Differentiation of Bacillus cereus and Bacillus thuringiensis Using Genome-Guided MALDI-TOF MS Based on Variations in Ribosomal Proteins
Abstract
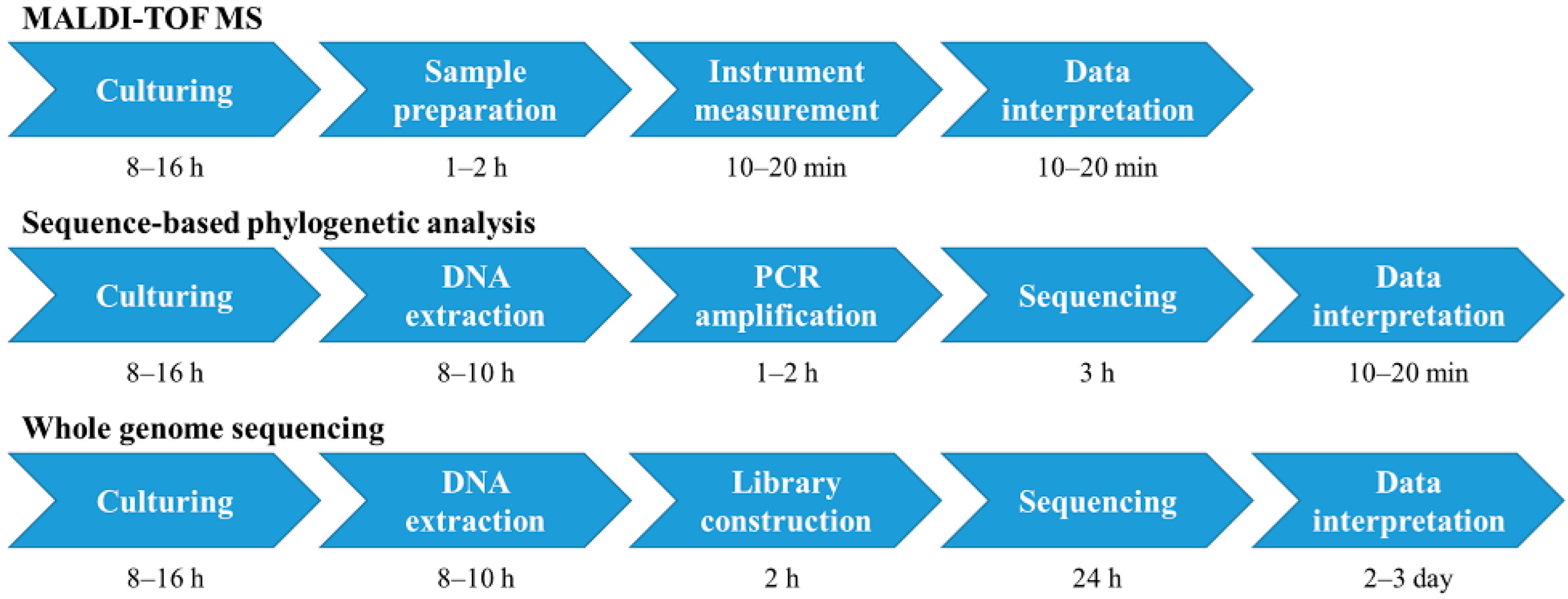
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Genomic Data Mining
2.3. Species Identification Based on Phylogenetic Tree of pycA
2.4. MALDI-TOF MS Analysis
2.5. Data Processing and Identification of Biomarker Proteins
3. Results
3.1. Genome Analysis
3.2. Identification of B. cereus and B. thuringiensis Based on the pycA Gene
3.3. Discovery and Identification of Biomarkers in MALDI-TOF MS
3.4. Assessment of Practical Application of MALDI-TOF MS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EFSA Panel on Biological Hazards (BIOHAZ). Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. Including Bacillus thuringiensis in foodstuffs. EFSA J. 2016, 14, 4524. [Google Scholar] [CrossRef]
- Stenfors Arnesen, L.P.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouzeau-Szynalski, K.; Stollewerk, K.; Messelhäusser, U.; Ehling-Schulz, M. Why be serious about emetic Bacillus cereus: Cereulide production and industrial challenges. Food Microbiol. 2020, 85, 103279. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Paudyal, N.; Pan, H.; Liao, X.; Zhang, X.; Li, X.; Fang, W.; Yue, M. A meta-analysis of major foodborne pathogens in Chinese food commodities between 2006 and 2016. Foodborne Pathog. Dis. 2018, 15, 187–197. [Google Scholar] [CrossRef]
- de Maagd, R.A.; Bravo, A.; Berry, C.; Crickmore, N.; Schnepf, H.E. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu. Rev. Genet. 2003, 37, 409–433. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, B.C.; Yu, Z.; Sun, M. Structural insights into Bacillus thuringiensis Cry, Cyt and parasporin toxins. Toxins 2014, 6, 2732–2770. [Google Scholar] [CrossRef] [Green Version]
- Armada, E.; Probanza, A.; Roldán, A.; Azcón, R. Native plant growth promoting bacteria Bacillus thuringiensis and mixed or individual mycorrhizal species improved drought tolerance and oxidative metabolism in Lavandula dentata plants. J. Plant Physiol. 2016, 192, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nayak, P.S.; Arakha, M.; Kumar, A.; Asthana, S.; Mallick, B.C.; Jha, S. An approach towards continuous production of silver nanoparticles using Bacillus thuringiensis. RSC Adv. 2016, 6, 8232–8242. [Google Scholar] [CrossRef]
- Chen, S.; Deng, Y.; Chang, C.; Lee, J.; Cheng, Y.; Cui, Z.; Zhou, J.; He, F.; Hu, M.; Zhang, L.H. Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19. Sci. Rep. 2015, 5, 8784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, I.; Lee, K.; Goodfellow, M.; Chun, J. Comparative genomic and phylogenomic analyses clarify relationships within and between Bacillus cereus and Bacillus thuringiensis: Proposal for the recognition of two Bacillus thuringiensis genomovars. Front. Microbiol. 2019, 10, 1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo-Esparza, J.F.; Hernández-González, I.; Ibarra, J.E. Search for Cry proteins expressed by Bacillus spp. genomes, using hidden Markov model profiles. 