Identification and Validation of Toxoplasma gondii Mitoribosomal Large Subunit Components
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Searches
2.2. Parasite Culture
2.3. DNA Cloning and Parasite Genetic Manipulation
2.4. Immunofluorescent Assay
2.5. Growth Assay
2.6. Native and Denaturing Gels, in Gel Activity Assay and Western Blot
2.7. Immuno-Precipitation
2.8. RTqPCR
3. Results
3.1. Identification of New Toxoplasma LSU Component Candidates
3.2. Endogenous Tagging Confirms the Mitochondrial Localization of TguL24m and Provides Evidence for Its Association with the Toxoplasma Mitoribosome
3.3. Depletion of Each of TgbL35m, TgbL36m and TgbL28m Results in Toxoplasma Growth Defect
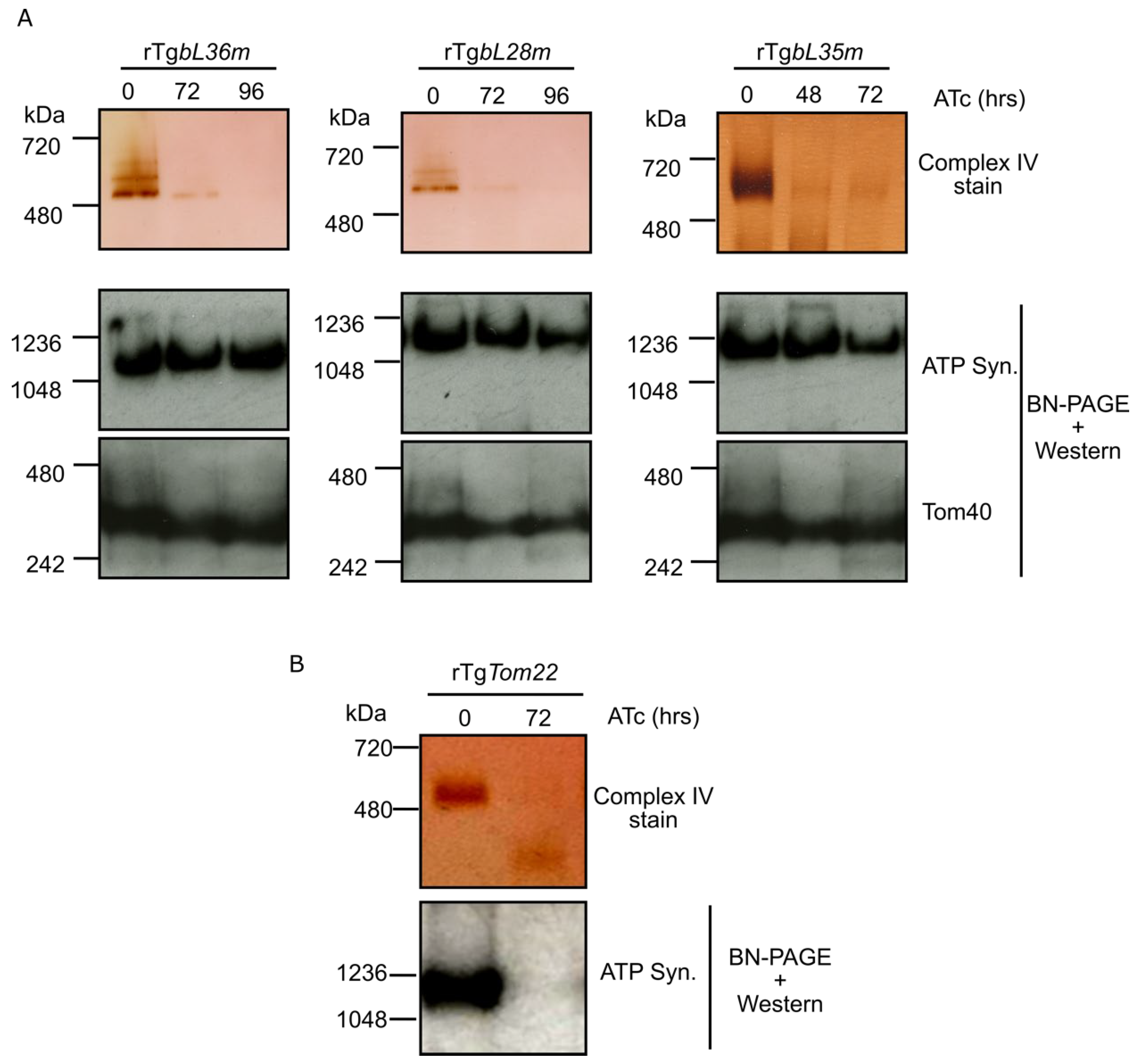
3.4. Depletion of Each of TgbL35m, TgbL36m and TgbL28m Results in a Mitochondrial Translation Defect
3.5. A Genetic Interaction Occurs between Genes Encoding LSU Components in Toxoplasma
4. Discussion
4.1. Identification of New Apicomplexan Mitoribosomal Proteins in This Study
4.2. Identification of New Apicomplexan Mitoribosomal Proteins with No Known Homologs
4.3. Proof of Principle for Methods to Assay Apicomplexan Mitoribosome Assembly and Function
4.4. Validation That TguL24m, a Homolog of a Key Component of Nascent Chain Protection in Human Mitoribosome, Is Part of the Toxoplasma Mitoribosome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ling, L.; Mulaka, M.; Munro, J.; Dass, S.; Mather, M.W.; Riscoe, M.K.; Llinás, M.; Zhou, J.; Ke, H. Genetic Ablation of the Mitoribosome in the Malaria Parasite Plasmodium Falciparum Sensitizes It to Antimalarials That Target Mitochondrial Functions. J. Biol. Chem. 2020, 295, 7235–7248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, H.; Dass, S.; Morrisey, J.M.; Mather, M.W.; Vaidya, A.B. The Mitochondrial Ribosomal Protein L13 Is Critical for the Structural and Functional Integrity of the Mitochondrion in Plasmodium Falciparum. J. Biol. Chem. 2018, 293, 8128–8137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacombe, A.; Maclean, A.E.; Ovciarikova, J.; Tottey, J.; Mühleip, A.; Fernandes, P.; Sheiner, L. Identification of the Toxoplasma Gondii Mitochondrial Ribosome, and Characterisation of a Protein Essential for Mitochondrial Translation. Mol. Microbiol. 2019, 112, 1235–1252. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.D.; Pasaje, C.F.A.; Kennedy, K.; McFadden, G.I.; Ralph, S.A. Targeting Protein Translation in Organelles of the Apicomplexa. Trends Parasitol. 2016, 32, 953–965. [Google Scholar] [CrossRef]
- Dass, S.; Mather, M.W.; Ke, H. Divergent Mitochondrial Ribosomes in Unicellular Parasitic Protozoans. Trends Parasitol. 2020, 36, 318–321. [Google Scholar] [CrossRef]
- De Carvalho, L.P.; Kreidenweiss, A.; Held, J. Drug Repurposing: A Review of Old and New Antibiotics for the Treatment of Malaria: Identifying Antibiotics with a Fast Onset of Antiplasmodial Action. Molecules 2021, 26, 2304. [Google Scholar] [CrossRef]
