Responses of Cyanobacterial Crusts and Microbial Communities to Extreme Environments of the Stratosphere
Abstract
:1. Introduction
2. Materials and Methods
2.1. Natural Cyanobacterial Crusts Collection
2.2. Experiment Design
2.3. Physiological and Biochemical Indicators Measurement
2.4. Microbial Community Analysis
2.5. Statistical Analysis
3. Results
3.1. Physiological and Biochemical Changes of Cyanobacterial Crusts
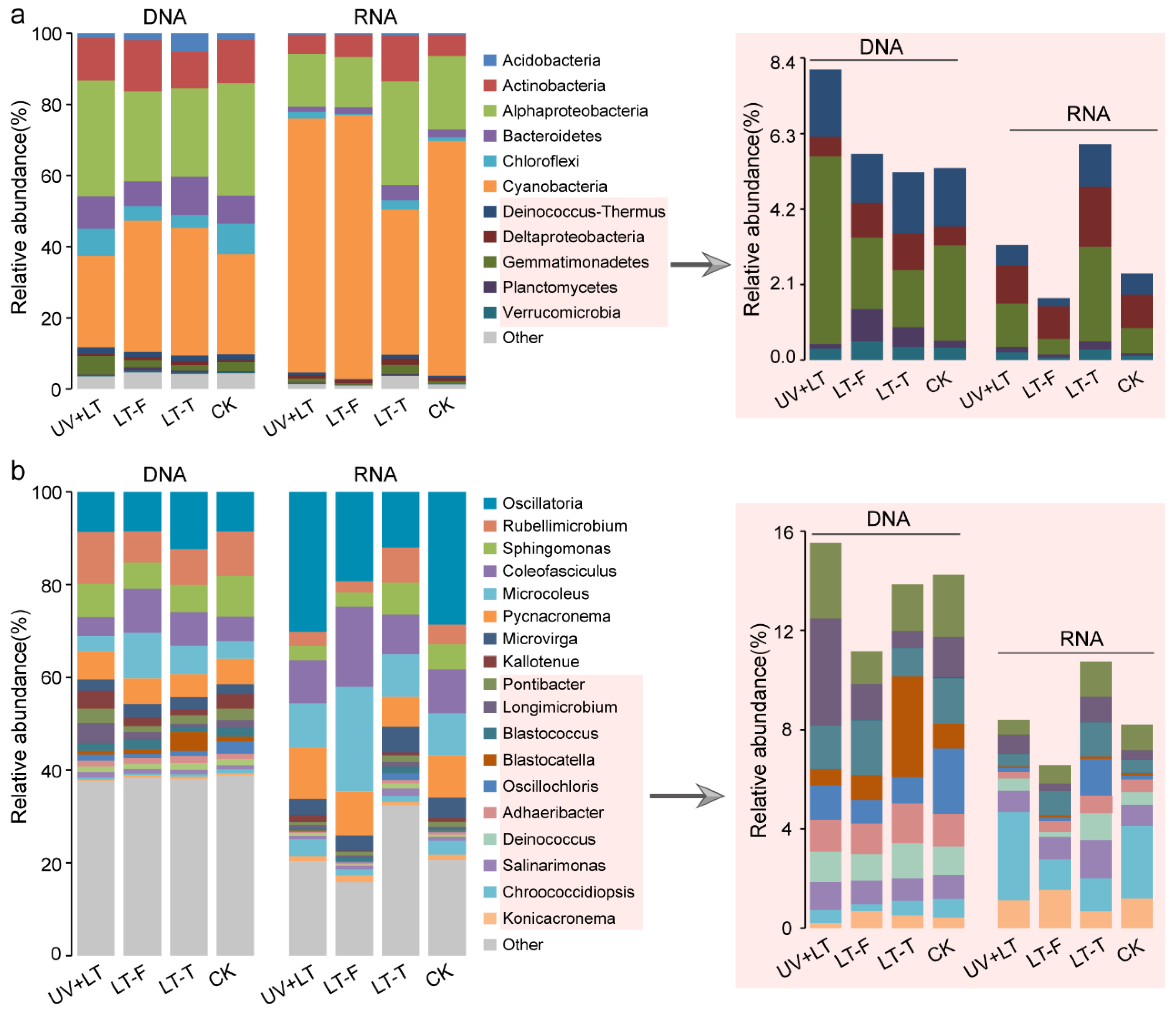
3.2. Changes in Bacteria Community
3.3. Changes in Relative Abundance of Archaea
4. Discussion
4.1. Damage Degree and Tolerance Mechanisms of Cyanobacterial Crusts
4.2. Regulation of Microbial Communities by Stratospheric Environment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DasSarma, P.; DasSarma, S. Survival of microbes in Earth’s stratosphere. Curr. Opin. Microbiol. 2018, 43, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Bryan, N.C.; Christner, B.C.; Guzik, T.G.; Granger, D.J.; Stewart, M.F. Abundance and survival of microbial aerosols in the troposphere and stratosphere. ISME J. 2019, 13, 2789–2799. [Google Scholar] [CrossRef] [PubMed]
- Lin, W. Life in the near space and implications for astrobiology. Chin. Sci. Bull. 2020, 65, 1297–1304. [Google Scholar] [CrossRef]
- D’Oliveira, F.A.; Melo, F.C.L.; Devezas, T.C. High-altitude platforms—present situation and technology trends. J. Aerosp. Technol. Manag. 2016, 8, 249–262. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, W.; Huang, C. The latest development of stratospheric aerocraft technology. Spacecr. Recove. Remot. Sens. 2019, 40, 1–13. [Google Scholar] [CrossRef]
- Wainwright, M.; Wickramasinghe, N.C.; Narlikar, J.V.; Rajaratnam, P. Microorganisms cultured from stratospheric air samples obtained at 41 km. FEMS Microbiol. Lett. 2003, 218, 161–165. [Google Scholar] [CrossRef]
- Griffin, D.W. Terrestrial microorganisms at an altitude of 20,000 m in Earth’s atmosphere. Aerobiologia 2004, 20, 135–140. [Google Scholar] [CrossRef]
- Shivaji, S.; Chaturvedi, P.; Suresh, K.; Reddy, G.S.N.; Dutt, C.B.S.; Wainwright, M.; Narlikar, J.V.; Bhargava, P.M. Bacillus aerius sp. nov., Bacillus aerophilus sp. nov., Bacillus stratosphericus sp. nov. and Bacillus altitudinis sp. nov., isolated from cryogenic tubes used for collecting air samples from high altitudes. Int. J. Syst. Evol. Microbiol. 2006, 56, 1465–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Itoh, T.; Yokobori, S.I.; Shimada, H.; Itahashi, S. Deinococcus aetherius sp. nov., isolated from the stratosphere. Int. J. Syst. Evol. Microbiol. 2010, 60, 776–779. [Google Scholar] [CrossRef] [Green Version]
- DasSarma, P.; Autunes, A.; Simoes, M.F.; DasSarma, S. Earth’s stratosphere and microbial life. Curr. Issues. Mol. Biol. 2020, 38, 197–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.J.; Thakrar, P.J.; Bharrat, A.E.; Dokos, A.; Kinney, T.K.; James, L.M.; Lane, M.; Khodadad, C.; Maguire, F.; Maloney, P.R.; et al. A balloon-based payload for Exposing Microorganisms in the Stratosphere (E-MIST). Gravit. Space Res. 2014, 2, 70–80. [Google Scholar] [CrossRef]
