Azospirillum spp. from Plant Growth-Promoting Bacteria to Their Use in Bioremediation
Abstract
:1. Introduction
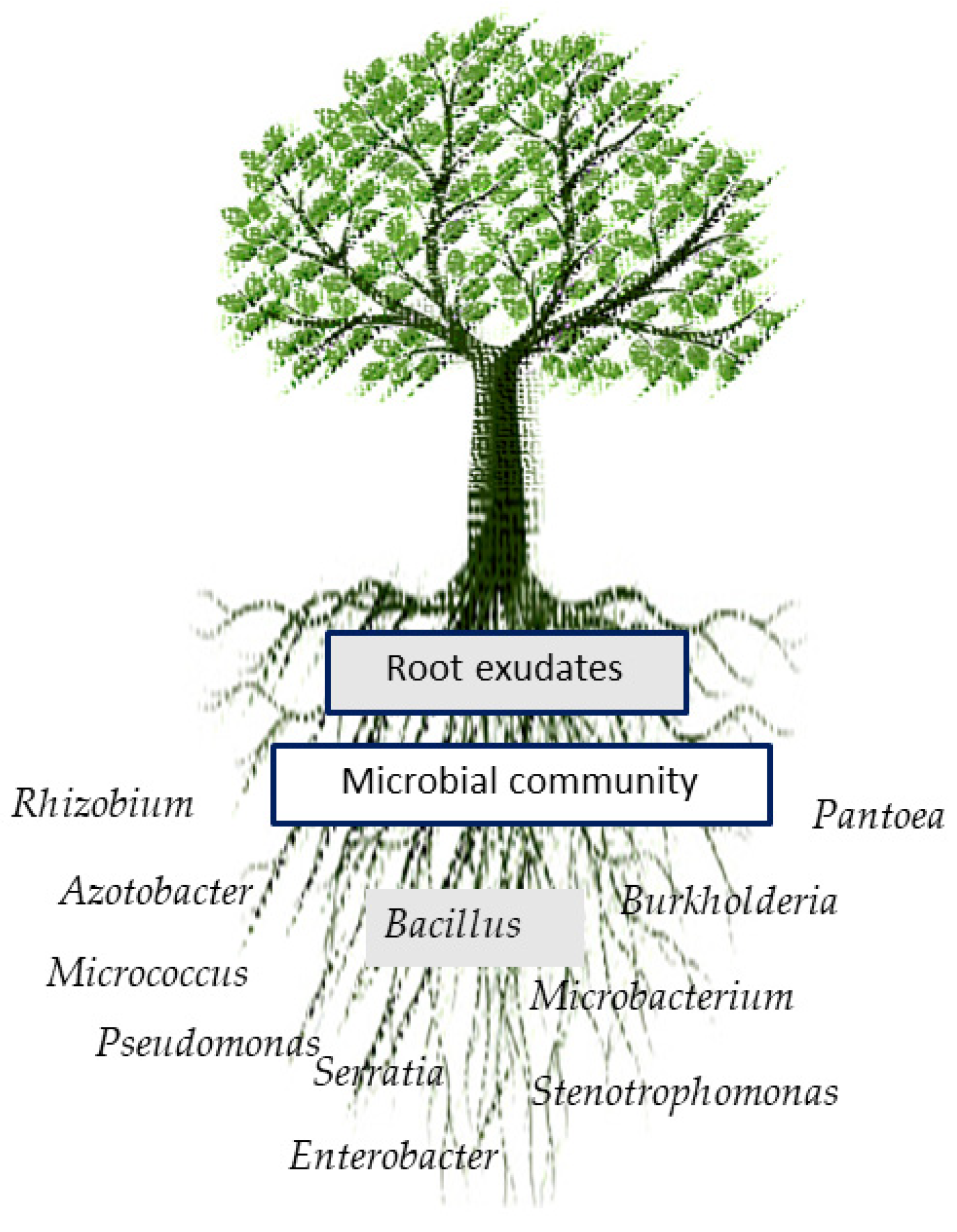
2. Plant Growth Promotion
3. Azospirillum
4. Genetics of Azospirillum Species
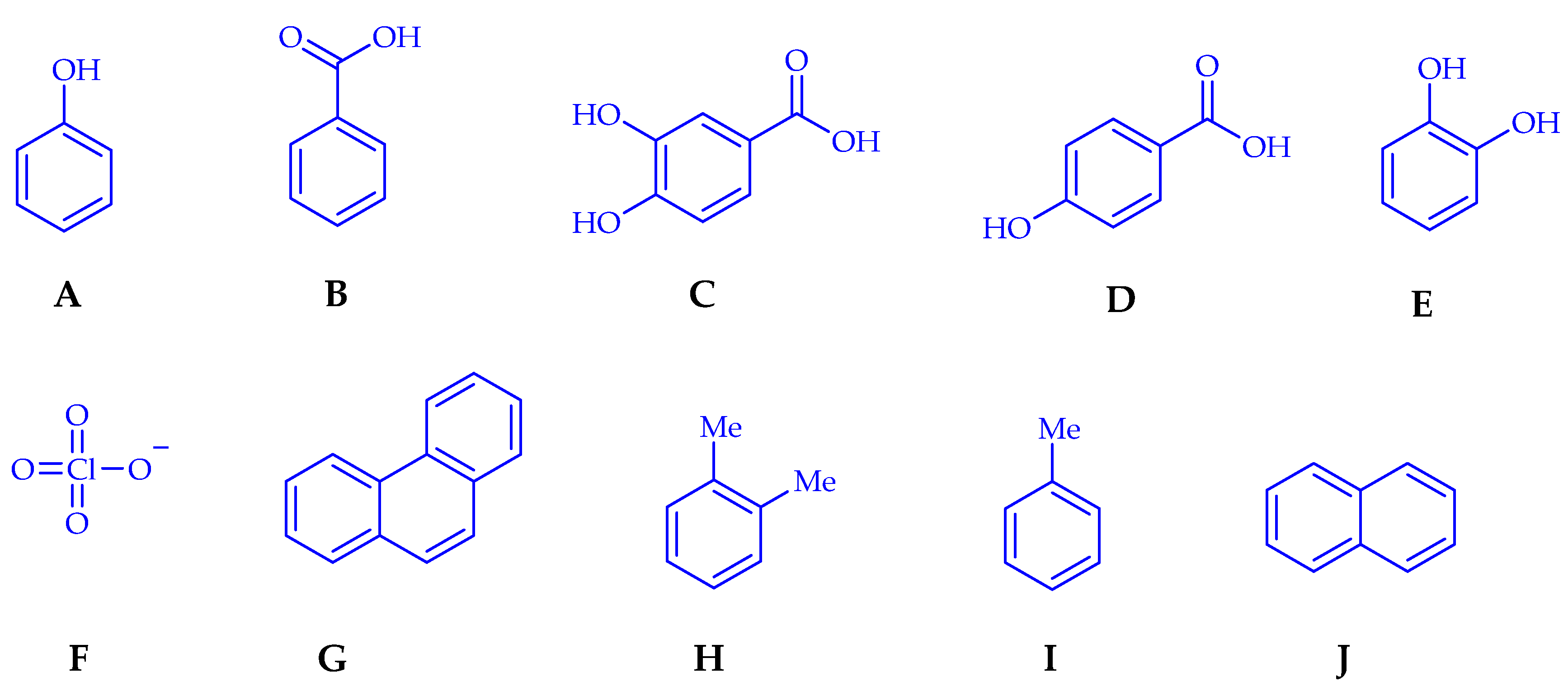
5. Degradation of Xenobiotics by Azospirillum Species
6. Tolerance of Heavy Metals in Azospirillum Species
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Asad, S.A.; Farooq, M.; Afzal, A.; West, H. Integrated phytobial heavy metal remediation strategies for a sustainable clean environment—A review. Chemosphere 2019, 217, 925–941. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.H.; Zhang, S.A.; Mo, X.D.; Li, Y.C.; Fu, Y.Q.; Liu, Z.G. Effects of plant growth-promoting rhizobacteria and N source on plant growth and N and P uptake by tomato grown on calcareous soils. Pedosphere 2017, 27, 1027–1036. [Google Scholar] [CrossRef]
- Núñez, R.R.; Lorenzo, M.; Ortiz, E.; Oramas, J. Biorremediación de la contaminación de petróleo en el mar. Rev. Electron. Agencia Medio Ambiente 2010, 10, 1–6. [Google Scholar]
- Wuana, R.A.; Okieimen, F.E. Heavy metals in contaminated soils: A review of sources, chemistry, risks and best available strategies for remediation. ISRN Ecol. 2011, 2011, 402647. [Google Scholar] [CrossRef] [Green Version]
- Chun, S.J.; Kim, Y.J.; Cui, Y.; Nam, K.H. Ecological network analysis reveals distinctive microbial modules associated with heavy metal contamination of abandoned mine soils in Korea. Environ. Pollut. 2021, 289, 117851. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Zhu, H.; Bing, H.; Tian, X.; Wang, Z.; Wang, X.; Wu, Y. Contamination, sources and health risk of heavy metals in soil and dust from different functional areas in an industrial city of Panzhihua City, Southwest China. J. Hazard. Mater. 2021, 420, 126638. [Google Scholar] [CrossRef]
- Sun, Z.; Mou, X.; Tong, C.; Wang, C.; Xie, Z.; Song, H.; Sun, W.; Lv, Y. Spatial variations and bioaccumulation of heavy metals in intertidal zone of the Yellow River estuary, China. Catena 2015, 126, 43–52. [Google Scholar] [CrossRef]
- Xu, F.; Qiu, L.; Cao, Y.; Huang, J.; Liu, Z.; Tian, X.; Li, A.; Yin, X. Trace metals in the surface sediments of the intertidal Jiaozhou Bay, China: Sources and contamination assessment. Mar. Pollut. Bull. 2016, 104, 371–378. [Google Scholar] [CrossRef]