3 Biotech 2019, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Méric, G.; Mageiros, L.; Pascoe, B.; Woodcock, D.J.; Mourkas, E.; Lamble, S.; Bowden, R.; Jolley, K.A.; Raymond, B.; Sheppard, S.K. Lineage-specific plasmid acquisition and the evolution of specialized pathogens in Bacillus thuringiensis and the Bacillus cereus group. Mol. Ecol. 2018, 27, 1524–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacchi, C.T.; Whitney, A.M.; Mayer, L.W.; Morey, R.; Steigerwalt, A.; Boras, A.; Weyant, R.S.; Popovic, T. Sequencing of 16S rRNA gene: A rapid tool for identification of Bacillus anthracis. Emerg. Infect. Dis. 2002, 8, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Shou, Y.; Yoshida, T.M.; Marrone, B.L. Differentiation of Bacillus anthracis, B. cereus, and B. thuringiensis by using pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 2007, 73, 3446–3449. [Google Scholar] [CrossRef] [Green Version]
- Chelliah, R.; Wei, S.; Park, B.J.; Rubab, M.; Banan-Mwine Dalirii, E.; Barathikannan, K.; Jin, Y.G.; Oh, D.H. Whole genome sequence of Bacillus thuringiensis ATCC 10792 and improved discrimination of Bacillus thuringiensis from Bacillus cereus group based on novel biomarkers. Microb. Pathog. 2019, 129, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Park, B.J.; Chelliah, R.; Wei, S.; Park, J.H.; Forghani, F.; Park, Y.S.; Cho, M.S.; Park, D.S.; Oh, D.H. Unique biomarkers as a potential predictive tool for differentiation of Bacillus cereus group based on real-time PCR. Microb. Pathog. 2018, 115, 131–137. [Google Scholar] [CrossRef]
- Wei, S.; Chelliah, R.; Park, B.J.; Kim, S.H.; Forghani, F.; Cho, M.S.; Park, D.S.; Jin, Y.G.; Oh, D.H. Differentiation of Bacillus thuringiensis from Bacillus cereus group using a unique marker based on real-time PCR. Front. Microbiol. 2019, 10, 883. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Rainey, F.A. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014, 64, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Lai, Q.; Göker, M.; Meier-Kolthoff, J.P.; Wang, M.; Sun, Y.; Wang, L.; Shao, Z. Genomic insights into the taxonomic status of the Bacillus cereus group. Sci. Rep. 2015, 5, 14082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattal, C.; Oberoi, J.K.; Goel, N.; Raveendran, R.; Khanna, S. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Gulati, V.; Fox, E.M.; Karpe, A.; Beale, D.J.; Sevior, D.; Bhave, M.; Palombo, E.A. Rapid identification and source-tracking of Listeria monocytogenes using MALDI-TOF mass spectrometry. Int. J. Food Microbiol. 2015, 202, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Manukumar, H.M.; Umesha, S. MALDI-TOF-MS based identification and molecular characterization of food associated methicillin-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 11414. [Google Scholar] [CrossRef] [PubMed]