- Khodadad, C.L.; Wong, G.M.; James, L.M.; Thakrar, P.J.; Lane, M.A.; Catechis, J.A.; Smith, D.J. Stratosphere conditions inactivate bacterial endospores from a Mars spacecraft assembly facility. Astrobiology 2017, 17, 337–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DasSarma, P.; Laye, V.J.; Harvey, J.; Reid, C.; Shultz, J.; Yarborough, A.; Lamb, A.; Koske-Phillips, A.; Herbst, A.; Molina, F.; et al. Survival of halophilic archaea in earth’s cold stratosphere. Int. J. Astrobiol. 2017, 16, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Pulschen, A.A.; de Araujo, G.G.; Carvalho, A.C.S.R.; Cerini, M.F.; Fonseca, L.D.; Galante, D.; Rodrigues, F. Survival of extremophilic yeasts in the stratospheric environment during balloon flights and in laboratory simulations. Appl. Environ. Microbiol. 2018, 84, e01942-18. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Liu, Y.; Song, L.; Huang, Z. Species composition and distribution of algae in semi-desert algal crusts. Chin. J. Appl. Ecol. 2000, 11, 61–65. [Google Scholar] [CrossRef]
- Hu, C.; Liu, Y.; Zhang, D.; Huang, Z. Cementing mechanism of algal crusts from desert area. Chin. Sci. Bull. 2002, 47, 1361–1368. [Google Scholar] [CrossRef]
- Steven, B.; Gallegos-Graves, V.; Yeager, C.; Belnap, J.; Kuske, C.R. Common and distinguishing features of the bacterial and fungal communities in biological soil crusts and shrub root zone soils. Soil Biol. Biochem. 2014, 69, 302–312. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Tamm, A.; Hassenrück, C.; Al-Rawahi, A.N.; Rodriguez-Caballero, E.; Fiedler, S.; Maier, S.; Weber, B. Habitat-dependent composition of bacterial and fungal communities in biological soil crusts from Oman. Sci. Rep. 2019, 9, 6468. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.; Yuan, S.; Li, Z.; Sun, J.; Li, X. Development of archaeal communities in biological soil crusts along a revegetation chronosequence in the Tengger Desert, north central China. Soil Till. Res. 2020, 196, 104443. [Google Scholar] [CrossRef]
- Wang, G.; Deng, S.; Li, C.; Liu, Y.; Chen, L.; Hu, C. Damage to DNA caused by UV-B radiation in the desert cyanobacterium Scytonema javanicum and the effects of exogenous chemicals on the process. Chemosphere 2012, 88, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Deng, S.; Liu, J.; Ye, C.; Zhou, X.; Chen, L. Cell damage caused by ultraviolet B radiation in the desert cyanobacterium Phormidium tenue and its recovery process. Ecotox. Environ. Safe 2017, 144, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Hui, R.; Li, X.; Zhao, R.; Liu, L.; Gao, Y.; Wei, Y. UV-B radiation suppresses chlorophyll fluorescence, photosynthetic pigment and antioxidant systems of two key species in soil crusts from the Tengger Desert, China. J. Arid Environ. 2015, 113, 6–15. [Google Scholar] [CrossRef]