- Martijn, J.; Vosseberg, J.; Guy, L.; Offre, P.; Ettema, T.J.G. Deep Mitochondrial Origin Outside the Sampled Alphaproteobacteria. Nature 2018, 557, 101–105. [Google Scholar] [CrossRef]
- Amunts, A.; Brown, A.; Toots, J.; Scheres, S.H.W.; Ramakrishnan, V. The Structure of the Human Mitochondrial Ribosome. Science 2015, 348, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Greber, B.J.; Bieri, P.; Leibundgut, M.; Leitner, A.; Aebersold, R.; Boehringer, D.; Ban, N. The Complete Structure of the 55S Mammalian Mitochondrial Ribosome. Science 2015, 348, 303–308. [Google Scholar] [CrossRef] [Green Version]
- Desai, N.; Brown, A.; Amunts, A.; Ramakrishnan, V. The Structure of the Yeast Mitochondrial Ribosome. Science 2017, 355, 528–531. [Google Scholar] [CrossRef] [Green Version]
- Amunts, A.; Brown, A.; Bai, X.-C.; Llácer, J.L.; Hussain, T.; Emsley, P.; Long, F.; Murshudov, G.; Scheres, S.H.W.; Ramakrishnan, V. Structure of the Yeast Mitochondrial Large Ribosomal Subunit. Science 2014, 343, 1485–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waltz, F.; Nguyen, T.-T.; Arrivé, M.; Bochler, A.; Chicher, J.; Hammann, P.; Kuhn, L.; Quadrado, M.; Mireau, H.; Hashem, Y.; et al. Small Is Big in Arabidopsis Mitochondrial Ribosome. Nat. Plants 2019, 5, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Waltz, F.; Soufari, H.; Bochler, A.; Giegé, P.; Hashem, Y. Cryo-EM Structure of the RNA-Rich Plant Mitochondrial Ribosome. Nat. Plants 2020, 6, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Waltz, F.; Salinas-Giegé, T.; Englmeier, R.; Meichel, H.; Soufari, H.; Kuhn, L.; Pfeffer, S.; Förster, F.; Engel, B.D.; Giegé, P.; et al. How to Build a Ribosome from RNA Fragments in Chlamydomonas Mitochondria. Nat. Commun. 2021, 12, 7176. [Google Scholar] [CrossRef]
- Ramrath, D.J.F.; Niemann, M.; Leibundgut, M.; Bieri, P.; Prange, C.; Horn, E.K.; Leitner, A.; Boehringer, D.; Schneider, A.; Ban, N. Evolutionary Shift toward Protein-Based Architecture in Trypanosomal Mitochondrial Ribosomes. Science 2018, 362, eaau7735. [Google Scholar] [CrossRef]
- Soufari, H.; Waltz, F.; Parrot, C.; Durrieu-Gaillard, S.; Bochler, A.; Kuhn, L.; Sissler, M.; Hashem, Y. Structure of the Mature Kinetoplastids Mitoribosome and Insights into Its Large Subunit Biogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 29851–29861. [Google Scholar] [CrossRef]
- Jacot, D.; Lourido, S.; Meissner, M.; Sheiner, L.; Soldati-Favre, D.; Striepen, B. Genetic Manipulation of Toxoplasma Gondii, 3rd ed.; Weiss, L.M., Kim, K.B.T.-T., Eds.; Academic Press: Cambridge, MA, USA, 2020; Chapter 20; pp. 897–940. [Google Scholar] [CrossRef]
- Feagin, J.E.; Harrell, M.I.; Lee, J.C.; Coe, K.J.; Sands, B.H.; Cannone, J.J.; Tami, G.; Schnare, M.N.; Gutell, R.R. The Fragmented Mitochondrial Ribosomal RNAs of Plasmodium Falciparum. PLoS ONE 2012, 7, e38320. [Google Scholar] [CrossRef]