- Sala-Comorera, L.; Vilaró, C.; Galofré, B.; Blanch, A.R.; García-Aljaro, C. Use of matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry for bacterial monitoring in routine analysis at a drinking water treatment plant. Int. J. Hyg. Environ. Health 2016, 219, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Mangmee, S.; Reamtong, O.; Kalambaheti, T.; Roytrakul, S.; Sonthayanon, P. MALDI-TOF mass spectrometry typing for predominant serovars of non-typhoidal Salmonella in a Thai broiler industry. Food Control 2020, 113, 107188. [Google Scholar] [CrossRef]
- Ojima-Kato, T.; Yamamoto, N.; Suzuki, M.; Fukunaga, T.; Tamura, H. Discrimination of Escherichia coli O157, O26 and O111 from other serovars by MALDI-TOF MS based on the S10-GERMS method. PLoS ONE 2014, 9, e113458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojima-Kato, T.; Yamamoto, N.; Takahashi, H.; Tamura, H. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) can precisely discriminate the lineages of Listeria monocytogenes and species of Listeria. PLoS ONE 2016, 11, e0159730. [Google Scholar] [CrossRef] [Green Version]
- Camoez, M.; Sierra, J.M.; Dominguez, M.A.; Ferrer-Navarro, M.; Vila, J.; Roca, I. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 2016, 22, 161.E1–161.E7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Jo, H.; Choi, E.; Kim, Y.; Kim, J.; Cho, H. Reliable identification of Bacillus cereus group species using low mass biomarkers by MALDI-TOF MS. J. Microbiol. Biotechnol. 2019, 29, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Manzulli, V.; Rondinone, V.; Buchicchio, A.; Serrecchia, L.; Cipolletta, D.; Fasanella, A.; Parisi, A.; Difato, L.; Iatarola, M.; Aceti, A.; et al. Discrimination of Bacillus cereus group members by MALDI-TOF mass spectrometry. Microorganisms 2021, 9, 1202. [Google Scholar] [CrossRef] [PubMed]
- Rothen, J.; Pothier, J.F.; Foucault, F.; Blom, J.; Nanayakkara, D.; Li, C.; Ip, M.; Tanner, M.; Vogel, G.; Pflüger, V.; et al. Subspecies typing of Streptococcus agalactiae based on ribosomal subunit protein mass variation by MALDI-TOF MS. Front. Microbiol. 2019, 10, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corver, J.; Sen, J.; Hornung, B.V.H.; Mertens, B.J.; Berssenbrugge, E.K.L.; Harmanus, C.; Sanders, I.M.J.G.; Kumar, N.; Lawley, T.D.; Kuijper, E.J.; et al. Identification and validation of two peptide markers for the recognition of Clostridioides difficile MLST-1 and MLST-11 by MALDI-MS. Clin. Microbiol. Infect. 2019, 25, 904.E1–904.E7. [Google Scholar] [CrossRef] [PubMed]
- Emele, M.F.; Možina, S.S.; Lugert, R.; Bohne, W.; Masanta, W.O.; Riedel, T.; Groß, U.; Bader, O.; Zautner, A.E. Proteotyping as alternate typing method to differentiate Campylobacter coli clades. Sci. Rep. 2019, 9, 4244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
- Gao, T.; Ding, Y.; Wu, Q.; Wang, J.; Zhang, J.; Yu, S.; Yu, P.; Liu, C.; Kong, L.; Feng, Z.; et al. Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Bacillus cereus isolated from pasteurized milk in China. Front. Microbiol. 2018, 9, 533. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Yu, P.; Yu, S.; Wang, J.; Zhang, J.; Zhang, Y.; Liao, X.; Wu, S.; Ye, Q.; Yang, X.; et al. Incidence, toxin gene profiling, antimicrobial susceptibility, and genetic diversity of Bacillus cereus isolated from quick-frozen food in China. LWT-Food Sci. Technol. 2021, 140, 110824. [Google Scholar] [CrossRef]
