Performance Evaluation of the Quantamatrix QMAC-dRAST System for Rapid Antibiotic Susceptibility Testing Directly from Blood Cultures
Abstract
1. Introduction
2. Methods
3. Results
3.1. QMAC-dRAST Performance
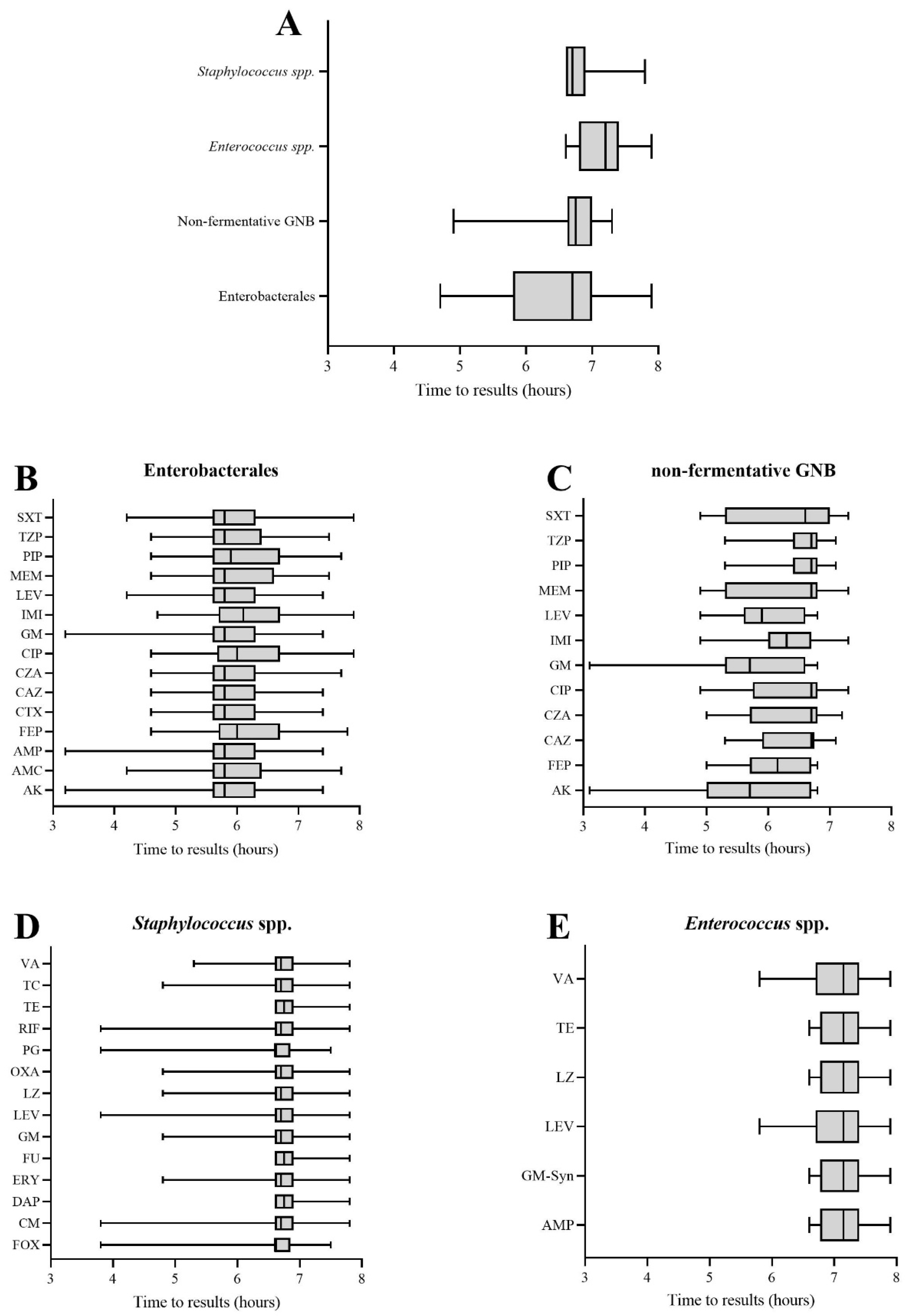
3.2. Time to AST Results
3.3. Repeatability and Reproducibility
4. Discussion
4.1. Main Findings
4.2. Limitation
4.3. Implication
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Goto, M.; Al-Hasan, M.N. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2013, 19, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.J.; Moehring, R.W.; Sloane, R.; Schmader, K.E.; Weber, D.J.; Fowler, V.G., Jr.; Smathers, E.; Sexton, D.J. Bloodstream infections in community hospitals in the 21st century: A multicenter cohort study. PLoS ONE 2014, 9, e91713. [Google Scholar] [CrossRef] [PubMed]
- Lambregts, M.M.C.; Wijnakker, R.; Bernards, A.T.; Visser, L.G.; Cessie, S.L.; Boer, M.G.J. Mortality after Delay of Adequate Empiric Antimicrobial Treatment of Bloodstream Infection. J. Clin. Med. 2020, 9, 1378. [Google Scholar] [CrossRef]
- Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and Costs of Sepsis in the United States-An Analysis Based on Timing of Diagnosis and Severity Level. Crit. Care Med. 2018, 46, 1889–1897. [Google Scholar] [CrossRef] [PubMed]
- Meda, M.; Clayton, J.; Varghese, R.; Rangaiah, J.; Grundy, C.; Dashti, F.; Garner, D.; Groves, K.; Fitzmaurice, K.; Hutley, E. What are the critical steps in processing blood cultures? A prospective audit evaluating current practice of reporting blood cultures in a centralised laboratory serving secondary care hospitals. J. Clin. Pathol. 2017, 70, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Tabak, Y.P.; Vankeepuram, L.; Ye, G.; Jeffers, K.; Gupta, V.; Murray, P.R. Blood Culture Turnaround Time in U.S. Acute Care Hospitals and Implications for Laboratory Process Optimization. J. Clin. Microbiol. 2018, 56, e00500-18. [Google Scholar] [CrossRef]
- Edmiston, C.E.; Garcia, R.; Barnden, M.; DeBaun, B.; Johnson, H.B. Rapid diagnostics for bloodstream infections: A primer for infection preventionists. Am. J. Infect. Control 2018, 46, 1060–1068. [Google Scholar] [CrossRef]
- Prod’hom, G.; Durussel, C.; Greub, G. A simple blood-culture bacterial pellet preparation for faster accurate direct bacterial identification and antibiotic susceptibility testing with the VITEK 2 system. J. Med. Microbiol. 2013, 62, 773–777. [Google Scholar] [CrossRef]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef]