- Gerhardt, K.E.; MacNeill, G.J.; Gerwing, P.D.; Greenberg, B.M. Phytoremediation of Salt-Impacted Soils and Use of Plant Growth-Promoting Rhizobacteria (PGPR) to Enhance Phytoremediation. In Phytoremediation; Ansari, A., Gill, S., Guy, R., Lanza, G., Newman, L., Eds.; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Pandey, S.; Ghoshb, S.; Misra, A.K. Synthesis of a Trisaccharide and a Tetrasaccharide from the Cell-Wall Lipopolysaccharides of Azospirillum brasilense S17. Synthesis 2009, 15, 2584–2590. [Google Scholar] [CrossRef]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fert. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Umesha, S.; Singh, P.K.; Singh, R.P. Microbial Biotechnology and Sustainable Agriculture. In Biotechnology for Sustainable Agriculture; Chapter 6; Singh, R.L., Monda, S., Eds.; Woodhead Publishing: Cambridge, UK; Elsevier: Chennai, India, 2018; pp. 185–205. [Google Scholar] [CrossRef]
- Lu, Q.; Weng, Y.; You, Y.; Xu, Q.; Li, H.; Li, Y.; Liu, H.; Du, S. Inoculation with abscisic acid (ABA)-catabolizing bacteria can improve phytoextraction of heavy metal in contaminated soil. Environ. Poll. 2020, 257, 113497. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Glick, B.R. The role of plant growth-promoting bacteria in metal phytoremediation. Adv. Microb. Physiol. 2017, 71, 97–132. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growthpromoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [Green Version]
- Kannapiran, E.; Ramkumar, V.S. Inoculation effect of nitrogen-fixing and phosphate-solubilizing bacteria to promote growth of black gram (Phaseolus mungo Roxb; Eng). Ann. Biol. Res. 2011, 2, 615–621. [Google Scholar]
- Mishra, P.K.; Joshi, P.; Suyal, P.; Bisht, J.K.; Bhatt, J.C. Potential of Phosphate Solubilising Microorganisms in Crop Production. In Bioresources for Sustainable Plant Nutrient Management; Satish Serial Publishing House: New Delhi, India, 2014; pp. 201–222. [Google Scholar]
- Mamta, R.P.; Pathania, V.; Gulati, A.; Singh, B.; Bhanwra, R.K.; Tewari, R. Stimulatory effect of phosphate-solubilizing bacteria on plant growth, stevioside and rebaudioside-A contents of Stevia rebaudiana Bertoni. Appl. Soil Ecol. 2010, 46, 222–229. [Google Scholar] [CrossRef]
- Sulbarán, M.; Pérez, E.; Ball, M.M.; Bahsas, A.; Yarzábal, L.A. Characterization of the mineral phosphate-solubilizing activity of Pantoea aglomerans MMB051 isolated from an iron-rich soil in southeastern Venezuela (Bolívar State). Curr. Microbiol. 2009, 58, 378–383. [Google Scholar] [CrossRef] [Green Version]
- Santos, S.; Neto, I.F.F.; Machado, M.D.; Soares, H.M.V.M.; Soares, E.V. Siderophore production by Bacillus megaterium: Effect of growth phase and cultural conditions. Appl. Biochem. Biotechnol. 2014, 172, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Mehnaz, S. Secondary Metabolites of Pseudomonas aurantiaca and Their Role in Plant Growth Promotion. In Plant Microbe Symbiosis: Fundamentals and Advances; Arora, N.K., Ed.; Springer: New Delhi, India, 2013; pp. 373–393. [Google Scholar]
