Effects of Chlorella vulgaris Enhancement on Endogenous Microbial Degradation of Marine Oil Spills and Community Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Crude Oil and Chemicals
2.2. Preparation of Inoculum and Experimental Medium
2.3. Biodegradation of Petroleum Hydrocarbons
2.4. Biological Activity Assay
2.5. PCR Amplification and Illumina Miseq Sequencing
2.6. Statistical Analysis
3. Results
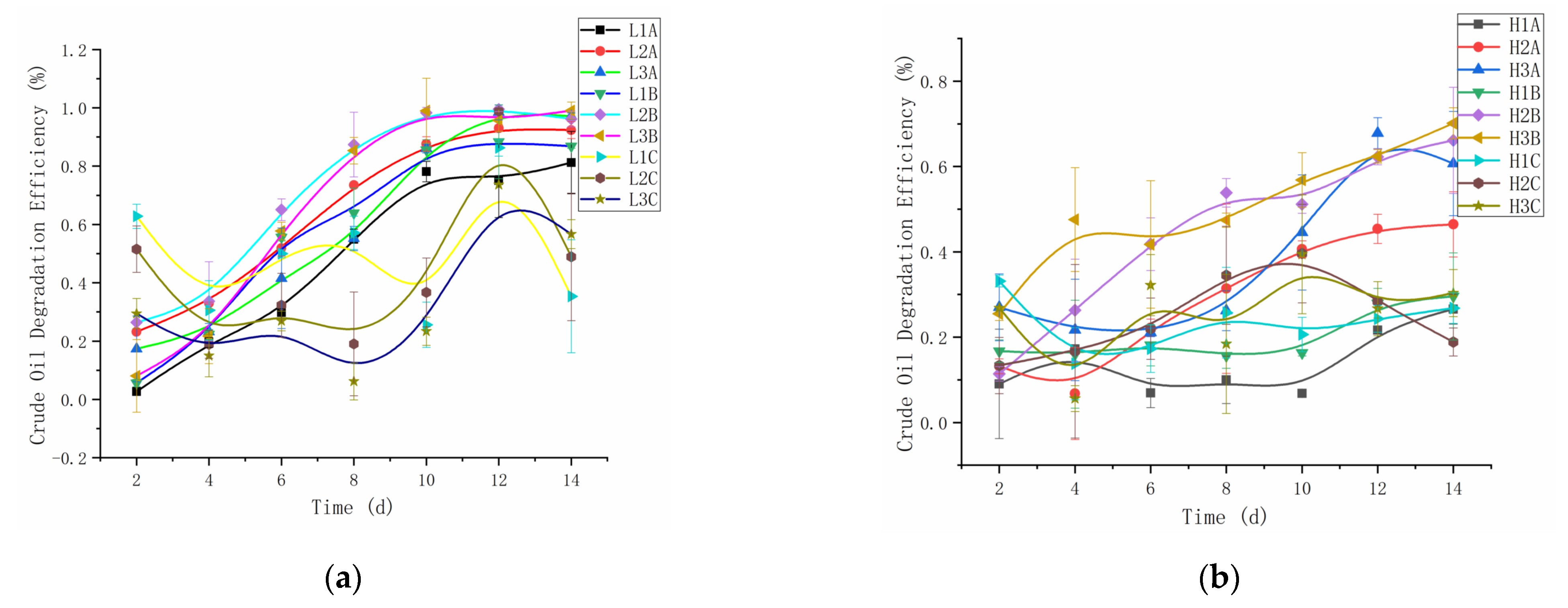
3.1. Crude Oil Biodegradation
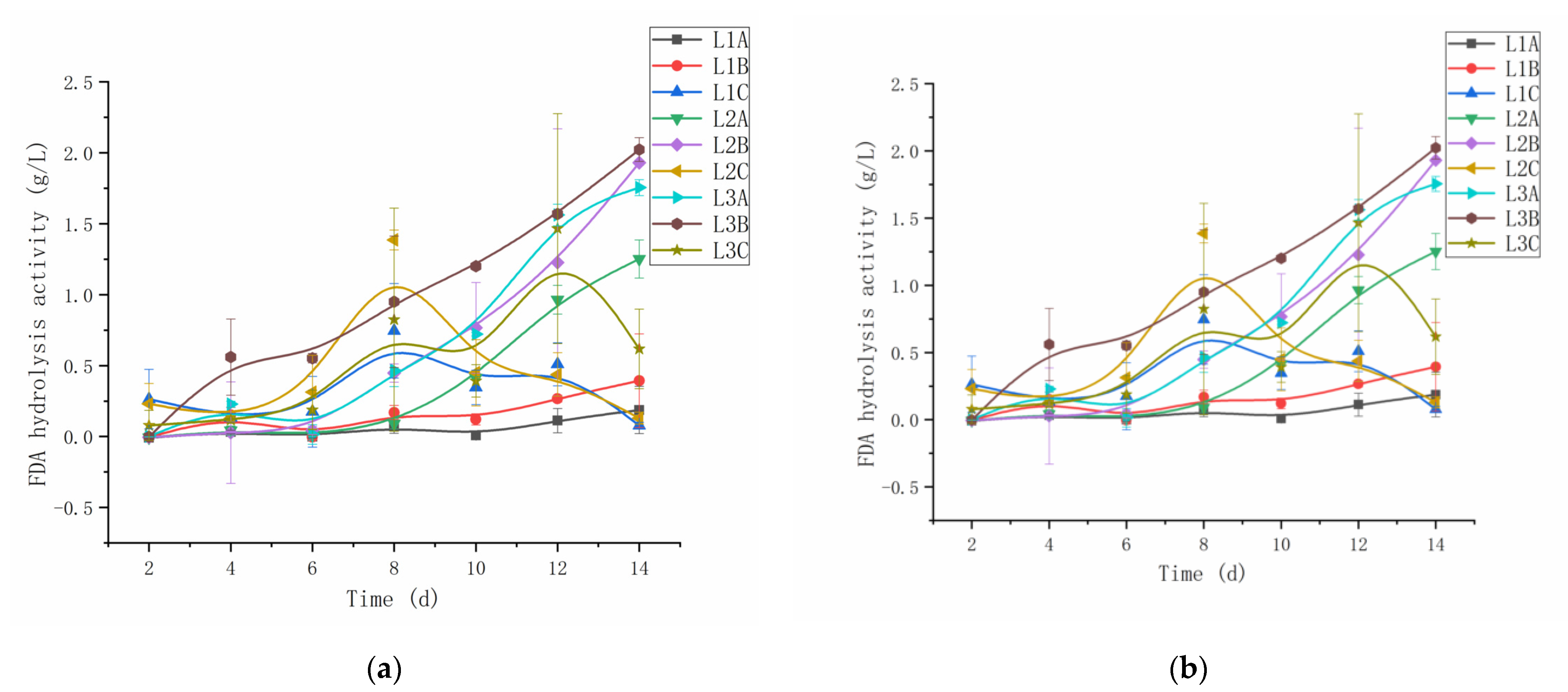
3.2. FDA Enzyme Activity
3.3. OTU Cluster Analysis
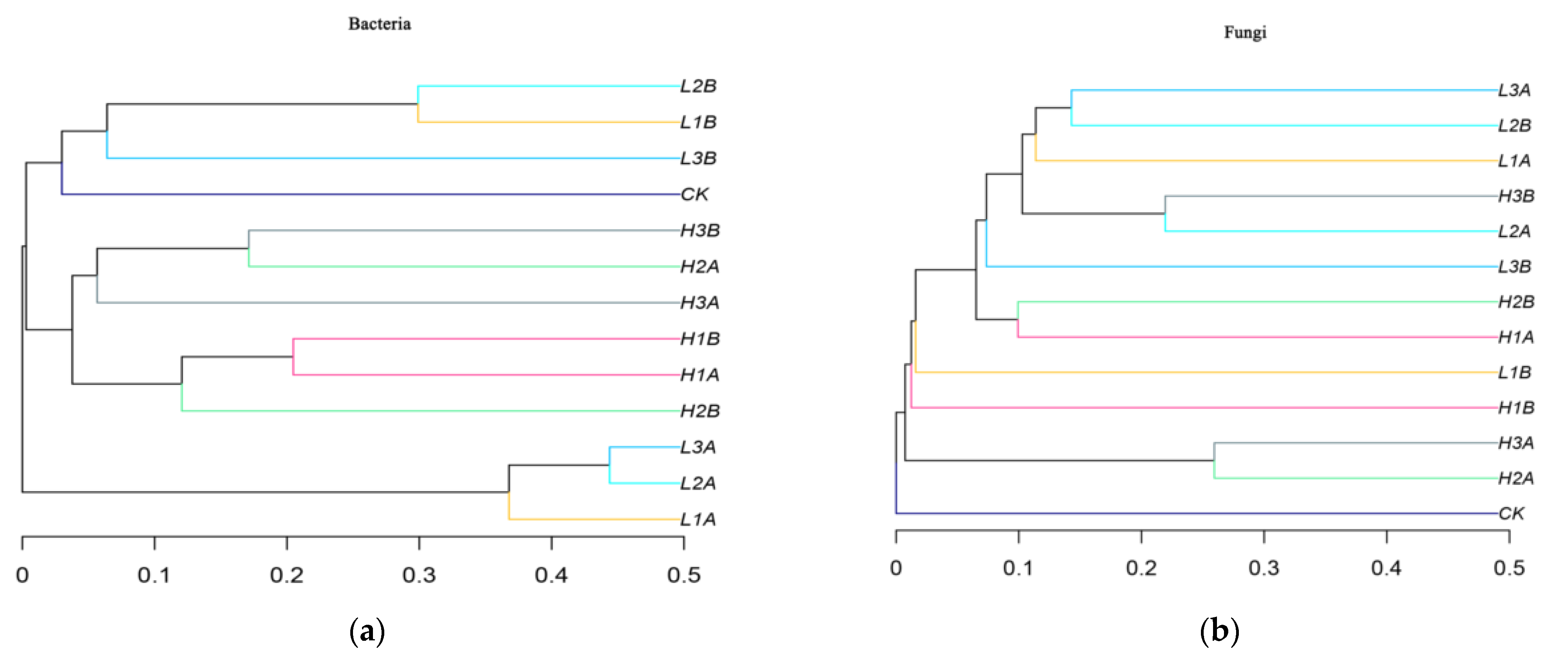
3.4. Changes in Bacterial and Fungal Diversity under the Influence of Chlorella
3.5. Effects of Chlorella on Bacterial Community Composition and Diversity
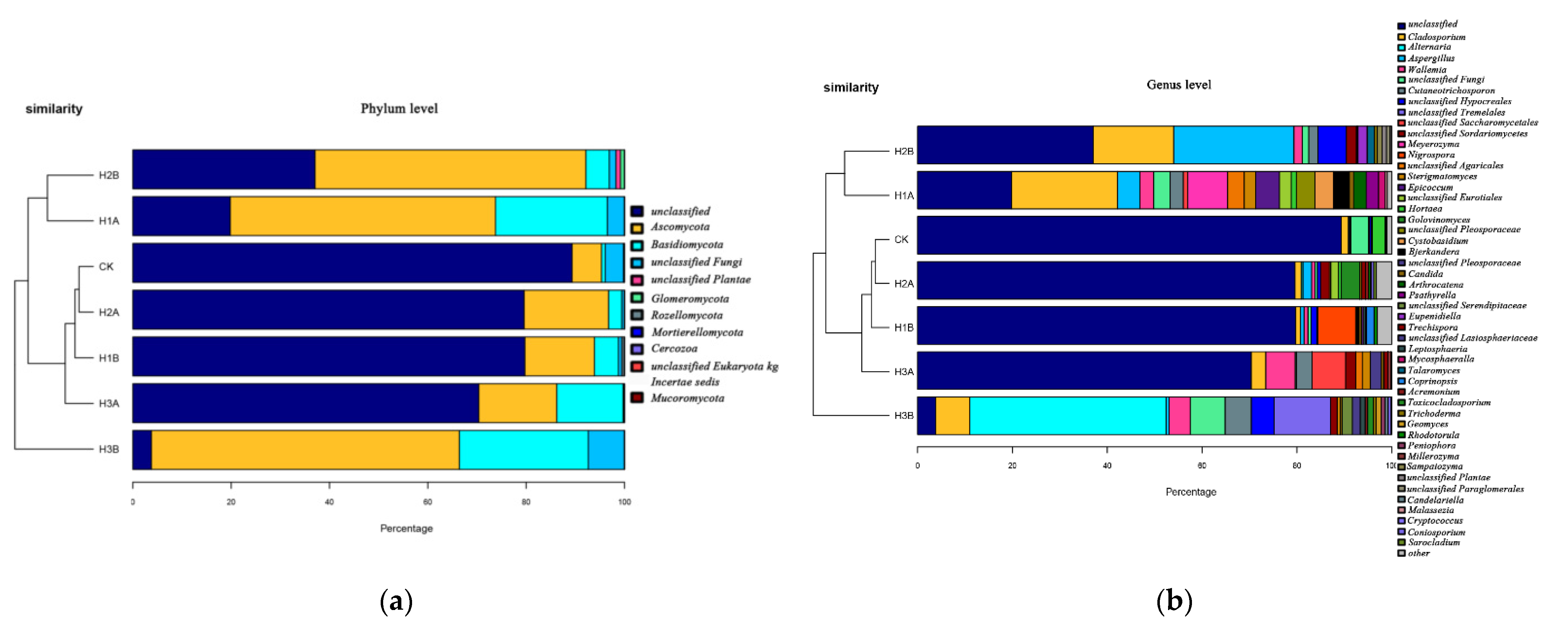
3.6. Effects of Chlorella on Fungal Community Composition and Diversity
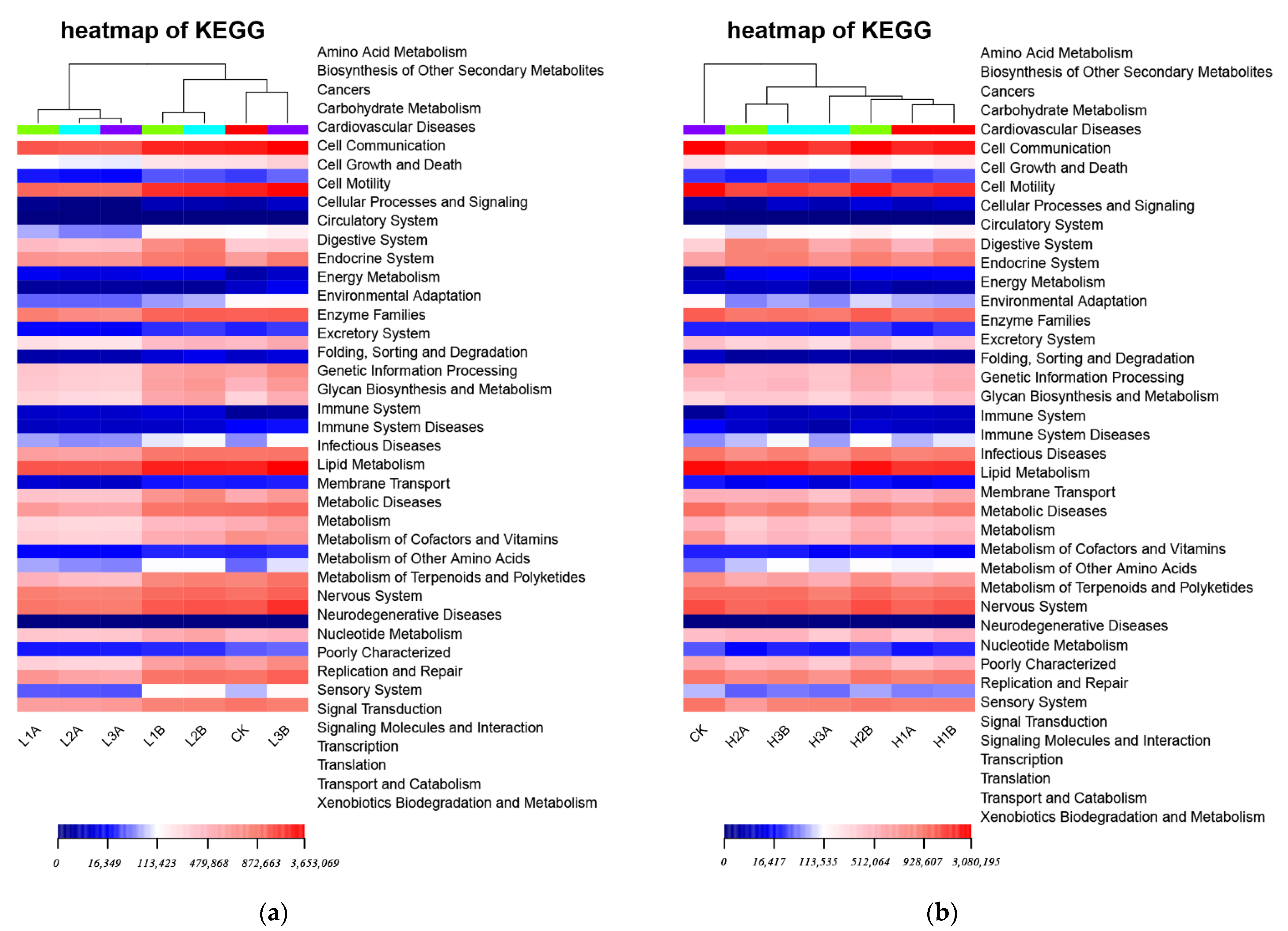
3.7. KEGG Statistics and Difference Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Q.; Bao, B.; Li, Y.; Liu, M.; Zhu, B.; Mu, J.; Chen, Z. Effects of marine oil pollution on microbial diversity in coastal waters and stimulating indigenous microorganism bioremediation with nutrients. Reg. Stud. Mar. Sci. 2020, 39, 101395. [Google Scholar] [CrossRef]
- Moradi, A.; Mosaddeghi, M.R.; Chavoshi, E.; Safadoust, A.; Soleimani, M. Effect of crude oil-induced water repellency on transport of Escherichia coli and bromide through repacked and physically-weathered soil columns. Int. J. Environ. Pollut. 2019, 255, 113230. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, L.A. Individual Stress, Collective Trauma, and social capital in the wake of the Exxon Valdez oil spill. Sociol. Inq. 2012, 82, 187–211. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Maddela, N.R.; Megharaj, M.; Venkateswarlu, K. Ecological Impacts of Total Petroleum Hydrocarbons. In Total Petroleum Hydrocarbons; Springer: Cham, Switzerland, 2020; pp. 95–138. [Google Scholar]