- Choi, J.; Jeong, H.Y.; Lee, G.Y.; Han, S.; Han, S.; Jin, B.; Lim, T.; Kim, S.; Kim, D.Y.; Kim, H.C.; et al. Direct, rapid antimicrobial susceptibility test from positive blood cultures based on microscopic imaging analysis. Sci. Rep. 2017, 7, 1148. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.; Kim, D.; Kang, D.Y.; Park, B.Y.; Yang, S.; Yoon, E.J.; Lee, H.; Jeong, S.H. Evaluation of the BD Phoenix M50 Automated Microbiology System for Antimicrobial Susceptibility Testing with Clinical Isolates in Korea. Microb. Drug Resist. 2019, 25, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Junkins, A.D.; Lockhart, S.R.; Heilmann, K.P.; Dohrn, C.L.; Von Stein, D.L.; Winokur, P.L.; Doern, G.V.; Richter, S.S. BD Phoenix and Vitek 2 detection of mecA-mediated resistance in Staphylococcus aureus with cefoxitin. J. Clin. Microbiol. 2009, 47, 2879–2882. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.W.; Munier, G.K.; Johnson, C.L. Direct comparison of the BD phoenix system with the MicroScan WalkAway system for identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative gram-negative organisms. J. Clin. Microbiol. 2008, 46, 2327–2333. [Google Scholar] [CrossRef]
- Croxatto, A.; Coste, A.T.; Pillonel, T.; Bertelli, C.; Greub, G.; Prod’hom, G. Evaluation of the BD Phoenix CPO Detect Test for the detection of carbapenemase producers. Clin. Microbiol. Infect. 2020, 26, 644.e9–644.e15. [Google Scholar] [CrossRef]
- Jacot, D.; Sarton-Loheac, G.; Coste, A.T.; Bertelli, C.; Greub, G.; Prod’hom, G.; Croxatto, A. Performance evaluation of the Becton Dickinson Kiestra IdentifA/SusceptA. Clin. Microbiol. Infect. 2021, 27, 1167.e9–1167.e17. [Google Scholar] [CrossRef]
- Yonezawa, T.; Watari, T.; Ashizawa, K.; Hanada, D.; Yanagiya, T.; Watanabe, N.; Terada, T.; Tomoda, Y.; Fujii, S. Development of an improved rapid BACpro® protocol and a method for direct identification from blood-culture-positive bottles using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Microbiol. Methods 2018, 148, 138–144. [Google Scholar] [CrossRef]
- Hanaki, H.; Kubo, R.; Nakano, T.; Kurihara, M.; Sunagawa, K. Characterization of HMRZ-86: A novel chromogenic cephalosporin for the detection of extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 2004, 53, 888–889. [Google Scholar] [CrossRef]
- Tijet, N.; Boyd, D.; Patel, S.N.; Mulvey, M.R.; Melano, R.G. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4578–4580. [Google Scholar] [CrossRef]
- FDA. Guidance for Industry and FDA. Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems; US Department of Health and Human Services: Silver Spring, MD, USA, 2009. [Google Scholar]