- Jiang, Z.; Chen, M.; Yu, X.; Xie, Z. 7-Hydroxytropolone produced and utilized as an iron-scavenger by Pseudomonas donghuensis. BioMetals 2016, 29, 817–826. [Google Scholar] [CrossRef]
- Cruz-Hernández, M.A.; Reyes-Peralta, J.; Mendoza-Herrera, A.; Rivera, G.; Bocanegra-García, V. Characterization of a Microbacterium sp. Strain isolated from soils contaminated with hydrocarbons in the Burgos basin, Mexico. Rev. Int. Contam. Ambient. 2021, 37, 227–235. [Google Scholar] [CrossRef]
- Lalitha, S. Plant Growth–Promoting Microbes: A Boon for Sustainable Agriculture. In Sustainable Agriculture towards Food Security; Dhanarajan, A., Ed.; Springer: Singapore, 2017; pp. 125–158. [Google Scholar] [CrossRef]
- Kaushal, M.; Wani, S.P. Plant-growth-promoting rhizobacteria: Drought stress alleviators to ameliorate crop production in drylands. Ann. Microbiol. 2016, 66, 35–42. [Google Scholar] [CrossRef]
- Vasseur-Coronado, M.; Dupré du Boulois, H.; Pertot, I.; Puopolo, G. Selection of plant growth promoting rhizobacteria sharing suitable features to be commercially developed as biostimulant products. Microbiol. Res. 2021, 245, 126672. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R.A. Comprehensive Review of Aliphatic Hydrocarbon Biodegradation by Bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef]
- Bashan, Y.; Bashan, L.E. How the Plant Growth-Promoting Bacterium Azospirillum Promotes Plant Growth—A Critical Assessment. In Advances in Agronomy; Sparks, D.L., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 108, pp. 78–122. [Google Scholar] [CrossRef]
- Souza, R.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Dos Santos, J.; Maranho, L. Rhizospheric microorganisms as a solution for the recovery of soils contaminated by petroleum: A review. J. Environ. Manag. 2018, 210, 104–113. [Google Scholar] [CrossRef]
- Díaz-Zorita, M.; Fernandez-Canigia, M.V.; Bravo, O.A.; Berger, A.; Satorre, E.H. Field Evaluation of Extensive Crops Inoculated with Azospirillum sp. In Handbook for Azospirillum, Technical Issues and Protocols; Cassan, F.D., Okon, Y., Creus, C.M., Eds.; Springer: Cham, Switzerland, 2015; pp. 435–445. [Google Scholar] [CrossRef]
- García, J.E.; Maroniche, G.; Creus, C.; Suárez-Rodríguez, R.; Ramírez-Trujillo, J.A.; Groppa, M.D. In vitro PGPR properties and osmotic tolerance of different Azospirillum native strains and their effects on growth of maize under drought stress. Microbiol. Res. 2017, 202, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Machado, H.B.; Funayama, S.; Rigo, L.U.; Pedrosa, F.O. Excretion of ammonium by Azospirillum brasilense mutants resistant to ethylenediamine. Can. J. Microbial. 1991, 37, 549–553. [Google Scholar] [CrossRef]