- Naz, S.; Iqbal, M.F.; Mahmood, I.; Allam, M. Marine oil spill detection using Synthetic Aperture Radar over Indian Ocean. Mar. Pollut. Bull. 2021, 162, 111921. [Google Scholar] [CrossRef] [PubMed]
- Perez-Cadahia, B.; Lafuente, A.; Cabaleiro, T.; Pasaro, E.; Mendez, J.; Laffon, B. Initial study on the effects of Prestige oil on human health. Environ. Int. 2007, 33, 176–185. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Q.; Zhang, Y.; Zheng, C.; Cai, B.; Liu, Y. Research on transport and weathering of oil spills in Jiaozhou Bight, China. Reg. Stud. Mar. Sci. 2022, 51, 102197. [Google Scholar] [CrossRef]
- Costa, A.S.; Romao, L.P.; Araujo, B.R.; Lucas, S.C.; Maciel, S.T.; Wisniewski, A., Jr.; Alexandre, M.R. Environmental strategies to remove volatile aromatic fractions (BTEX) from petroleum industry wastewater using biomass. Bioresour. Technol. 2012, 105, 31–39. [Google Scholar] [CrossRef]
- Souza, E.C.; Vessoni-Penna, T.C.; De Souza Oliveira, R.P. Biosurfactant-enhanced hydrocarbon bioremediation: An overview. Int. Biodeterior. Biodegradation. 2014, 89, 88–94. [Google Scholar] [CrossRef]
- Muthukumar, B.; Al Salhi, M.S.; Narenkumar, J.; Devanesan, S.; Rao, T.N.; Kim, W.; Rajasekar, A. Characterization of two novel strains of Pseudomonas aeruginosa on biodegradation of crude oil and its enzyme activities. Environ. Pollut. 2022, 304, 119223. [Google Scholar] [CrossRef]
- Li, J.; Chen, Q.; Bao, B.; Liu, M.; Bao, M.; Liu, J.; Mu, J. RNA-seq analysis reveals the significant effects of different light conditions on oil degradation by marine Chlorella vulgaris. Mar. Pollut. Bull. 2018, 137, 267–276. [Google Scholar] [CrossRef]
- Vural, C.; Vural, C.; Ozdemir, G. Monitoring of the degradation of aromatic hydrocarbons by bioaugmented activated sludge. J. Chem. Technol. Biotechnol. 2019, 95, 52–62. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, X.; Chen, X.; Kong, D.; Liu, X.; Shen, S. Synergistic degradation of crude oil by indigenous bacterial consortium and exogenous fungus Scedosporium boydii. Bioresour. Technol. 2018, 264, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Madueño, L.; Coppotelli, B.M.; Alvarez, H.M.; Morelli, I.S. Isolation and characterization of indigenous soil bacteria for bioaugmentation of PAH contaminated soil of semiarid Patagonia, Argentina. Int. Biodeterior. Biodegradation. 2011, 65, 345–351. [Google Scholar] [CrossRef]
