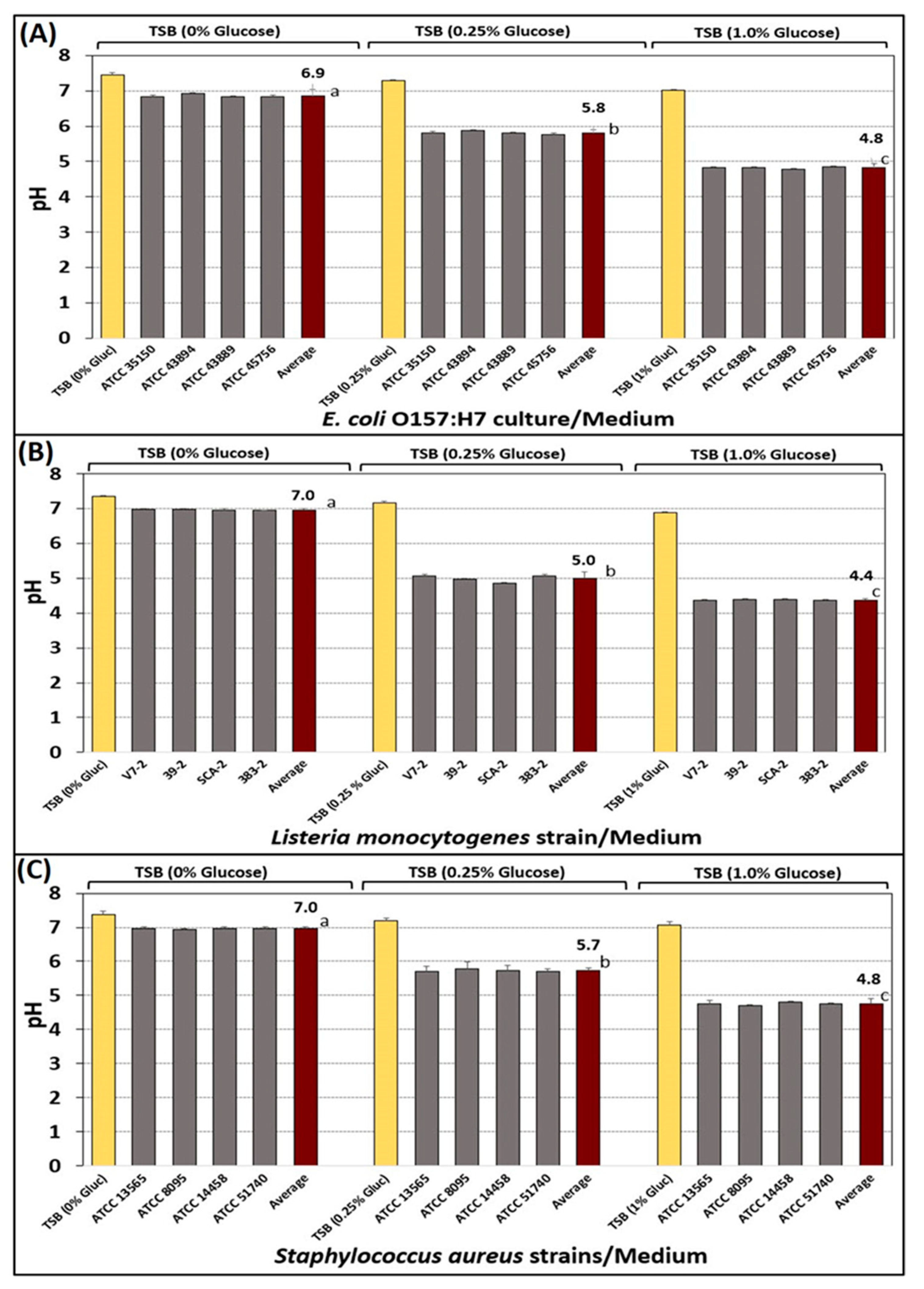
3.1. Acid Adaptation of Bacterial Cultures
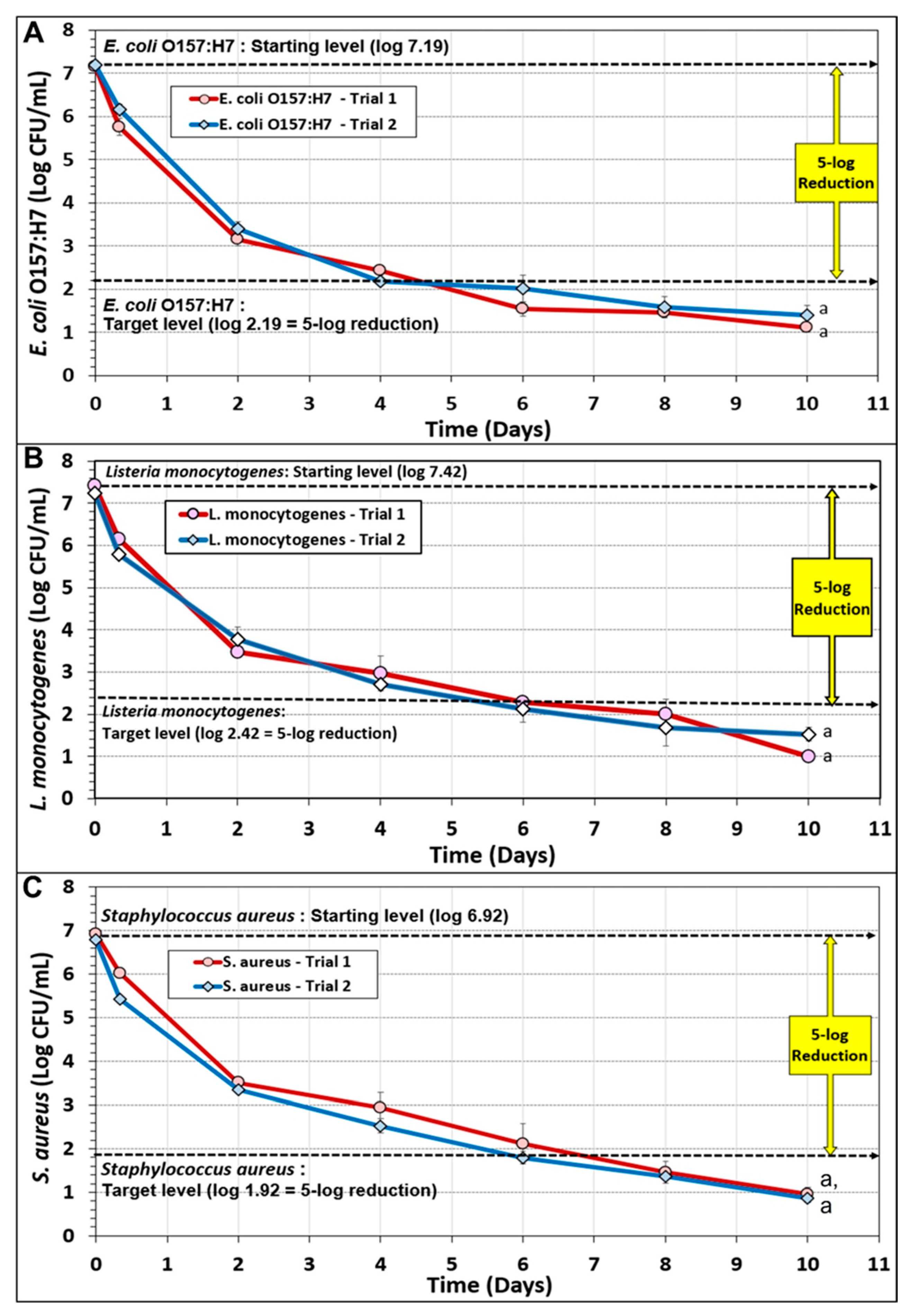
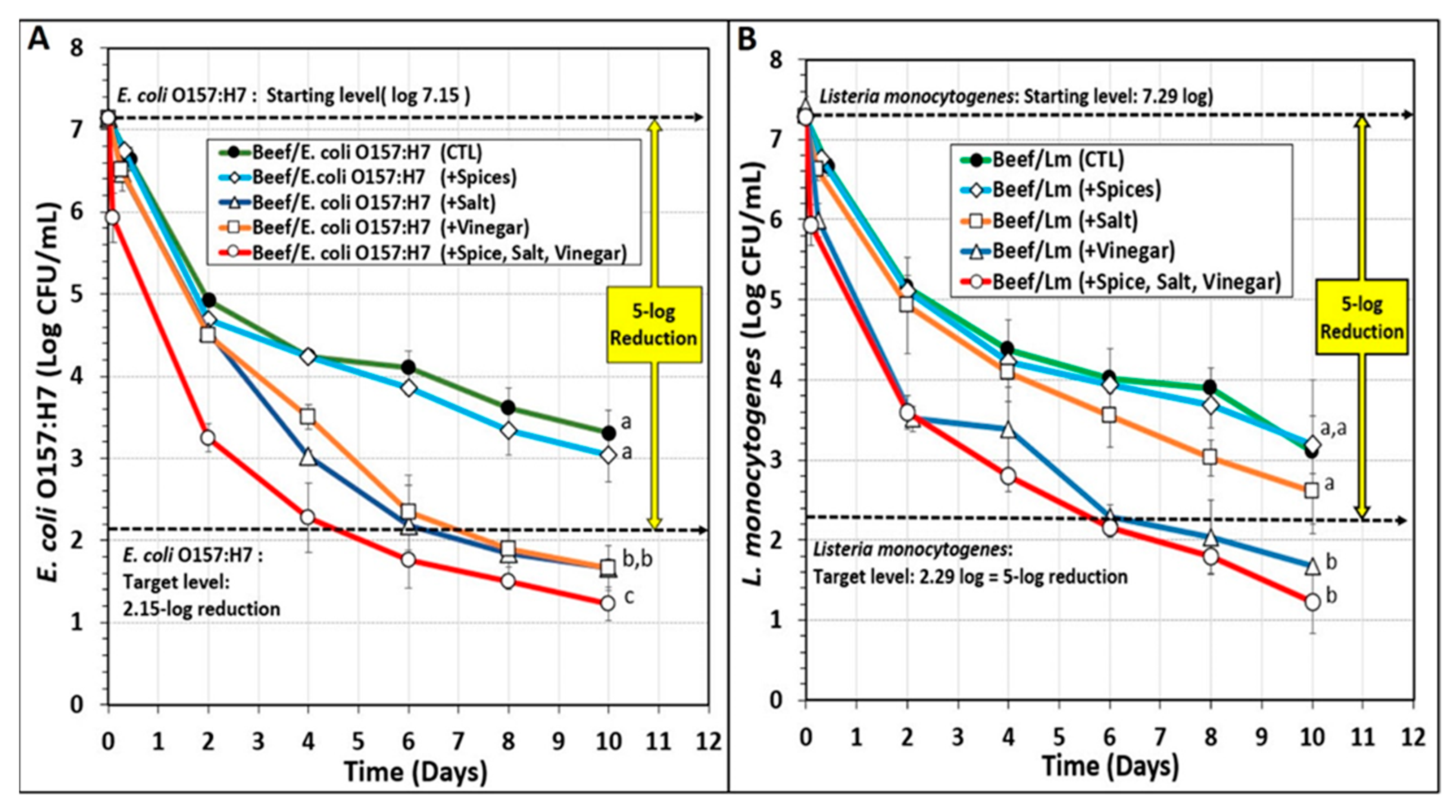
3.2. Biltong Process Reduction and Contribution of the Individual Components of the Marinade
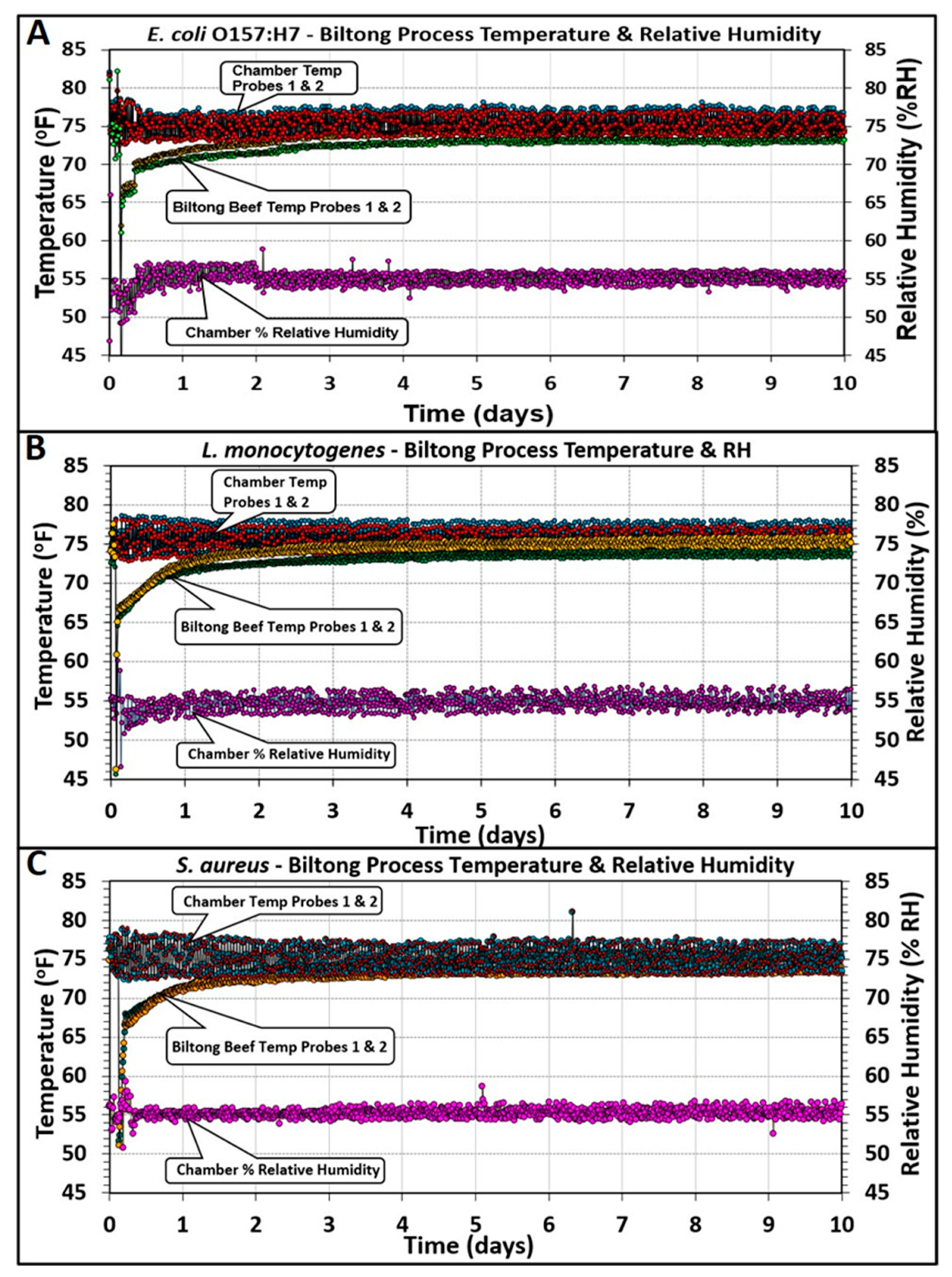
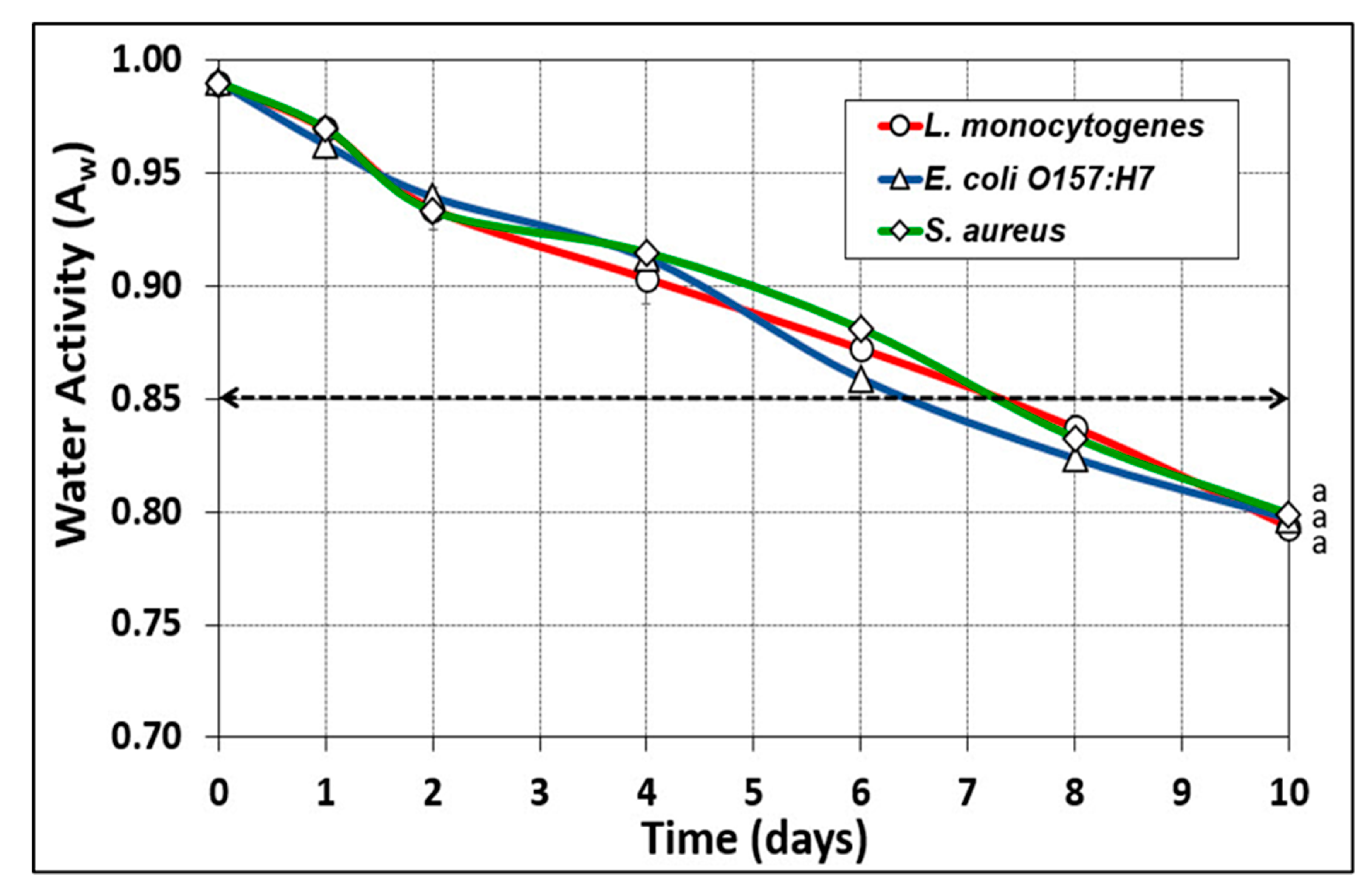
3.3. Biltong Process Temperature, Relative Humidity, and Water Activity (Aw) Measurements