- Santos, K.F.D.N.; Moure, V.R.; Hauer, V.; Santos, A.R.S.; Donatti, L.; Galvao, C.W.; Pedrosa, F.O.; Souza, E.M.; Wassem, R.; Steffens, M.B.R. Wheat colonization by an Azospirillum brasilense ammonium-excreting strain reveals upregulation of nitrogenase and superior plant growth promotion. Plant Soil 2017, 415, 245–255. [Google Scholar] [CrossRef]
- Ben Dekhil, S.; Cahill, M.; Stackebrandt, E.; Sly, L.I. Transfer of Conglomeromonas largomobilis subsp. largomobilis to the genus Azospirillum as Azospirillum largomobile comb. nov., and elevation of Conglomeromonas largomobilis subsp. parooensis to the new type species of Conglomeromonas, Conglomeromonas parooensis sp. nov. Syst. Appl. Microbiol. 1997, 20, 72–77. [Google Scholar] [CrossRef]
- Xie, C.-H.; Yokota, A. Azospirillum oryzae sp. nov., a nitrogen-fixing bacterium isolated from the roots of the rice plant Oryza sativa. Int. J. Syst. Evol. Microbiol. 2005, 55, 1435–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beijerinck, M.W. Uber ein Spirillum, welches frein Stickstoff binden kann? Zentralbl. Bakteriol. Parasitenkd. Infektionskr. 1925, 63, 353–359. [Google Scholar]
- Khammas, K.M.; Ageron, E.; Grimont, P.A.D.; Kaiser, P. Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and. Res. Microbiol. 1991, 140, 679–693. [Google Scholar] [CrossRef]
- Lin, S.Y.; Shen, F.T.; Young, L.S.; Zhu, Z.L.; Chen, W.M.; Young, C.C. Azospirillum formosense sp. nov., a diazotroph from agricultural soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 1185–1190. [Google Scholar] [CrossRef]
- Lavrinenko, K.; Chernousova, E.; Gridneva, E.; Dubinina, G.; Akimov, V.; Kuever, J.; Lysenko, A.; Grabovich, M. Azospirillum thiophilum sp. nov., a diazotrophic bacterium isolated from a sulfide spring. Int. J. Syst. Evol. Microbiol. 2010, 60, 2832–2837. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Zhang, R.; Feng, J.; Wang, C.; Chen, J. Azospirillum griseum sp. nov., isolated from lakewater. Int. J. Syst. Evol. Microbiol. 2019, 69, 3676–3681. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, X.J.; Liu, H.C.; Zhou, Y.G.; Wu, X.L.; Nie, Y.; Kang, Y.Q.; Cai, M. Azospirillum oleiclasticum sp. nov, a nitrogen-fixing and heavy oil degrading bacterium isolated from an oil production mixture of Yumen Oilfield. Syst. Appl. Microbiol. 2021, 44, 126171. [Google Scholar] [CrossRef]