- Liu, C.; Yu, P.; Yu, S.; Wang, J.; Guo, H.; Zhang, Y.; Zhang, J.; Liao, X.; Li, C.; Wu, S.; et al. Assessment and molecular characterization of Bacillus cereus isolated from edible fungi in China. BMC Microbiol. 2020, 20, 310. [Google Scholar] [CrossRef]
- Yu, S.; Yu, P.; Wang, J.; Li, C.; Guo, H.; Liu, C.; Kong, L.; Yu, L.; Wu, S.; Lei, T.; et al. A study on prevalence and characterization of Bacillus cereus in ready-to-eat foods in China. Front. Microbiol. 2020, 10, 3043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Fasolato, L.; Cardazzo, B.; Carraro, L.; Fontana, F.; Novelli, E.; Balzan, S. Edible processed insects from e-commerce: Food safety with a focus on the Bacillus cereus group. Food Microbiol. 2018, 76, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibb, S.; Strimmer, K. MALDIquant: A versatile R package for the analysis of mass spectrometry data. Bioinformatics 2012, 28, 2270–2271. [Google Scholar] [CrossRef]
- López-Fernández, H.; Santos, H.M.; Capelo, J.L.; Fdez-Riverola, F.; Glez-Peña, D.; Reboiro-Jato, M. Mass-Up: An all-in-one open software application for MALDI-TOF mass spectrometry knowledge discovery. BMC Bioinform. 2015, 16, 318. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, N.; Nagai, S.; Fujita, A.; Ido, Y.; Kato, K.; Saito, A.; Moriya, Y.; Tomimatsu, Y.; Kaneta, N.; Tsujimoto, Y.; et al. Discrimination of psychrotolerant Bacillus cereus group based on MALDI-TOF MS analysis of ribosomal subunit proteins. Food Microbiol. 2020, 91, 103542. [Google Scholar] [CrossRef]
- Welker, M.; Van Belkum, A.; Girard, V.; Charrier, J.P.; Pincus, D. An update on the routine application of MALDI-TOF MS in clinical microbiology. Expert Rev. Proteom. 2019, 16, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Grenga, L.; Pible, O.; Armengaud, J. Pathogen proteotyping: A rapidly developing application of mass spectrometry to address clinical concerns. Clin. Mass Spectrom. 2019, 14, 9–17. [Google Scholar] [CrossRef]
- Lieberman, T.D.; Michel, J.B.; Aingaran, M.; Potter-Bynoe, G.; Roux, D.; Davis, M.R., Jr.; Skurnik, D.; Leiby, N.; LiPuma, J.J.; Goldberg, J.B.; et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat. Genet. 2011, 43, 1275–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Yang, J.; Duan, J.; Liu, S.; Huang, X.; Wen, X.; Huang, X.; Fu, C.; Li, J.; Dou, Q.; et al. Assessing molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae (CR-KP) with MLST and MALDI-TOF in central China. Sci. Rep. 2019, 9, 2271. [Google Scholar] [CrossRef]
- Fiedoruk, K.; Daniluk, T.; Fiodor, A.; Drewicka, E.; Buczynska, K.; Leszczynska, K.; Bideshi, D.K.; Swiecicka, I. MALDI-TOF MS portrait of emetic and non-emetic Bacillus cereus group members. Electrophoresis 2016, 37, 2235–2247. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.R.; Magnabosco, C.; Momper, L.; Gold, D.A.; Bosak, T.; Fournier, G.P. An expanded ribosomal phylogeny of Cyanobacteria supports a deep placement of plastids. Front. Microbiol. 2019, 10, 1612. [Google Scholar] [CrossRef] [Green Version]