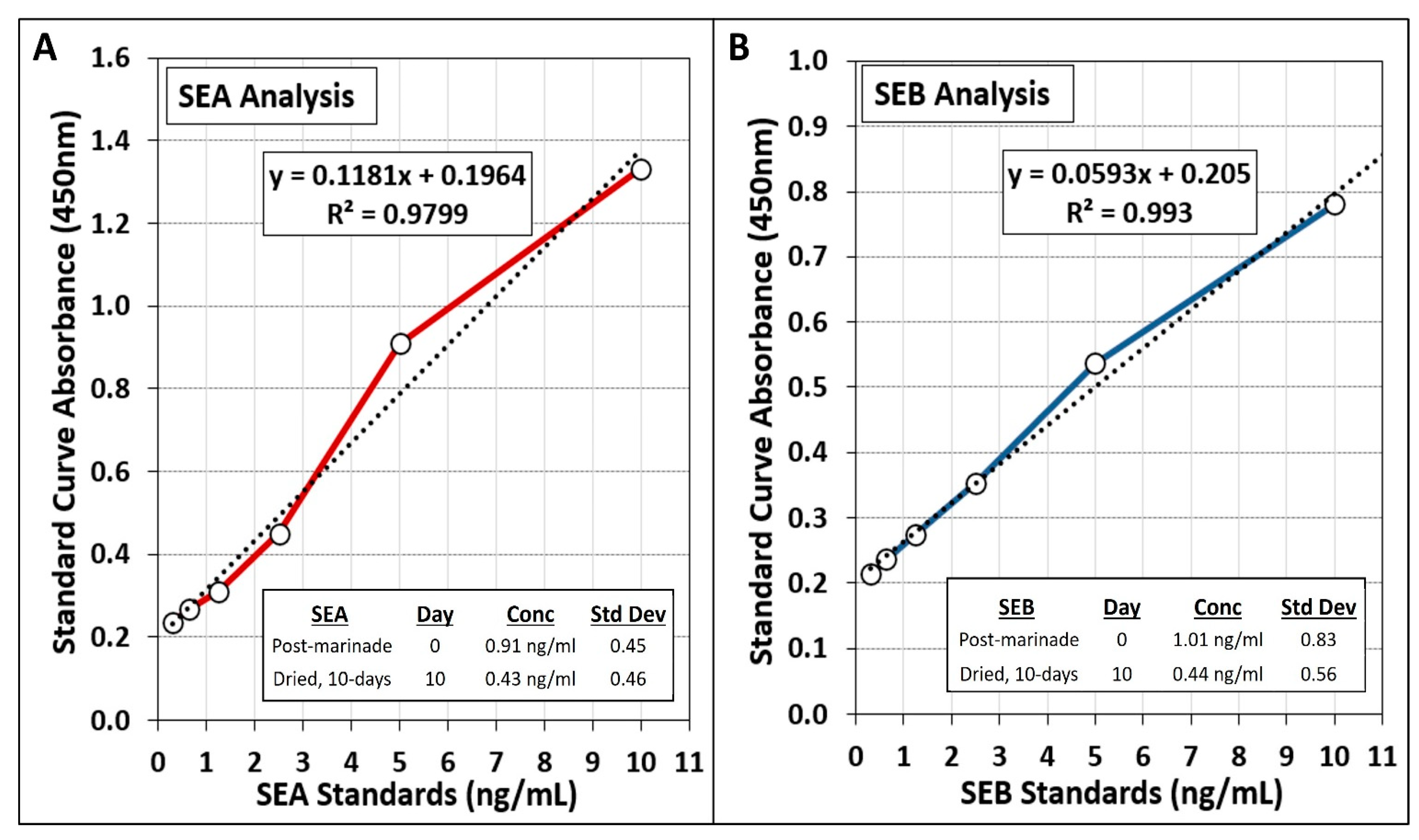
3.4. Analysis of Staphylococcal Enterotoxin A (SEA) and B (SEB) during Biltong Processing with S. aureus-Inoculated Beef
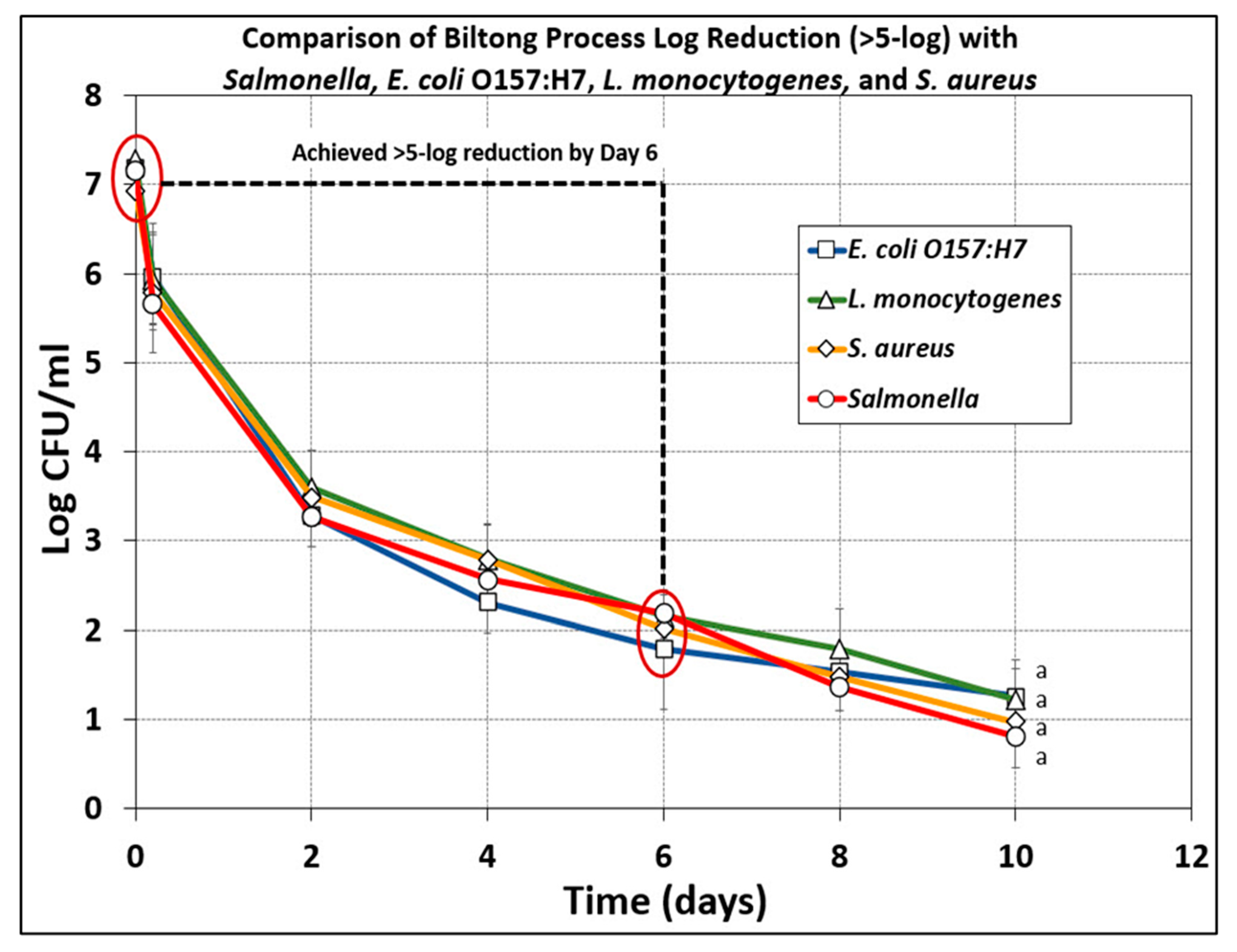
3.5. Comparison of Microbial Reduction of E. coli O157:H7, L. monocytogenes, S. aureus, and Salmonella serovars during Biltong Processing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wentworth, E.N. Dried