- Bowker, M.A.; Maestre, F.T.; Escolar, C. Biological crusts as a model system for examining the biodiversity-ecosystem function relationship in soils. Soil Biol. Biochem. 2010, 42, 405–417. [Google Scholar] [CrossRef]
- Bowker, M.A.; Maestre, F.T.; Eldridge, D.; Belnap, J.; Castillo-Monroy, A.; Escolar, C.; Soliveres, S. Biological soil crusts (biocrusts) as a model system in community, landscape and ecosystem ecology. Biodivers. Conserv. 2014, 23, 1619–1637. [Google Scholar] [CrossRef]
- Pushkareva, E.; Johansen, J.R.; Elster, J. A review of the ecology, ecophysiology and biodiversity of microalgae in Arctic soil crusts. Polar Biol. 2016, 39, 2227–2240. [Google Scholar] [CrossRef]
- Xu, H.; Raanan, H.; Dai, G.; Oren, N.; Berkowicz, S.; Murik, O.; Kaplan, A.; Qiu, B. Reading and surviving the harsh conditions in desert biological soil crust: The cyanobacterial viewpoint. FEMS Microbiol. Rev. 2021, 6, fuab036. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Q.; Ouyang, H.; Lan, S.; Hu, C. Pyrosequencing reveals significant changes in microbial communities along the ecological succession of biological soil crusts in the Tengger Desert of China. Pedosphere 2018, 28, 350–362. [Google Scholar] [CrossRef]
- Lan, S.; Thomas, A.D.; Tooth, S.; Wu, L.; Hu, C. Small-Scale spatial heterogeneity of photosynthetic fluorescence associated with biological soil crust succession in the Tengger Desert, China. Microb. Ecol. 2019, 78, 936–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Pichel, F.; Castenholz, R.W. Characterization and biological implications of scytonemin, a cyanobacterial sheath pigments. J. Phycol. 1991, 27, 395–409. [Google Scholar] [CrossRef]
- Rest, M.V.D.; Gingeas, G. The pigment complement of the photosynthetic reaction center isolated from Rhodospirillum rubrum. Biol. Chem. 1974, 249, 6446–6453. [Google Scholar] [CrossRef]
- Colica, G.; Li, H.; Rossi, F.; Li, D.; Liu, Y.; De Philippis, R. Microbial secreted exopolysaccharides affect the hydrological behavior of induced biological soil crusts in desert sandy soils. Soil Biol. Biochem. 2014, 68, 62–70. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.; Wang, H.; Gai, X. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil. Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Takai, K.; Horikoshi, K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microb. 2000, 66, 5066–5072. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Smith, D.J.; Griffin, D.W.; McPeters, R.D.; Ward, P.D.; Schuerger, A.C. Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia 2011, 27, 319–332. [Google Scholar] [CrossRef]
- Baque, M.; Verseux, C.; Boettger, U.; Rabbow, E.; de Vera, J.P.P.; Billi, D. Preservation of biomarkers from Cyanobacteria mixed with Mars like regolith under simulated martian atmosphere and UV flux. Orig. Life Evol. B 2016, 46, 289–310. [Google Scholar] [CrossRef]