- Semrany, S.; Favier, L.; Djelal, H.; Taha, S.; Amrane, A. Bioaugmentation: Possible solution in the treatment of Bio-Refractory Organic Compounds (Bio-ROCs). Biochem. Eng. J. 2012, 69, 75–86. [Google Scholar] [CrossRef]
- Cerqueira, V.S.; Hollenbach, E.B.; Maboni, F.; Vainstein, M.H.; Camargo, F.A.; Do Carmo, R.P.M.; Bento, F.M. Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour. Technol. 2011, 102, 11003–11010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hesnawi, R.M.; Adbeib, M.M. Effect of Nutrient Source on Indigenous Biodegradation of Diesel Fuel Contaminated Soil. APCBEE Procedia 2013, 5, 557–561. [Google Scholar] [CrossRef] [Green Version]
- Hoof, L.V. Co-management: An alternative to enforcement? ICES J. Mar. Sci. 2010, 67, 395–401. [Google Scholar] [CrossRef] [Green Version]
- Tao, K.; Liu, X.; Chen, X.; Hu, X.; Cao, L.; Yuan, X. Biodegradation of crude oil by a defined co-culture of indigenous bacterial consortium and exogenous Bacillus subtilis. Bioresour. Technol. 2017, 224, 327–332. [Google Scholar] [CrossRef]
- Stephens, E.; Ross, I.L.; King, Z.; Mussgnug, J.H.; Kruse, O.; Posten, C.; Borowitzka, M.A.; Hankamer, B. An economic and technical evaluation of microalgal biofuels. Nat. Biotechnol. 2010, 28, 126–128. [Google Scholar] [CrossRef] [Green Version]
- Xaaldi Kalhor, A.; Movafeghi, A.; Mohammadi-Nassab, A.D.; Abedi, E.; Bahrami, A. Potential of the green alga Chlorella vulgaris for biodegradation of crude oil hydrocarbons. Mar. Pollut. Bull. 2017, 123, 286–290. [Google Scholar] [CrossRef]
- Zhou, C.; Zhou, Q.; Zhang, X. Transformation of acetaminophen in natural surface water and the change of aquatic microbes. Water. Res. 2019, 148, 133–141. [Google Scholar] [CrossRef]