- Woods, C.R. Macrolide-inducible resistance to clindamycin and the D-test. Pediatric Infect. Dis. J. 2009, 28, 1115–1118. [Google Scholar] [CrossRef]
- Huh, H.J.; Song, D.J.; Shim, H.J.; Kwon, W.K.; Park, M.S.; Ryu, M.R.; Cho, E.H.; Oh, J.; Yoo, I.Y.; Lee, N.Y. Performance evaluation of the QMAC-dRAST for staphylococci and enterococci isolated from blood culture: A comparative study of performance with the VITEK-2 system. J. Antimicrob. Chemother. 2018, 73, 1267–1271. [Google Scholar] [CrossRef] [PubMed]
- Grohs, P.; Rondinaud, E.; Fourar, M.; Rouis, K.; Mainardi, J.L.; Podglajen, I. Comparative evaluation of the QMAC-dRAST V2.0 system for rapid antibiotic susceptibility testing of Gram-negative blood culture isolates. J. Microbiol. Methods 2020, 172, 105902. [Google Scholar] [CrossRef] [PubMed]
- Fattouh, R.; Tijet, N.; McGeer, A.; Poutanen, S.M.; Melano, R.G.; Patel, S.N. What Is the Appropriate Meropenem MIC for Screening of Carbapenemase-Producing Enterobacteriaceae in Low-Prevalence Settings? Antimicrob. Agents Chemother. 2015, 60, 1556–1559. [Google Scholar] [CrossRef] [PubMed]
- Oueslati, S.; Nordmann, P.; Poirel, L. Heterogeneous hydrolytic features for OXA-48-like β-lactamases. J. Antimicrob. Chemother. 2015, 70, 1059–1063. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Garnacho-Montero, J.; Gutiérrez-Pizarraya, A.; Escoresca-Ortega, A.; Corcia-Palomo, Y.; Fernández-Delgado, E.; Herrera-Melero, I.; Ortiz-Leyba, C.; Márquez-Vácaro, J.A. De-escalation of empirical therapy is associated with lower mortality in patients with severe sepsis and septic shock. Intensive Care Med. 2014, 40, 32–40. [Google Scholar] [CrossRef]
- Heenen, S.; Jacobs, F.; Vincent, J.-L. Antibiotic strategies in severe nosocomial sepsis: Why do we not de-escalate more often? Crit. Care Med. 2012, 40, 1404–1409. [Google Scholar] [CrossRef]
- Morel, J.; Casoetto, J.; Jospé, R.; Aubert, G.; Terrana, R.; Dumont, A.; Molliex, S.; Auboyer, C. De-escalation as part of a global strategy of empiric antibiotherapy management. A retrospective study in a medico-surgical intensive care unit. Crit. Care 2010, 14, R225. [Google Scholar] [CrossRef]
| Bacterial Species | N° Bacteria | Resistance Mechanisms | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spiking | |||||||||||||
| No | Yes | ||||||||||||
| All | 56 | 194 | Wild Type | AmpC | ESBL | AmpC ESBL | AmpC Carbapenemase | Probable K1 | Probable SHV-1 | Carbapenemase | VRE | Methicillin-R | Total |