- Young, C.C.; Hupfer, H.; Siering, C.; Ho, M.J.; Arun, A.B.; Lai, W.A.; Rekha, P.D.; Shen, F.T.; Hung, M.H.; Chen, W.M.; et al. Azospirillum rugosum sp. nov., isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2008, 58, 959–963. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.Y.; Young, C.C.; Hupfer, H.; Siering, C.; Arun, A.B.; Chen, W.M.; Lai, W.A.; Shen, F.T.; Rekha, P.D.; Yassin, A.F. Azospirillum picis sp. nov., isolated from discarded tar. Int. J. Syst. Evol. Microbiol. 2009, 59, 761–765. [Google Scholar] [CrossRef]
- Lin, S.Y.; Liu, Y.C.; Hameed, A.; Hsu, Y.H.; Lai, W.A.; Shen, F.T.; Young, C.C. Azospirillum fermentarium sp. nov., a nitrogen-fixing species isolated from a fermenter. Int. J. Syst. Evol. Microbiol. 2013, 63, 3762–3768. [Google Scholar] [CrossRef]
- Zhou, S.; Han, L.; Wang, Y.; Yang, G.; Li, Z.; Hu, P. Azospirillum humicireducens sp. nov., a nitrogen-fixing bacterium isolated from a microbial fuel cell. Int. J. Syst. Evol. Microbiol. 2013, 63, 2618–2624. [Google Scholar] [CrossRef] [Green Version]
- Tarrand, J.J.; Krieg, N.R.; Dobereiner, J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azosporillum gen. nov. and two species, Azospirillum lipoferum (Reijerinckia) comb., nov. and Azospirillumbrasilense sp. nov. Can. J. Microbiol. 1979, 24, 967–980. [Google Scholar] [CrossRef]
- Reinhold, B.; Hurek, T.; Fendrik, I.; Pot, B.; Gillis, M.; Kersters, K.; Thielemans, S.; De Ley, J. Azospirillum halopraeferens sp. nov., a nitrogen-fixing organism associated with roots of kallar grass (Leptochloa fusca (L.) kunth). Int. J. Syst. Bacteriol. 1987, 37, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Eckert, B.; Weber, O.B.; Kirchhof, G.; Halbritter, A.; Stoffels, M.; Hartmann, A.; Kirchhof, G.; Halbritter, A.; Eckert, B. Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int. J. Syst. Evol. Microbiol. 2001, 51, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Peng, G.; Wang, H.; Zhang, G.; Hou, W.; Liu, Y.; Wang, E.T.; Tan, Z. Azospirillum melinis sp. nov., a group of diazotrophs isolated from tropical molasses grass. Int. J. Syst. Evol. Microbiol. 2006, 56, 1263–1271. [Google Scholar] [CrossRef] [Green Version]
- Mehnaz, S.; Weselowski, B.; Lazarovits, G. Azospirillum canadense sp. nov., a nitrogen-fixing bacterium isolated from corn rhizosphere. Int. J. Syst. Evol. Microbiol. 2007, 57, 620–624. [Google Scholar] [CrossRef]