- Liu, R.; Wang, L.; Liu, Q.; Wang, Z.; Li, Z.; Fang, J.; Zhang, L.; Luo, M. Depth-Resolved Distribution of Particle-Attached and Free-Living Bacterial Communities in the Water Column of the New Britain Trench. Front. Microbiol. 2018, 9, 625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-Z.; Li, J.; Yi, Y.; Nobu, M.K.; Narihiro, T.; Tang, Y.-Q. Response to inhibitory conditions of acetate-degrading methanogenic microbial community. J. Biosci. Bioeng. 2020, 129, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef]
- Tang, Y.-Q.; Ji, P.; Lai, G.-L.; Chi, C.-Q.; Liu, Z.-S.; Wu, X.-L. Diverse microbial community from the coalbeds of the Ordos Basin, China. Int. J. Coal Geol. 2012, 90–91, 21–33. [Google Scholar] [CrossRef]
- Pi, Y.-R.; Bao, M.-T. Investigation of kinetics in bioaugmentation of crude oil via high-throughput sequencing: Enzymatic activities, bacterial community composition and functions. Pet. Sci. 2022. [Google Scholar] [CrossRef]
- Wang, H.; Wang, B.; Dong, W.; Hu, X. Co-acclimation of bacterial communities under stresses of hydrocarbons with different structures. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; He, L.Y.; Tao, X.Q.; Dang, Z.; Guo, C.L.; Lu, G.N.; Yi, X.Y. Construction of an artificial microalgal-bacterial consortium that efficiently degrades crude oil. J. Hazard. Mater. 2010, 181, 1158–1162. [Google Scholar] [CrossRef]
- Daccò, C.; Girometta, C.; Asemoloye, M.D.; Carpani, G.; Picco, A.M.; Tosi, S. Key fungal degradation patterns, enzymes and their applications for the removal of aliphatic hydrocarbons in polluted soils: A review. Int. Biodeterior. Biodegrad. 2020, 147, 104866. [Google Scholar] [CrossRef]
- Jacques, R.J.S.; Okeke, B.C.; Bento, F.M.; Teixeira, A.S.; Peralba, M.C.R.; Camargo, F.A. Microbial consortium bioaugmentation of a polycyclic aromatic hydrocarbons contaminated soil. Bioresour. Technol. 2008, 99, 2637–2643. [Google Scholar] [CrossRef]