- Namasivayam, S.; Baptista, R.P.; Xiao, W.; Hall, E.M.; Doggett, J.S.; Troell, K.; Kissinger, J.C. A Novel Fragmented Mitochondrial Genome in the Protist Pathogen Toxoplasma Gondii and Related Tissue Coccidia. Genome Res. 2021, 31, 852–865. [Google Scholar] [CrossRef]
- Feagin, J.E.; Werner, E.; Gardner, M.J.; Williamson, D.H.; Wilson, R.J. Homologies between the Contiguous and Fragmented RRNAs of the Two Plasmodium Falciparum Extrachromosomal DNAs Are Limited to Core Sequences. Nucleic Acids Res. 1992, 20, 879–887. [Google Scholar] [CrossRef] [Green Version]
- Feagin, J.E.; Mericle, B.L.; Werner, E.; Morris, M. Identification of Additional RRNA Fragments Encoded by the Plasmodium Falciparum 6 Kb Element. Nucleic Acids Res. 1997, 25, 438–446. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Curt-Varesano, A.; Braun, L.; Ranquet, C.; Hakimi, M.-A.; Bougdour, A. The Aspartyl Protease TgASP5 Mediates the Export of the Toxoplasma GRA16 and GRA24 Effectors into Host Cells. Cell. Microbiol. 2016, 18, 151–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheiner, L.; Demerly, J.L.; Poulsen, N.; Beatty, W.L.; Lucas, O.; Behnke, M.S.; White, M.W.; Striepen, B. A Systematic Screen to Discover and Analyze Apicoplast Proteins Identifies a Conserved and Essential Protein Import Factor. PLOS Pathog. 2011, 7, e1002392. [Google Scholar] [CrossRef] [Green Version]
- Van Dooren, G.G.; Yeoh, L.M.; Striepen, B.; McFadden, G.I. The Import of Proteins into the Mitochondrion of Toxoplasma Gondii. J. Biol. Chem. 2016, 291, 19335–19350. [Google Scholar] [CrossRef] [Green Version]
- Maclean, A.E.; Bridges, H.R.; Silva, M.F.; Ding, S.; Ovciarikova, J.; Hirst, J.; Sheiner, L. Complexome Profile of Toxoplasma Gondii Mitochondria Identifies Divergent Subunits of Respiratory Chain Complexes Including New Subunits of Cytochrome Bc1 Complex. PLoS Pathog. 2021, 17, e1009301. [Google Scholar] [CrossRef]
- Gupta, A.; Shah, P.; Haider, A.; Gupta, K.; Siddiqi, M.I.; Ralph, S.A.; Habib, S. Reduced Ribosomes of the Apicoplast and Mitochondrion of Plasmodium spp. and Predicted Interactions with Antibiotics. Open Biol. 2014, 4, 140045. [Google Scholar] [CrossRef] [Green Version]
- Barylyuk, K.; Koreny, L.; Ke, H.; Butterworth, S.; Crook, O.M.; Lassadi, I.; Gupta, V.; Tromer, E.; Mourier, T.; Stevens, T.J.; et al. A Comprehensive Subcellular Atlas of the Toxoplasma Proteome via HyperLOPIT Provides Spatial Context for Protein Functions. Cell Host Microbe 2020, 28, 752–766.e9. [Google Scholar] [CrossRef]
- Itoh, Y.; Andréll, J.; Choi, A.; Richter, U.; Maiti, P.; Best, R.B.; Barrientos, A.; Battersby, B.J.; Amunts, A. Mechanism of Membrane-Tethered Mitochondrial Protein Synthesis. Science 2021, 371, 846–849. [Google Scholar] [CrossRef]
- Huynh, M.-H.; Carruthers, V.B. Tagging of Endogenous Genes in a Toxoplasma Gondii Strain Lacking Ku80. Eukaryot. Cell 2009, 8, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Sidik, S.M.; Huet, D.; Ganesan, S.M.; Huynh, M.-H.; Wang, T.; Nasamu, A.S.; Thiru, P.; Saeij, J.P.J.; Carruthers, V.B.; Niles, J.C.; et al. A Genome-Wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 2016, 166, 1423–1435.e12. [Google Scholar] [CrossRef] [Green Version]