meat: Early man’s travel ration. Agric. Hist. 1956, 30, 2–10. [Google Scholar]
- Bermúdez-Aguirre, D.; Tapia, M.S.; Welti-Chanes, J. Specialty Foods; DEStech Publications, Inc.: Lancaster, PA, USA, 2008; pp. 447–461. [Google Scholar]
- Challa, H.J.; Bandlamudi, M.; Uppaluri, K.R. Paleolithic Diet. In StatPearls; StatPearls Publishing, LLC.: Treasure Island, FL, USA, 2021. [Google Scholar]
- Kussaga, J.B.; Jacxsens, L.; Tiisekwa, B.P.; Luning, P.A. Food safety management systems performance in African food processing companies: A review of deficiencies and possible improvement strategies. J. Sci. Food Agric. 2014, 94, 2154–2169. [Google Scholar] [CrossRef]
- Burfoot, D.; Everis, L.; Mulvey, L.; Wood, A.; Betts, R. Literature Review on Microbiological Hazards Associated with Biltong and Similar Dried Meats. Available online: https://www.food.gov.uk/sites/default/files/media/document/574-1-1007_B13015_Final_Report.pdf (accessed on 13 March 2022).
- BC-CDC. South African RTE dehydrated meats (biltong, chili bites, droëwors). In Food Issue, Notes from the Field; British Columbia Centre for Disease Control: Vancouver, BC, Canada, 2020; pp. 1–7. [Google Scholar]
- USDA-FSIS. FSIS Compliance Guideline for Meat and Poultry Jerky Produced by Small and very Small Establishments; U.S. Food Safety and Inspection Service: Washington, DC, USA, 2014; pp. 1–54.
- USDA-FSIS. Salmonella Compliance Guidelines for Small and very Small Meat and Poultry Establishments That Produce Ready-To-Eat (RTE) Products and Revised Appendix A, June 2017; U.S. Food Safety and Inspection Service: Washington, DC, USA, 2017; pp. 1–37.