- Wang, F.; Li, C.; Wang, H.; Chen, W.; Huang, Q. Characterization of a phenanthrene-degrading microbial consortium enriched from petrochemical contaminated environment. Int. Biodeterior. Biodegrad. 2016, 115, 286–292. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A Comprehensive Review of Aliphatic Hydrocarbon Biodegradation by Bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Meckenstock, R.U.; Boll, M.; Mouttaki, H.; Koelschbach, J.S.; Cunha Tarouco, P.; Weyrauch, P.; Dong, X.; Himmelberg, A.M. Anaerobic Degradation of Benzene and Polycyclic Aromatic Hydrocarbons. J. Mol. Microbiol. Biotechnol. 2016, 26, 92–118. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.K.; Harayama, S. Biodegradation of n-alkylcycloalkanes and n-alkylbenzenes via new pathways in Alcanivorax sp. strain MBIC 4326. Appl. Environ. Microbiol. 2001, 67, 1970–1974. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, T.; Aitken, M.D. Role of methylotrophs in the degradation of hydrocarbons during the Deepwater Horizon oil spill. ISME J. 2014, 8, 2543–2545. [Google Scholar] [CrossRef] [Green Version]
- Ali Khan, A.H.; Tanveer, S.; Anees, M.; Muhammad, Y.S.; Iqbal, M.; Yousaf, S. Role of nutrients and illuminance in predicting the fate of fungal mediated petroleum hydrocarbon degradation and biomass production. J. Environ. Manag. 2016, 176, 54–60. [Google Scholar] [CrossRef]
- Shinde, V.L.; Suneel, V.; Shenoy, B.D. Diversity of bacteria and fungi associated with tarballs: Recent developments and future prospects. Mar. Pollut. Bull. 2017, 117, 28–33. [Google Scholar] [CrossRef]
- Brown, L.M.; Gunasekera, T.S.; Bowen, L.L.; Ruiz, O.N. Draft Genome Sequence of Rhodovulum sp. Strain NI22, a Naphthalene-Degrading Marine Bacterium. Genome. Announc. 2015, 3, 11–12. [Google Scholar] [CrossRef]
- Zhou, J.; Li, H.; Chen, X.; Wan, D.; Mai, W.; Sun, C. Cometabolic degradation of low-strength coking wastewater and the bacterial community revealed by high-throughput sequencing. Bioresour. Technol. 2017, 245, 379–385. [Google Scholar] [CrossRef]