- Mehnaz, S.; Weselowski, B.; Lazarovits, G. Azospirillum zeae sp. nov., a diazotrophic bacterium isolated from rhizosphere soil of Zea mays. Int. J. Syst. Evol. Microbiol. 2007, 57, 2805–2809. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wei, W.; Wang, X.; Xu, L.; Lai, R. Azospirillum palatum sp. nov. isolated from forest soil in Zhejiang province, China. J. Gen. Appl. Microbiol. 2009, 55, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.Y.; Hameed, A.; Liu, Y.C.; Hsu, Y.H.; Lai, W.A.; Shen, F.T.; Young, C.C. Azospirillum soli sp. nov., a nitrogen-fixing species isolated from agricultural soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 4601–4607. [Google Scholar] [CrossRef]
- Young, C.C.; Lin, S.Y.; Hameed, A.; Liu, Y.C.; Hsu, Y.H.; Huang, H.I.; Lai, W.A. Azospirillum agricola sp. nov., a nitrogen-fixing species isolated from cultivated soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1453–1458. [Google Scholar] [CrossRef]
- Tikhonova, E.N.; Grouzdev, D.S.; Kravchenko, I.K. Azospirillum palustre sp. nov., a methylotrophic nitrogen-fixing species isolated from raised bog. Int. J. Syst. Evol. Microbiol. 2019, 69, 2787–2793. [Google Scholar] [CrossRef] [PubMed]
- Anandham, R.; Heo, J.; Krishnamoorthy, R.; SenthilKumar, M.; Gopal, N.O.; Kim, S.J.; Kwon, S.W. Azospirillum ramasamyi sp. nov., a novel diazotrophic bacterium isolated from fermented bovine products. Int. J. Syst. Evol. Microbiol. 2019, 69, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Ferreira, N.; Hayashi Sant’ Anna, F.; Massena Reis, V.; Ambrosini, A.; Gazolla Volpiano, C.; Rothballer, M.; Schwab, S.; Baura, V.A.; Balsanelli, E.; Pedrosa, F.O.; et al. Genome-based reclassification of Azospirillum brasilense Sp245 as the type strain of Azospirillum baldaniorum sp. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 6203–6212. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.L.; Ming, H.; Ding, C.L.; Ji, W.L.; Cheng, L.J.; Niu, M.M.; Zhang, Y.M.; Zhang, L.Y.; Meng, X.L.; Nie, G.X. Azospirillum thermophilum sp. nov., isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2020, 70, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Bashan, Y. Ecology and application of Azospirillum and other plant growth-promoting bacteria (PGPB)—Special issue. Eur. J. Soil Biol. 2009, 45, 1–2. [Google Scholar] [CrossRef]
- Martin-Didonet, C.C.G.; Chubatsu, L.S.; Souza, E.M.; Kleina, M.; Rego, F.G.; Rigo, M.L.U.; Yates, M.G.; Pedrosa, F.O. Genome structure of the genus Azospirillum. J. Bacteriol. 2000, 182, 4113–4116. [Google Scholar] [CrossRef] [Green Version]
- Wisniewski-Dyé, F.; Lozano, L.; Acosta-Cruz, E.; Borland, S.; Drogue, B.; Prigent-Combaret, C.; Rouy, Z.; Barbe, V.; Mendoza Herrera, A.; González, V.; et al. Genome Sequence of Azospirillum brasilense CBG497 and Comparative Analyses of Azospirillum Core and Accessory Genomes provide Insight into Niche Adaptation. Genes 2012, 3, 576–602. [Google Scholar] [CrossRef]