- Emele, M.F.; Joppe, F.M.; Riedel, T.; Overmann, J.; Rupnik, M.; Cooper, P.; Kusumawati, R.L.; Berger, F.K.; Laukien, F.; Zimmermann, O.; et al. Proteotyping of Clostridioides difficile as alternate typing method to ribotyping is able to distinguish the ribotypes RT027 and RT176 from other ribotypes. Front. Microbiol. 2019, 10, 2087. [Google Scholar] [CrossRef]
- Flores-Treviño, S.; Garza-González, E.; Mendoza-Olazarán, S.; Morfín-Otero, R.; Camacho-Ortiz, A.; Rodríguez-Noriega, E.; Martínez-Meléndez, A.; Bocanegra-Ibarias, P. Screening of biomarkers of drug resistance or virulence in ESCAPE pathogens by MALDI-TOF mass spectrometry. Sci. Rep. 2019, 9, 18945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.Y.; Chung, C.R.; Wang, Z.; Li, S.; Chu, B.Y.; Horng, J.T.; Lu, J.J.; Lee, T.Y. A large-scale investigation and identification of methicillin-resistant Staphylococcus aureus based on peaks binning of matrix-assisted laser desorption ionization-time of flight MS spectra. Brief. Bioinform. 2020, 22, bbaa138. [Google Scholar] [CrossRef] [PubMed]


| Gene | Annotation | Post-Translational Modification | Theoretical Molecular Weight (Da) | Sensitivity (%) | ||
|---|---|---|---|---|---|---|
| B. cereus | B. thuringiensis | B. cereus | B. thuringiensis | |||
| rpmD | 50S ribosomal protein L30 | methionine removed | 6424.60 | 6438.62 | 100 (106/106) | 100 (175/175) |
| rpmE2 | 50S ribosomal protein L31 type B | - | 9184.33 | 9157.30 | 98.11 (104/106) | 100 (175/175) |
| rpsT | 30S ribosomal protein S20 | methionine removed | 9210.58 | 9226.58 | 100 (106/106) | 99.43 (174/175) |
| rpsF | 30S ribosomal protein S6 | - | 11,298.99 | 11,284.92 | 100 (106/106) | 98.86 (173/175) |
| rpsJ | 30S ribosomal protein S10 | methionine removed | 11,552.45 | 11,566.48 | 100 (106/106) | 100 (175/175) |
| rplR | 50S ribosomal protein L18 | - | 13,105.97 | 13,093.92 | 100 (106/106) | 100 (175/175) |
| rpsM | 30S ribosomal protein S13 | methionine removed | 13,687.83 | 13,661.79 | 100 (106/106) | 100 (175/175) |
| rplO | 50S ribosomal protein L15 | - | 15,477.74 | 15,521.79 | 100 (106/106) | 100 (175/175) |
| rplM | 50S ribosomal protein L13 | - | 16,427.96 | 16,457.98 | 100 (106/106) | 100 (175/175) |
| rpsG | 30S ribosomal protein S7 | methionine removed | 17,779.61 | 17,914.77 | 99.06 (105/106) | 100 (175/175) |
| rplF | 50S ribosomal protein L6 | methionine removed | 19,384.34 | 19,341.32 | 99.06 (105/106) | 100 (175/175) |
| rplE | 50S ribosomal protein L5 | - | 20,165.48 | 20,136.44 | 100 (106/106) | 100 (175/175) |
| rplC | 50S ribosomal protein L3 | methionine removed | 22,560.99 | 22,544.99 | 99.06 (105/106) | 99.43 (174/175) |
| Experimental m/z | Presence of Peak (%) | Protein Name | Post-Translational Modification | Theoretical m/z | Amino Acid Substitution | |
|---|---|---|---|---|---|---|
| B. cereus | B. thuringiensis | |||||
| 3211 | 97.22 (35/36) | 0.00 (0/45) | 50S ribosomal protein L30 * | methionine removed | 3213 | V → L |