- Chebbi, A.; Hentati, D.; Zaghden, H.; Baccar, N.; Rezgui, F.; Chalbi, M.; Sayadi, S.; Chamkha, M. Polycyclic aromatic hydrocarbon degradation and biosurfactant production by a newly isolated Pseudomonas sp. strain from used motor oil-contaminated soil. Int. Biodeterior. Biodegrad. 2017, 122, 128–140. [Google Scholar] [CrossRef]
- Rabodonirina, S.; Rasolomampianina, R.; Krier, F.; Drider, D.; Merhaby, D.; Net, S.; Ouddane, B. Degradation of fluorene and phenanthrene in PAHs-contaminated soil using Pseudomonas and Bacillus strains isolated from oil spill sites. J. Environ. Manag. 2019, 232, 1–7. [Google Scholar] [CrossRef] [PubMed]



| Classification | Sample | Barcode | Raw_num | Mean_len | Clean_num | Mean_len |
|---|---|---|---|---|---|---|
| Bacteria | CK | GTCCCA | 54,361 | 447.82 | 54,241 | 405.60 |
| L1A | ATCGCA | 86,397 | 465.70 | 86,238 | 424.52 | |
| L1B | TTACGA | 113,931 | 453.07 | 113,690 | 411.30 | |
| L2A | TGTTAT | 92,096 | 467.21 | 91,959 | 426.35 | |
| L2B | GCCATC | 133,024 | 451.07 | 132,736 | 409.36 | |
| L3A | TGTGTT | 126,295 | 467.90 | 126,121 | 427.23 | |
| L3B | TAGGAC | 68,698 | 451.40 | 68,644 | 410.60 | |
| H1A | TGGACG | 45,333 | 459.60 | 45,296 | 418.81 | |
| H1B | CGATGT | 59,305 | 458.41 | 59,250 | 417.59 | |
| H2A | TCCTGT | 113,531 | 464.18 | 113,409 | 422.99 | |
| H2B | GAAGGC | 44,009 | 449.19 | 43,957 | 407.94 | |
| H3A | ATGTCA | 44,843 | 458.82 | 44,805 | 417.50 | |
| H3B | CGGTTA | 83,506 | 456.44 | 83,394 | 415.26 | |
| Fungi | CK | TACGACA | 90,066 | 326.10 | 89,182 | 283.14 |
| L1A | TGTGCTA | 161,561 | 162.87 | 75,719 | 236.57 | |
| L1B | TCACTCG | 67,333 | 344.81 | 66,047 | 305.03 | |
| L2A | TACTCTG | 94,398 | 235.68 | 77,000 | 232.07 | |
| L2B | TGGACTC | 99,821 | 284.55 | 98,355 | 243.28 | |
| L3A | TCTGTAG | 95,305 | 265.14 | 88,416 | 237.09 | |
| L3B | TACACTC | 89,859 | 251.21 | 81,584 | 224.43 | |
| H1A | TGATCGC | 84,263 | 271.11 | 80,680 | 235.59 | |
| H1B | TCGTCAT | 116,317 | 263.11 | 80,979 | 288.47 | |
| H2A | TATCAGC | 97,088 | 247.41 | 89,636 | 217.09 | |
| H2B | TGCGACG | 85,628 | 265.37 | 77,701 | 236.30 | |
| H3A | ATACAGA | 134,335 | 206.03 | 116,955 | 178.19 | |
| H3B | ATGTCTC | 93,896 | 264.41 | 90,499 | 227.13 |