- Baque, M.; Scalzi, G.; Rabbow, E.; Rettberg, P.; Billi, D. Biofilm and planktonic lifestyles differently support the resistance of the desert cyanobacterium Chroococcidiopsis under space and Martian simulations. Orig. Life Evol. B 2013, 43, 377–389. [Google Scholar] [CrossRef]
- Campbell, S.E. Soil stabilization by a prokaryotic desert crust: Implications for Precambrian land biota. Orig. Life Evol. B 1979, 9, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Hirota, M.; Tang, Y.; Cui, X.; Zhao, X.; Oikawa, T. Strong temperature dependence and no moss photosynthesis in winter CO2 flux for a Kobresia meadow on the Qinghai–Tibetan plateau. Soil Biol. Biochem. 2005, 37, 1966–1969. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Nowak, R.; Wu, P.; Gao, Y.; Zhao, Y.; Huang, L.; Hu, Y.; Jia, R. Effect of sand-stabilizing shrubs on soil respiration in a temperate desert. Plant Soil 2013, 367, 449–463. [Google Scholar] [CrossRef]
- Xie, J.; Zha, T.; Jia, X.; Qian, D.; Wu, B.; Zhang, Y.; Bourque, C.P.A.; Chen, J.; Sun, G.; Peltola, H. Irregular precipitation events in control of seasonal variations in CO2 exchange in a cold desert-shrub ecosystem in northwest China. J. Arid Environ. 2015, 120, 33–41. [Google Scholar] [CrossRef]
- Yang, F.; Huang, J.; He, Q.; Zheng, X.; Zhou, C.; Pan, H.; Huo, W.; Yu, H.; Liu, X.; Meng, L.; et al. Impact of differences in soil temperature on the desert carbon sink. Geoderma 2020, 379, 114636. [Google Scholar] [CrossRef]
- Soule, T.; Shipe, D.; Lothamer, J. Extracellular polysaccharide production in a scytonemin-deficient mutant of Nostoc punctiforme under UVA and oxidative stress. Curr. Microbiol. 2016, 73, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Andrew, M.; Jayaraman, G. Structural features of microbial exopolysaccharides in relation to their antioxidant activity. Carbohyd. Res. 2020, 487, 107881. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Li, X.; Zhao, X.; Wang, Z. Effects of Enhanced UV-B on the photosynthesis of algal crusts. J. Desert Res. 2011, 31, 889–893. [Google Scholar]
- Rastogi, R.P.; Sonani, R.P.; Madamwar, D. Effects of PAR and UV Radiation on the structural and functional integrity of phycocyanin, phycoerythrin and allophycocyanin isolated from the marine Cyanobacterium Lyngbya sp. A09DM. Photochem. Photobiol. 2015, 91, 837–844. [Google Scholar] [CrossRef]
- Yang, H.; Wang, T.; Yu, X.; Yang, Y.; Wang, C.F.; Yang, Q.H.; Wang, X. Enhanced sugar accumulation and regulated plant hormone signaling genes contribute to cold tolerance in hypoploid Saccharum spontaneum. BMC Genom. 2020, 21, 507. [Google Scholar] [CrossRef]
- Wiley, E. Do carbon reserves increase tree survival during stress and following disturbance? Curr. For. Rep. 2020, 110, 393–401. [Google Scholar] [CrossRef]