| 3218 | 0.00 (0/36) | 100.00 (45/45) | 3220 | |||
| 6427 | 100.00 (36/36) | 0.00 (0/45) | 50S ribosomal protein L30 | 6426 | ||
| 6441 | 0.00 (0/36) | 100.00 (45/45) | 6440 | |||
| 9160 | 0.00 (0/36) | 100.00 (45/45) | 50S ribosomal protein L31 type B | - | 9158 | N → S, L → I |
| 9188 | 97.22 (35/36) | 0.00 (0/45) | 9185 | |||
| 9214 | 100.00 (36/36) | 0.00 (0/45) | 30S ribosomal protein S20 | methionine removed | 9212 | A → S |
| 9229 | 0.00 (0/36) | 100.00 (45/45) | 9228 | |||
| Bacterial Species | Strain | m/z of Biomarkers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 3211 | 6427 | 9188 | 9214 | 3218 | 6441 | 9160 | 9229 | ||
| Bacillus cereus | ATCC 14579 | + | + | + | + | − | − | − | − |
| Bacillus thuringiensis | ATCC 10792 | − | − | − | − | + | + | + | + |
| Bacillus megaterium | ATCC 14581 | − | − | − | − | − | − | − | − |
| Escherichia coli | ATCC 25922 | − | − | − | − | − | − | − | + |
| Escherichia coli | ATCC 8739 | − | − | − | − | − | − | − | + |
| Salmonella Enteritidis | CMCC 50335 | − | − | − | − | − | − | − | − |
| Salmonella Typhimurium | ATCC 14028 | − | − | − | − | − | − | − | − |
| Vibrio parahaemolyticus | ATCC 33847 | + | + | − | − | − | − | − | − |
| Listeria monocytogenes | ATCC 19115 | − | − | − | − | − | − | − | − |
| Staphylococcus aureus | ATCC 25923 | + | + | − | − | − | − | − | − |
| Cronobacter sakazakii | ATCC 29544 | − | − | − | − | − | − | − | − |
| Pseudomonas aeruginosa | ATCC 15442 | − | − | − | − | − | − | − | − |
| Campylobacter jejuni | ATCC 33291 | − | − | − | − | + | − | + | − |
| Yersinia enterocolitica | CMCC 52204 | − | − | − | − | − | − | − | − |
| Klebsiella pneumoniae | ATCC 700603 | − | − | − | − | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Wei, X.; Zhang, J.; Zhou, H.; Chen, N.; Wang, J.; Feng, Y.; Yu, S.; Zhang, J.; Wu, S.; et al. Differentiation of Bacillus cereus and Bacillus thuringiensis Using Genome-Guided MALDI-TOF MS Based on Variations in Ribosomal Proteins. Microorganisms 2022, 10, 918. https://doi.org/10.3390/microorganisms10050918
Chen M, Wei X, Zhang J, Zhou H, Chen N, Wang J, Feng Y, Yu S, Zhang J, Wu S, et al. Differentiation of Bacillus cereus and Bacillus thuringiensis Using Genome-Guided MALDI-TOF MS Based on Variations in Ribosomal Proteins. Microorganisms. 2022; 10(5):918. https://doi.org/10.3390/microorganisms10050918
Chicago/Turabian StyleChen, Minling, Xianhu Wei, Junhui Zhang, Huan Zhou, Nuo Chen, Juan Wang, Ying Feng, Shubo Yu, Jumei Zhang, Shi Wu, and et al. 2022. "Differentiation of Bacillus cereus and Bacillus thuringiensis Using Genome-Guided MALDI-TOF MS Based on Variations in Ribosomal Proteins" Microorganisms 10, no. 5: 918. https://doi.org/10.3390/microorganisms10050918
APA StyleChen, M., Wei, X., Zhang, J., Zhou, H., Chen, N., Wang, J., Feng, Y., Yu, S., Zhang, J., Wu, S., Ye, Q., Pang, R., Ding, Y., & Wu, Q. (2022). Differentiation of Bacillus cereus and Bacillus thuringiensis Using Genome-Guided MALDI-TOF MS Based on Variations in Ribosomal Proteins. Microorganisms, 10(5), 918. https://doi.org/10.3390/microorganisms10050918