- Karolenko, C.; Muriana, P. Quantification of process lethality (5-log reduction) of Salmonella and salt concentration during sodium replacement in biltong marinade. Foods 2020, 9, 1570. [Google Scholar] [CrossRef] [PubMed]
- Karolenko, C.E.; Bhusal, A.; Nelson, J.L.; Muriana, P.M. Processing of biltong (dried beef) to achieve USDA-FSIS 5-log reduction of Salmonella without a heat lethality step. Microorganisms 2020, 8, 791. [Google Scholar] [CrossRef] [PubMed]
- Nickelson, R.; Luchansky, J.B.; Kaspar, C.; Johnson, E. Dry Fermented Sausage and Escherichia coli O157:H7 Validation Research. An Executive Summary Prepared by The Blue Ribbon Task Force of the National Cattlemen’s Beef Association; Research Report No. 11-316.; NCBA: Chicago, IL, USA, 1996. [Google Scholar]
- Matsheka, M.I.; Mpuchane, S.; Gashe, B.A.; Allotey, J.; Khonga, E.B.; Coetzee, S.H.; Murindamombe, G. Microbial quality assessment and predominant microorganism of biltong produced in butcheries in Gaborone, Botswana. Food Nutr. Sci. 2014, 5, 11. [Google Scholar] [CrossRef][Green Version]
- Naidoo, K.; Lindsay, D. Pathogens associated with biltong product and their in vitro survival of hurdles used during production. Food Prot. Trends 2010, 30, 532–538. [Google Scholar]
- USDA-FSIS. Food Science Research Priorities; Data Gaps. Available online: https://www.fsis.usda.gov/science-data/research-priorities (accessed on 5 March 2022).
- Laufer, A.S.; Grass, J.; Holt, K.; Whichard, J.M.; Griffin, P.M.; Gould, L.H. Outbreaks of Salmonella infections attributed to beef --United States, 1973-2011. Epidemiol. Infect. 2015, 143, 2003–2013. [Google Scholar] [CrossRef]
- CDC. Outbreak of Salmonellosis associated with beef jerky--New Mexico, 1995. MMWR Morb. Mortal. Wkly. Rep. 1995, 44, 785–788. [Google Scholar]
- Eidson, M.; Swell, C.M.; Graves, G.; Olson, R. Beef jerky gastroenteritis outbreaks. J. Environ. Health 2000, 62, 9. [Google Scholar]
- Omer, M.K.; Álvarez-Ordoñez, A.; Prieto, M.; Skjerve, E.; Asehun, T.; Alvseike, O.A. A systematic review of bacterial foodborne outbreaks related to red meat and meat products. Foodborne Pathog. Dis. 2018, 15, 598–611. [Google Scholar] [CrossRef]
- Mhlambi, S.G.; Naidoo, K.; Lindsay, D. Enterotoxin-producing Staphylococcus strains associated with South African biltong at point of sale. J. Food Safety 2010, 30, 307–317. [Google Scholar] [CrossRef]
- Naidoo, K.; Lindsay, D. Survival of Listeria monocytogenes, and enterotoxin-producing Staphylococcus aureus and Staphlyococcus pasteruri, during two types of biltong-manufacturing process. Food Cont. 2010, 21, 1042–1050. [Google Scholar] [CrossRef]
- Muriana, P.M.; Quimby, W.; Davidson, C.A.; Grooms, J. Postpackage pasteurization of ready-to-eat deli meats by submersion heating for reduction of Listeria monocytogenes. J. Food Prot. 2002, 65, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Muriana, P.M.; Eager, J.; Wellings, B.; Morgan, B.; Nelson, J.; Kushwaha, K. Evaluation of antimicrobial interventions against E. coli O157:H7 on the surface of raw beef to reduce bacterial translocation during blade tenderization. Foods 2019, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Berry, E.D.; Cutter, C.N. Effects of acid adaptation of Escherichia coli O157:H7 on efficacy of acetic acid spray washes to decontaminate beef carcass tissue. Appl. Environ. Microbiol. 2000, 66, 1493–1498. [Google Scholar] [CrossRef]
- Panneerseelan, L.; Muriana, P.M. An immunomagnetic PCR signal amplification assay for sensitive detection of Staphylococcus aureus enterotoxins in foods. J. Food Prot. 2009, 72, 2538–2546. [Google Scholar] [CrossRef]
- Mehrotra, M.; Wang, G.; Johnson, W.M. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 2000, 38, 1032–1035. [Google Scholar] [CrossRef]
- Wilde, S.; Jørgensen, F.; Campbell, A.; Rowbury, R.; Humphrey, T. Growth of Salmonella enterica Serovar Enteritidis PT4 in media containing glucose results in enhanced RpoS-independent heat and acid tolerance but does not affect the ability to survive air-drying on surfaces. Food Microbiol 2000, 17, 679–686. [Google Scholar] [CrossRef]
- Karolenko, C.E.; Bhusal, A.; Gautam, D.; Muriana, P.M. Selenite cystine agar for enumeration of inoculated Salmonella serovars recovered from stressful conditions during antimicrobial validation studies. Microorganisms 2020, 8, 338. [Google Scholar] [CrossRef]
- NACMCF. Parameters for determining inoculated pack/challenge study protocols (National Advisory Committee on the Microbiological Criteria for Foods). J. Food Prot. 2010, 73, 140–202. [Google Scholar] [CrossRef]
- USDA-FSIS. FSIS Compliance Guideline HACCP Systems Validation (April 2015); USDA-FSIS, Ed.; USDA: Washington, DC, USA, 2015; pp. 1–68.