- Huet, D.; Rajendran, E.; van Dooren, G.G.; Lourido, S. Identification of Cryptic Subunits from an Apicomplexan ATP Synthase. eLife 2018, 7, e38097. [Google Scholar] [CrossRef] [PubMed]
- Salunke, R.; Mourier, T.; Banerjee, M.; Pain, A.; Shanmugam, D. Highly Diverged Novel Subunit Composition of Apicomplexan F-Type ATP Synthase Identified from Toxoplasma Gondii. PLoS Biol. 2018, 16, e2006128. [Google Scholar] [CrossRef] [PubMed]
- Mühleip, A.; Kock Flygaard, R.; Ovciarikova, J.; Lacombe, A.; Fernandes, P.; Sheiner, L.; Amunts, A. ATP Synthase Hexamer Assemblies Shape Cristae of Toxoplasma Mitochondria. Nat. Commun. 2021, 12, 120. [Google Scholar] [CrossRef]
- Tomal, A.; Kwasniak-Owczarek, M.; Janska, H. An Update on Mitochondrial Ribosome Biology: The Plant Mitoribosome in the Spotlight. Cells 2019, 8, 1562. [Google Scholar] [CrossRef] [Green Version]
- Robles, P.; Quesada, V. Emerging Roles of Mitochondrial Ribosomal Proteins in Plant Development. Int. J. Mol. Sci. 2017, 18, 2595. [Google Scholar] [CrossRef] [Green Version]
- Hillebrand, A.; Matz, J.M.; Almendinger, M.; Müller, K.; Matuschewski, K.; Schmitz-Linneweber, C. Identification of Clustered Organellar Short (Cos) RNAs and of a Conserved Family of Organellar RNA-Binding Proteins, the Heptatricopeptide Repeat Proteins, in the Malaria Parasite. Nucleic Acids Res. 2018, 46, 10417–10431. [Google Scholar] [CrossRef]



| Gene ID (Name) | Previous Publication/Homology | Mitoprot Score | CRISPR Screen Score | Plaque Defect? |
|---|---|---|---|---|
| TGME49_226280 (TgbL28m) | [3,27] | 0.9971 | −4.91 | Yes |
| TGME49_222180 (TgbL36m) | homolog of Trypanosoma bL36m | 0.7884 | −3.96 | Yes |
| TGME49_320005 (TgbL35m) | homolog of Arabidopsis bL35m | 0.9989 | −4.99 | Yes |
| TGME49_216010 (TguL24m) | homology of human uL24m | 0.492 | −3.66 | -- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shikha, S.; Silva, M.F.; Sheiner, L. Identification and Validation of Toxoplasma gondii Mitoribosomal Large Subunit Components. Microorganisms 2022, 10, 863. https://doi.org/10.3390/microorganisms10050863
Shikha S, Silva MF, Sheiner L. Identification and Validation of Toxoplasma gondii Mitoribosomal Large Subunit Components. Microorganisms. 2022; 10(5):863. https://doi.org/10.3390/microorganisms10050863
Chicago/Turabian StyleShikha, Shikha, Mariana Ferreira Silva, and Lilach Sheiner. 2022. "Identification and Validation of Toxoplasma gondii Mitoribosomal Large Subunit Components" Microorganisms 10, no. 5: 863. https://doi.org/10.3390/microorganisms10050863
APA StyleShikha, S., Silva, M. F., & Sheiner, L. (2022). Identification and Validation of Toxoplasma gondii Mitoribosomal Large Subunit Components. Microorganisms, 10(5), 863. https://doi.org/10.3390/microorganisms10050863