- Yin, B.; Zhang, Y. Physiological regulation of Syntrichia caninervis Mitt. in different microhabitats during periods of snow in the Gurbantunggut Desert, northwestern China. J. Plant Physiol. 2016, 194, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Koponen, H.T.; Bååth, E. Soil bacteria growth after a freezing/thawing event. Soil Biol. Biochem. 2016, 100, 229–232. [Google Scholar] [CrossRef]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Costa, O.Y.A.; Raaijmakers, J.M.; Kuramae, E.E. Microbial extracellular polymeric substances: Ecological function and impact on soil aggregation. Front. Microbiol. 2018, 9, 1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baubin, C.; Farrell, A.M.; Šťovíčeka, A.; Ghazaryan, L.; Giladi, I.; Gillor, O. Seasonal and spatial variability in total and active bacterial communities from desert soil. Pedobiologia 2019, 74, 7–14. [Google Scholar] [CrossRef]
- Zhou, Y.; Leung, M.H.Y.; Tong, X.; Lai, Y.; Tong, J.C.K.; Ridley, I.A.; Lee, P.K.H. Profiling airborne microbiota in mechanically ventilated buildings across seasons in Hong Kong reveals higher metabolic activity in low-abundance bacteria. Environ. Sci. Technol. 2021, 55, 249–259. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Klatt, C.G.; Inskeep, W.P.; Herrgard, M.J.; Jay, Z.J.; Rusch, D.B.; Tringe, S.G.; Parenteau, M.N.; Ward, D.W.; Boomer, S.M.; Bryant, D.A.; et al. Community structure and function of high-temperature chlorophototrophic microbial mats inhabiting diverse geothermal environments. Front. Microbiol. 2013, 4, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrshad, M.; Salcher, M.M.; Okazaki, Y.; Nakano, S.; Simek, K.; Andrei, A.; Ghai, R. Hidden in plain sight—highly abundant and diverse planktonic freshwater Chloroflexi. Microbiome 2018, 6, 176. [Google Scholar] [CrossRef] [PubMed]
- Ballhausen, M.B.; Hewitt, R.; Rillig, M.C. Mimicking climate warming effects on Alaskan soil microbial communities via gradual temperature increase. Sci. Rep. 2020, 10, 8533. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Ward, L.M.; Fischer, W.W. Evolution of the 3-hydroxypropionate bicycle and recent transfer of anoxygenic photosynthesis into the Chloroflexi. Proc. Natl. Acad. Sci. USA 2017, 114, 10749–10754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochetkova, T.V.; Zayulina, K.S.; Zhigarkov, V.S.; Minaev, N.V.; Chichkov, B.N.; Novikov, A.A.; Toshchakov, S.V.; Elcheninov, A.G.; Kublanov, I.V. Tepidiforma bonchosmolovskayae gen. nov., sp nov., a moderately thermophilic Chloroflexi bacterium from a Chukotka hot spring (Arctic, Russia), representing a novel class, Tepidiformia, which includes the previously uncultivated lineage OLB14. Int. J. Syst. Evol. Microbiol. 2020, 70, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.K.; Gieler, B.A.; Heisler, D.L.; Palisoc, M.M.; Williams, A.J.; Dohnalkova, A.C.; Ming, H.; Yu, T.T.; Dodsworth, J.; Li, W.J.; et al. Kallotenue papyrolyticum gen. nov., sp. nov., a cellulolytic and filamentous thermophile that represents a novel lineage (Kallotenuales ord. nov., Kallotenuaceae fam. nov.) within the class Chloroflexia. Int. J. Syst. Evol. Microbiol. 