| Classification | Sample | Seq_num | OTU | Shannon | ACE | Chao1 | Coverage | Simpson |
|---|---|---|---|---|---|---|---|---|
| Bacteria | CK | 37,907 | 907 | 2.9526 | 1025.1650 | 958.0203 | 0.9954 | 0.2005 |
| H1A | 40,297 | 771 | 3.2891 | 1185.5860 | 1086.3910 | 0.9919 | 0.1139 | |
| H1B | 50,637 | 920 | 3.1865 | 1302.4010 | 1144.3850 | 0.9935 | 0.1328 | |
| H2A | 93,149 | 1226 | 2.5940 | 1594.5820 | 1461.0970 | 0.9958 | 0.2437 | |
| H2B | 20,090 | 571 | 3.8727 | 901.8986 | 808.0522 | 0.9884 | 0.0624 | |
| H3A | 36,790 | 574 | 2.3597 | 861.5187 | 760.2338 | 0.9935 | 0.1981 | |
| H3B | 57,011 | 881 | 3.2164 | 1270.6230 | 1127.2820 | 0.9940 | 0.1033 | |
| L1A | 80,987 | 888 | 2.3545 | 1183.1880 | 1135.9590 | 0.9964 | 0.2548 | |
| L1B | 75,251 | 897 | 3.7709 | 1113.4030 | 1038.3030 | 0.9969 | 0.0656 | |
| L2A | 86,414 | 854 | 1.7350 | 1109.2600 | 1045.9510 | 0.9969 | 0.3662 | |
| L2B | 63,471 | 703 | 3.7776 | 852.4927 | 811.2891 | 0.9974 | 0.0511 | |
| L3A | 120,588 | 915 | 1.4163 | 1173.5610 | 1069.3960 | 0.9978 | 0.4090 | |
| L3B | 57,488 | 944 | 2.8666 | 1303.8270 | 1195.3190 | 0.9943 | 0.1677 | |
| Fungi | CK | 87,785 | 400 | 3.2318 | 416.1471 | 405.1786 | 0.9997 | 0.0898 |
| H1A | 80,081 | 219 | 3.2326 | 563.8095 | 333.4872 | 0.9988 | 0.0588 | |
| H1B | 79,488 | 354 | 3.1260 | 370.2629 | 357.9545 | 0.9996 | 0.0946 | |
| H2A | 88,090 | 242 | 2.3100 | 290.8538 | 270.5577 | 0.9994 | 0.2681 | |
| H2B | 77,243 | 173 | 2.9895 | 322.7502 | 241.0556 | 0.9994 | 0.0969 | |
| H3A | 116,515 | 205 | 2.0583 | 260.2814 | 238.5490 | 0.9995 | 0.3007 | |
| H3B | 89,889 | 173 | 2.4210 | 246.1856 | 224.4839 | 0.9994 | 0.2019 | |
| L1A | 75,342 | 152 | 3.0662 | 352.5439 | 219.5357 | 0.9992 | 0.0722 | |
| L1B | 62,445 | 325 | 1.8302 | 359.2254 | 336.0588 | 0.9992 | 0.3602 | |
| L2A | 76,472 | 198 | 2.8612 | 303.8780 | 285.5676 | 0.9989 | 0.0950 | |
| L2B | 97,354 | 184 | 3.4065 | 229.1768 | 213.4000 | 0.9995 | 0.0540 | |
| L3A | 87,757 | 148 | 2.7871 | 228.3164 | 199.2069 | 0.9994 | 0.1173 | |