| Enterobacterales, total | 28 | 102 | 130 | ||||||||||
| Escherichia coli | 16 | 15 | 17 | 2 | 12 | 31 | |||||||
| Klebsiella pneumoniae | 5 | 16 | 8 | 6 | 3 | 4 | 21 | ||||||
| Enterobacter cloacae | 0 | 14 | 7 | 4 | 3 | 14 | |||||||
| Proteus mirabilis | 2 | 9 | 10 | 1 | 11 | ||||||||
| Klebsiella aerogenes | 1 | 9 | 10 | 10 | |||||||||
| Klebsiella oxytoca | 1 | 9 | 5 | 2 | 3 | 10 | |||||||
| Serratia marcescens | 0 | 10 | 10 | 10 | |||||||||
| Proteus vulgaris | 0 | 6 | 6 | 6 | |||||||||
| Proteus hauseri | 0 | 5 | 5 | 5 | |||||||||
| Citrobacter freundii | 1 | 3 | 3 | 1 | 4 | ||||||||
| Morganella morganii | 1 | 3 | 4 | 4 | |||||||||
| Citrobacter koseri | 0 | 3 | 3 | 3 | |||||||||
| Salmonella spp. | 1 | 0 | 1 | 1 | |||||||||
| Non-fermentative GNB, total | 5 | 15 | 20 | ||||||||||
| Pseudomonas aeruginosa | 4 | 9 | 11 | 2 | 13 | ||||||||
| Acinetobacter baumannii | 0 | 5 | 3 | 2 | 5 | ||||||||
| Acinetobacter spp. | 1 | 0 | 1 | 1 | |||||||||
| Stenotrophomonas maltophilia | 0 | 1 | 1 | 1 | |||||||||
| Staphylococcus spp., total | 20 | 49 | 69 | ||||||||||
| Staphylococcus aureus | 9 | 15 | 9 | 15 | 24 | ||||||||
| Staphylococcus epidermidis | 8 | 11 | 10 | 9 | 19 | ||||||||
| Staphylococcus hominis | 2 | 7 | 7 | 2 | 9 | ||||||||
| Staphylococcus capitis | 1 | 6 | 6 | 1 | 7 | ||||||||
| Staphylococcus haemolyticus | 0 | 5 | 4 | 1 | 5 | ||||||||
| Staphylococcus lugdunensis | 0 | 5 | 4 | 1 | 5 | ||||||||
| Enterococcus spp., total | 3 | 28 | 31 | ||||||||||
| Enterococcus faecium | 0 | 14 | 8 | 6 | 14 | ||||||||
| Enterococcus faecalis | 3 | 10 | 12 | 1 | 13 | ||||||||
| Enterococcus casseliflavus | 0 | 3 | 3 | 3 | |||||||||
| Enterococcus gallinarum | 0 | 1 | 1 | 1 | |||||||||
| Total | 125 | 42 | 20 | 5 | 4 | 3 | 3 | 8 | 11 | 29 | 250 | ||
| N° of Antibiotics Tested | CA | CA% | me | me% | ME | ME%S | VME | VME%R | S | S_% | R | R_% | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enterobacterales | |||||||||||||
| Amikacin | 130 | 130 | 100 | 130 | 100 | 0 | 0.0 | ||||||
| Amoxicillin–Clavulanate | 130 | 125 | 96.2 | 3 | 5.9 | 2 | 2.5 | 51 | 39.2 | 79 | 60.8 | ||
| Ampicillin | 130 | 130 | 100 | 16 | 12.3 | 114 | 87.7 | ||||||
| Ceftazidime | 130 | 114 | 87.7 | 11 | 8.5 | 5 | 5.1 | 98 | 75.4 | 32 | 24.6 | ||
| Ceftazidime–Avibactam | 129 | 129 | 100 | 129 | 100 | 0 | 0.0 | ||||||
| Ciprofloxacin | 129 | 123 | 95.3 | 5 | 3.9 | 1 | 1 | 104 | 80.6 | 25 | 19.4 | ||
| Cefepime | 130 | 116 | 89.2 | 9 | 6.9 | 5 | 4.6 | 108 | 83.1 | 22 | 16.9 | ||