- Álvarez-Ordóñez, A.; Prieto, M.; Bernardo, A.; Hill, C.; López, M. The acid tolerance response of Salmonella spp.: An adaptive strategy to survive in stressful environments prevailing in foods and the host. Food Res. Intl. 2012, 45, 482–492. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Fernández, A.; Bernardo, A.; López, M. A comparative study of thermal and acid inactivation kinetics in fruit juices of Salmonella enterica serovar Typhimurium and Salmonella enterica serovar Senftenberg grown at acidic conditions. Foodborne Pathog. Dis. 2009, 6, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Samelis, J.; Sofos, J.N.; Kendall, P.A.; Smith, G.C. Effect of acid adaptation on survival of Escherichia coli O157:H7 in meat decontamination washing fluids and potential effects of organic acid interventions on the microbial ecology of the meat plant environment. J. Food Prot. 2002, 65, 33–40. [Google Scholar] [CrossRef]
- Calicioglu, M.; Sofos, J.N.; Samelis, J.; Kendall, P.A.; Smith, G.C. Inactivation of acid-adapted and nonadapted Escherichia coli O157:H7 during drying and storage of beef jerky treated with different marinades. J. Food Prot. 2002, 65, 1394–1405. [Google Scholar] [CrossRef]
- Calicioglu, M.; Sofos, J.N.; Samelis, J.; Kendall, P.A.; Smith, G.C. Effect of acid adaptation on inactivation of Salmonella during drying and storage of beef jerky treated with marinades. Int. J. Food Microbiol. 2003, 89, 51–65. [Google Scholar] [CrossRef]
- Jones, M.; Arnaud, E.; Gouws, P.; Hoffman, L.C. Effects of the addition of vinegar, weight loss and packaging method on the physicochemical properties and microbiological profile of biltong. Meat Sci. 2019, 156, 214–221. [Google Scholar] [CrossRef]
- Burnham, G.M.; Hanson, D.J.; Koshick, C.M.; Ingham, S.C. Death of Salmonella serovars, Escherichia coli O157: H7, Staphylococcus aureus and Listeria monocytogenes during the drying of meat: A case study using Biltong and Droewors. J. Food Safety 2008, 28, 198–209. [Google Scholar] [CrossRef]
- Petit, T.; Caro, Y.; Petit, A.S.; Santchurn, S.J.; Collignan, A. Physicochemical and microbiological characteristics of biltong, a traditional salted dried meat of South Africa. Meat Sci. 2014, 96, 1313–1317. [Google Scholar] [CrossRef]

| Organism | Strain Designation | Culture Collection Designation | Source |
|---|---|---|---|
| L. monocytogenes | ATCC 49594 | PMM 264 | ScottA-2; Clinical isolate |
| L. monocytogenes | V7-2 | PMM 266 | Clinical isolate |
| L. monocytogenes | 39-2 | PMM 39 | Retail hotdogs |
| L. monocytogenes | 383-2 | PMM 383 | Retail ground beef |
| E. coli O157:H7 | ATCC 35150 | PMM 407 | Human feces |
| E. coli O157:H7 | ATCC 43889 | PMM 1111 | Human feces |
| E. coli O157:H7 | ATCC 43894 | PMM 405 | Human feces |
| E. coli O157:H7 | ATCC 45756 | PMM 715 | JB Luchansky, USDA-ARS |
| S. aureus | ATCC 8095 | PMM 323 | Cream pie |
| S. aureus | ATCC 13565 | PMM 318 | Ham, enterotoxin illness |
| S. aureus | ATCC 14458 | PMM 319 | Human feces, diarrhea |
| S. aureus | ATCC 51740 | PMM 678 | Margarine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gavai, K.; Karolenko, C.; Muriana, P.M. Effect of Biltong Dried Beef Processing on the Reduction of Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus, and the Contribution of the Major Marinade Components. Microorganisms 2022, 10, 1308. https://doi.org/10.3390/microorganisms10071308
Gavai K, Karolenko C, Muriana PM. Effect of Biltong Dried Beef Processing on the Reduction of Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus, and the Contribution of the Major Marinade Components. Microorganisms. 2022; 10(7):1308. https://doi.org/10.3390/microorganisms10071308
Chicago/Turabian StyleGavai, Kavya, Caitlin Karolenko, and Peter M. Muriana. 2022. "Effect of Biltong Dried Beef Processing on the Reduction of Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus, and the Contribution of the Major Marinade Components" Microorganisms 10, no. 7: 1308. https://doi.org/10.3390/microorganisms10071308
APA StyleGavai, K., Karolenko, C., & Muriana, P. M. (2022). Effect of Biltong Dried Beef Processing on the Reduction of Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus, and the Contribution of the Major Marinade Components. Microorganisms, 10(7), 1308. https://doi.org/10.3390/microorganisms10071308