| L3B | 80,723 | 202 | 3.4413 | 274.5173 | 242.5263 | 0.9993 | 0.0579 |
| Phylum | Genus | Abundance Ratio % | ||||||
|---|---|---|---|---|---|---|---|---|
| CK | L1A | L1B | L2A | L2B | L3A | L3B | ||
| Proteobacteria | Oceanicola | 1.14 | 0.09 | 17.59 | 0.02 | 11.04 | 0.01 | 1.62 |
| Rhodovulum | 0.13 | 0 | 9.74 | 0.01 | 9.71 | 0 | 0.34 | |
| Erythrobacter | 0.42 | 0.36 | 1.27 | 0 | 0.54 | 0.02 | 20.08 | |
| Methylophaga | 0 | 1.97 | 2.89 | 1.58 | 2.77 | 0.01 | 0.95 | |
| Pseudomonas | 1.59 | 70.12 | 3.27 | 84.21 | 4.57 | 89.77 | 0.23 | |
| Burkholderia | 0.04 | 1.42 | 0.1 | 2.2 | 0.17 | 2.73 | 0.01 | |
| Ralstonia | 0.01 | 0.77 | 0.06 | 1.06 | 0.08 | 1.41 | 0.01 | |
| Planctomycetes | Planctomicrobium | 0.02 | 0 | 9.72 | 0 | 13.78 | 0 | 3.17 |
| Gimesia | 0.03 | 0 | 3.63 | 0 | 9.74 | 0 | 0.71 | |
| Bacteroidetes | Phaeodactylibacter | 0.27 | 0.02 | 0.92 | 0.04 | 1.78 | 0.02 | 13.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Z.; Liu, M.; Bao, B.; Guo, J.; Tao, H.; Zhu, B.; Chen, Q. Effects of Chlorella vulgaris Enhancement on Endogenous Microbial Degradation of Marine Oil Spills and Community Diversity. Microorganisms 2022, 10, 905. https://doi.org/10.3390/microorganisms10050905
Song Z, Liu M, Bao B, Guo J, Tao H, Zhu B, Chen Q. Effects of Chlorella vulgaris Enhancement on Endogenous Microbial Degradation of Marine Oil Spills and Community Diversity. Microorganisms. 2022; 10(5):905. https://doi.org/10.3390/microorganisms10050905
Chicago/Turabian StyleSong, Zhao, Mei Liu, Bo Bao, Jian Guo, Hengcong Tao, Baikang Zhu, and Qingguo Chen. 2022. "Effects of Chlorella vulgaris Enhancement on Endogenous Microbial Degradation of Marine Oil Spills and Community Diversity" Microorganisms 10, no. 5: 905. https://doi.org/10.3390/microorganisms10050905
APA StyleSong, Z., Liu, M., Bao, B., Guo, J., Tao, H., Zhu, B., & Chen, Q. (2022). Effects of Chlorella vulgaris Enhancement on Endogenous Microbial Degradation of Marine Oil Spills and Community Diversity. Microorganisms, 10(5), 905. https://doi.org/10.3390/microorganisms10050905