2013, 63, 4675–4682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bay, S.K.; Dong, X.; Bradley, J.A.; Leung, P.M.; Grinter, R.; Jirapanjawat, T. Trace gas oxidizers are widespread and active members of soil microbial communities. Nat. Microbiol. 2021, 6, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Garcia-Lopez, M.; Bills, G.F.; Genilloud, O. Longimicrobium terrae gen. nov., sp nov., an oligotrophic bacterium of the under-represented phylum Gemmatimonadetes isolated through a system of miniaturized diffusion chambers. Int. J. Syst. Evol. Microbiol. 2016, 66, 1976–1985. [Google Scholar] [CrossRef] [PubMed]
- Stieglmeier, M.; Klingl, A.; Alves, R.J.E.; Rittmann, S.K.M.R.; Melcher, M.; Leisch, N.; Schleper, C. Nitrososphaera viennensis gen. nov., sp nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 2014, 64, 2738–2752. [Google Scholar] [CrossRef]
- Hezbri, K.; Louati, M.; Nouioui, I.; Gtari, M.; Rohde, M.; Sproeer, C.; Schumann, P.; Klenk, H.; Ghodhbane-Gtari, F.; Montero-Calasanz, M.D. Blastococcus capsensis sp nov., isolated from an archaeological Roman pool and emended description of the genus Blastococcus, B. aggregatus, B. saxobsidens, B. jejuensis and B. endophyticus. Int. J. Syst. Evol. Microbiol. 2016, 66, 4864–4872. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.F.; Nouioui, I.; Sangal, V.; Choi, S.; Yang, S.J.; Kim, B.Y.; Trujillo, M.E.; Riesco, R.; Montero-Calasanz, M.D.; Rahmani, T.P.D.; et al. Blastococcus atacamensis sp nov., a novel strain adapted to life in the Yungay core region of the Atacama Desert. Int. J. Syst. Evol. Microbiol. 2018, 68, 2712–2721. [Google Scholar] [CrossRef]
- An, M.; Mou, S.; Zhang, X.; Ye, N.H.; Zheng, Z.; Cao, S.N.; Xu, D.; Fan, X.; Wang, Y.T.; Miao, J.L. Temperature regulates fatty acid desaturases at a transcriptional level and modulates the fatty acid profile in the Antarctic microalga Chlamydomonas sp. ICE-L. Bioresour. Technol. 2013, 134, 151–157. [Google Scholar] [CrossRef]
- Rao, M.P.N.; Lou, Z.; Dong, Z.; Li, Q.; Liu, B.; Guo, S.; Nie, G.; Li, W. Metagenomic analysis further extends the role of Chloroflexi in fundamental biogeochemical cycles. Environ. Res. 2020, 209, 112888. [Google Scholar] [CrossRef]
- Csotonyi, J.; Swiderski, J.; Stackebrandt, E.; Yurkov, V. A new environment for aerobic anoxygenic phototrophic bacteria: Biological soil crusts. Env. Microbiol. Rep. 2010, 2, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Berg, I.A.; Keppen, O.I.; Krasil’nikova, E.N.; Ugol’kova, N.V.; Ivanovsky, R.N. Carbon metabolism of filamentous anoxygenic phototrophic bacteria of the family Oscillochloridaceae. Microbiology. 2005, 74, 258–264. [Google Scholar] [CrossRef]