| Gentamicin | 130 | 125 | 96.2 | 4 | 3.1 | 1 | 5.6 | 112 | 86.2 | 18 | 13.8 | ||
| Imipenem | 118 | 103 | 87.3 | 15 | 12.7 | 115 | 97.5 | 3 | 2.5 | ||||
| Levofloxacin | 130 | 120 | 92.3 | 10 | 7.7 | 114 | 87.7 | 16 | 12.3 | ||||
| Meropenem | 130 | 129 | 99.2 | 1 | 0.8 | 129 | 99.2 | 1 | 0.8 | ||||
| Piperacillin–Tazobactam | 130 | 122 | 93.8 | 6 | 4.6 | 1 | 1 | 1 | 3.4 | 101 | 77.7 | 29 | 22.3 |
| Trimethoprim–Sulfamethoxazole | 130 | 128 | 98.5 | 1 | 0.8 | 1 | 1.1 | 89 | 68.5 | 41 | 31.5 | ||
| Non-fermentative GNB | |||||||||||||
| Amikacin | 19 | 18 | 94.7 | 1 | 5.3 | 16 | 84.2 | 3 | 15.8 | ||||
| Ceftazidime | 13 | 13 | 100 | 8 | 61.5 | 5 | 38.5 | ||||||
| Ceftazidime–Avibactam | 12 | 12 | 100 | 9 | 75 | 3 | 25.0 | ||||||
| Ciprofloxacin | 19 | 17 | 89.5 | 2 | 10.5 | 14 | 73.7 | 5 | 26.3 | ||||
| Cefepime | 13 | 11 | 84.6 | 2 | 22.2 | 9 | 69.2 | 4 | 30.8 | ||||
| Gentamicin | 18 | 18 | 100 | 12 | 66.7 | 6 | 33.3 | ||||||
| Imipenem | 19 | 18 | 94.7 | 1 | 5.3 | 12 | 63.2 | 7 | 36.8 | ||||
| Levofloxacin | 19 | 17 | 89.5 | 2 | 25 | 11 | 57.9 | 8 | 42.1 | ||||
| Meropenem | 19 | 17 | 89.5 | 2 | 10.5 | 14 | 73.7 | 5 | 26.3 | ||||
| Piperacillin–Tazobactam | 13 | 11 | 84.6 | 2 | 20 | 10 | 76.9 | 3 | 23.1 | ||||
| Trimethoprim–Sulfamethoxazole | 7 | 4 | 57.1 | 2 | 28.6 | 1 | 16.7 | 6 | 85.7 | 1 | 14.3 | ||
| Staphylococcus spp. | |||||||||||||
| Clindamycin | 69 | 67 | 97.1 | 1 | 1.4 | 1 | 1.8 | 58 | 84.1 | 11 | 15.9 | ||
| Daptomycin | 69 | 68 | 98.6 | 1 | 1.4 | 69 | 100 | 0 | 0.0 | ||||
| Gentamicin | 69 | 67 | 97.1 | 1 | 2 | 1 | 5.6 | 51 | 73.9 | 18 | 26.1 | ||
| Linezolid | 69 | 69 | 100 | 69 | 100 | 0 | 0.0 | ||||||
| Levofloxacin | 69 | 60 | 87 | 1 | 1.4 | 8 | 17 | 48 | 69.6 | 21 | 30.4 | ||
| Oxacillin | 69 | 46 | 66.7 | 16 | 50 | 7 | 18.9 | 32 | 46.4 | 37 | 53.6 | ||
| Penicillin G | 24 | 24 | 100 | 1 | 4.2 | 23 | 95.8 | ||||||
| Teicoplanin | 69 | 68 | 98.6 | 1 | 1.5 | 65 | 94.2 | 4 | 5.8 | ||||
| Vancomycin | 69 | 69 | 100 | 69 | 100 | 0 | 0.0 | ||||||
| Enterococcus spp. | |||||||||||||
| Ampicillin | 31 | 26 | 83.9 | 3 | 9.7 | 2 | 8 | 25 | 80.6 | 6 | 19.4 | ||
| Gentamicin-Syn | 31 | 30 | 96.8 | 1 | 3.7 | 27 | 87.1 | 4 | 12.9 | ||||
| Linezolid | 31 | 30 | 96.8 | 1 | 100 | 30 | 96.8 | 1 | 3.2 | ||||
| Levofloxacin | 26 | 24 | 92.3 | 2 | 11.1 | 18 | 69.2 | 8 | 30.8 | ||||
| Teicoplanin | 31 | 31 | 100 | 24 | 77.4 | 7 | 22.6 | ||||||
| Vancomycin | 31 | 30 | 96.8 | 1 | 4.8 | 21 | 67.7 | 10 | 32.3 | ||||
| Total | 2604 | 2459 | 94.4 | 74 | 2.8 | 56 | 2.8 | 15 | 2.6 | 2025 | 77.77 | 579 | 22.2 |