- Keppen, O.; Ivanovsky, R.N.; Streshinskaya, G.M.; Lebedeva, N.V.; Shashkov, A.A.; Dmitrenok, A.; Grouzdev, D. The cell wall of the filamentous anoxygenic phototrophic bacterium Oscillochloris trichoides. Microbiology 2018, 164, 57–64. [Google Scholar] [CrossRef]
- Tahon, G.; Willems, A. Isolation and characterization of aerobic anoxygenic phototrophs from exposed soils from the Sof. Rondane Mountains, East Antarctica. Syst. Appl. Microbiol. 2017, 40, 357–369. [Google Scholar] [CrossRef]
- Busse, H.J.; Denner, E.B.M.; Buczolits, S.; Salkinoja-Salonen, M.; Bennasar, A.; Kampfer, P. Sphingomonas aurantiaca sp nov., Sphingomonas aerolata sp nov. and Sphingomonas faeni sp nov., air- and dustborne and Antarctic, orange-pigmented, psychrotolerant bacteria, and emended description of the genus Sphingomonas. Int. J. Syst. Evol. Microbiol. 2003, 53, 1253–1260. [Google Scholar] [CrossRef]
- Wang, N.F.; Zhang, T.; Zhang, F.; Wang, E.T.; He, J.F.; Ding, H.; Zhang, B.; Liu, J.; Ran, X.B. Diversity and structure of soil bacterial communities in the Fildes Region (maritime Antarctica) as revealed by 454 pyrosequencing. Front. Microbiol. 2015, 6, 1188. [Google Scholar] [CrossRef] [Green Version]




| DNA | RNA | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| UV + LT | LT-F | LT-T | CK | UV + LT | LT-F | LT-T | CK | ||
| Bacterial | OTUs | 812 ± 16 | 844 ± 74 | 859 ± 24 | 877 ± 8 | 618 ± 77 b | 565 ± 38 b | 749 ± 59 a | 582 ± 66 b |
| Shannon | 4.69 ± 0.23 | 4.67 ± 0.15 | 4.59 ± 0.27 | 4.72 ± 0.19 | 3.75 ± 0.49 | 3.65 ± 0.35 | 4.42 ± 0.45 | 3.82 ± 0.60 | |
| Chao1 | 947 ± 58 | 1040 ± 65 | 975 ± 23 | 953 ± 59 | 777 ± 84 b | 739 ± 16 b | 912 ± 49 a | 739 ± 29 b | |
| Archaea | OTUs | 499 ± 107 | 664 ± 178 | 592 ± 225 | 559 ± 158 | 611 ± 309 | 500 ± 323 | 457 ± 312 | 800 ± 88 |
| Shannon | 3.24 ± 0.54 | 3.89 ± 0.87 | 3.49 ± 1.04 | 4.09 ± 0.62 | 3.63 ± 1.31 | 3.05 ± 1.30 | 3.72 ± 0.49 | 4.52 ± 0.54 | |
| Chao1 | 670 ± 78 | 831 ± 200 | 734 ± 222 | 721 ± 140 | 787 ± 281 | 665 ± 340 | 681 ± 367 | 983 ± 67 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Hu, C.; Yang, H. Responses of Cyanobacterial Crusts and Microbial Communities to Extreme Environments of the Stratosphere. Microorganisms 2022, 10, 1252. https://doi.org/10.3390/microorganisms10061252
Li Q, Hu C, Yang H. Responses of Cyanobacterial Crusts and Microbial Communities to Extreme Environments of the Stratosphere. Microorganisms. 2022; 10(6):1252. https://doi.org/10.3390/microorganisms10061252
Chicago/Turabian StyleLi, Qi, Chunxiang Hu, and Haijian Yang. 2022. "Responses of Cyanobacterial Crusts and Microbial Communities to Extreme Environments of the Stratosphere" Microorganisms 10, no. 6: 1252. https://doi.org/10.3390/microorganisms10061252
APA StyleLi, Q., Hu, C., & Yang, H. (2022). Responses of Cyanobacterial Crusts and Microbial Communities to Extreme Environments of the Stratosphere. Microorganisms, 10(6), 1252. https://doi.org/10.3390/microorganisms10061252