| QMAC-dRAST MICs that Differed from the Mode Value by the Indicated Dilution | ||||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | 1 | 2 | >2 | ||
| Enterobacterales | n | 0 | 0 | 11 | 454 | 15 | 0 | 0 |
| % | 0 | 0 | 3.56 | 92.40 | 2.88 | 0.48 | 0 | |
| Pseudomonas aeruginosa | n | 0 | 2 | 7 | 101 | 0 | 0 | 0 |
| % | 0 | 1.82 | 6.36 | 91.82 | 0 | 0 | 0 | |
| Staphylococcus spp. | n | 0 | 1 | 14 | 336 | 10 | 5 | 4 |
| % | 0 | 0.27 | 3.78 | 90.81 | 2.70 | 1.35 | 1.08 | |
| Enterococcus spp. | n | 0 | 0 | 5 | 60 | 5 | 0 | 0 |
| % | 0 | 0 | 7.14 | 85.71 | 7.14 | 0 | 0 | |
| Total | n | 0 | 3 | 37 | 961 | 30 | 5 | 4 |
| % | 0 | 0.29 | 3.56 | 92.40 | 2.88 | 0.48 | 0.38 | |
| QM-dRAST MICs that Differed from the Mode Value by the Indicated Dilution | ||||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | 1 | 2 | >2 | ||
| Enterobacterales | n | 0 | 4 | 21 | 424 | 25 | 1 | 5 |
| % | 0 | 0.83 | 4.38 | 88.33 | 5.21 | 0.21 | 1.04 | |
| P. aeruginosa | n | 0 | 6 | 12 | 71 | 15 | 4 | 2 |
| % | 0 | 5.45 | 10.91 | 64.55 | 13.64 | 3.64 | 1.82 | |
| Staphylococcus spp. | n | 0 | 10 | 40 | 277 | 27 | 5 | 11 |
| % | 0 | 2.70 | 10.81 | 74.86 | 7.30 | 1.35 | 2.97 | |
| Enterococcus spp. | n | 0 | 3 | 6 | 57 | 4 | 0 | 0 |
| % | 0 | 4.29 | 8.57 | 81.43 | 5.71 | 0 | 0 | |
| Total | n | 0 | 23 | 79 | 829 | 71 | 10 | 18 |
| % | 0 | 2.23 | 7.67 | 80.49 | 6.89 | 0.97 | 1.75 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosselin, M.; Prod’hom, G.; Greub, G.; Croxatto, A. Performance Evaluation of the Quantamatrix QMAC-dRAST System for Rapid Antibiotic Susceptibility Testing Directly from Blood Cultures. Microorganisms 2022, 10, 1212. https://doi.org/10.3390/microorganisms10061212
Rosselin M, Prod’hom G, Greub G, Croxatto A. Performance Evaluation of the Quantamatrix QMAC-dRAST System for Rapid Antibiotic Susceptibility Testing Directly from Blood Cultures. Microorganisms. 2022; 10(6):1212. https://doi.org/10.3390/microorganisms10061212
Chicago/Turabian StyleRosselin, Manon, Guy Prod’hom, Gilbert Greub, and Antony Croxatto. 2022. "Performance Evaluation of the Quantamatrix QMAC-dRAST System for Rapid Antibiotic Susceptibility Testing Directly from Blood Cultures" Microorganisms 10, no. 6: 1212. https://doi.org/10.3390/microorganisms10061212
APA StyleRosselin, M., Prod’hom, G., Greub, G., & Croxatto, A. (2022). Performance Evaluation of the Quantamatrix QMAC-dRAST System for Rapid Antibiotic Susceptibility Testing Directly from Blood Cultures. Microorganisms, 10(6), 1212. https://doi.org/10.3390/microorganisms10061212