- Kwak, Y.; Shin, J.H. First Azospirillum genome from aquatic environments: Whole-genome sequence of Azospirillum thiophilum BV-S T, a novel diazotroph harboring a capacity of sulfur-chemolithotrophy from a sulfide spring. Mar. Genom. 2016, 25, 21–24. [Google Scholar] [CrossRef]
- García, J.E.; Labarthe, M.; Pagnussat, L.; Amenta, M.; Creus, C.M.; Maroniche, G.A. Signs of a phyllospheric lifestyle in the genome of the stress-tolerant strain Azospirillum brasilense Az19. Syst. Appl. Microbiol. 2020, 43, 126130. [Google Scholar] [CrossRef]
- Pérez Castañeda, L.M.; Cruz Hernández, M.A.; Mendoza Herrera, A. Variabilidad genética de aislamientos no-típicos de Azospirillum brasilense por análisis PCR-RFLP del ADN 16S ribosomal. Phyton 2011, 80, 27–34. [Google Scholar]
- Barkovskii, A.L.; Korshunova, V.E.; Pozdnyacova, L.I. Catabolism of phenol and benzoate by Azospirillum strains. Appl. Soil Ecol. 1995, 2, 17–24. [Google Scholar] [CrossRef]
- López-de-Victoria, G.; Lowell, C.R. Chemotaxis of Azospirillum species to aromatic compounds. Appl. Environ. Microbiol. 1993, 59, 2951–2955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckford, R.; Cook, D.; Saul, D.; Aislabie, J.; Foght, J. Free-Living Heterotrophic Nitrogen-Fixing Bacteria Isolated from Fuel-Contaminated Antarctic Soils. Appl. Environ. Microbiol. 2002, 68, 5181–5185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nozawa, M.; Scow, K.M.; Rolston, D.E. Reduction of Perchlorate and Nitrate by Microbial Communities in Vadose Soil. Appl. Environ. Microbiol. 2005, 71, 3928–3934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muratova, A.Y.; Turkovskaya, O.V.; Antonyuk, L.P.; Makarov, O.E.; Pozdnyakova, L.I.V.; Ignatov, V. Oil-Oxidizing Potential of Associative Rhizobacteria of the Genus Azospirillum. Microbiology 2005, 74, 210–215. [Google Scholar] [CrossRef]
- Miranda-Martínez, M.R.; Delgadillo-Martínez, J.; Alarcón, A.; Ferrera-Cerrato, R. Degradación de fenantreno por microorganismos en la rizósfera del pasto alemán. Terra Latinoam. 2007, 25, 25–33. [Google Scholar]
- Cruz-Hernández, M.A.; Jimenez-Andrade, J.M.; Herrera, A.M. Characterization of the degradation potential of xenobiotic compounds by the rhizobacteria Azospirillum brasilense. Mex. J. Biotechnol. 2019, 4, 10–22. [Google Scholar] [CrossRef]
- Johnsen, A.R.; Wick, L.Y.; Harms, H. Principles of microbial PAH-degradation in soil. Environ. Pollut. 2005, 133, 71–84. [Google Scholar] [CrossRef]
- Chong, H.; Li, Q. Degradación de fenantreno por microorganismos en la rizósfera del pasto alemán. Microb. Cell Fact. 2017, 16, 137. [Google Scholar] [CrossRef] [Green Version]
- Nievas, M.L.; Commendatore, M.G.; Esteves, J.L.; Bucalá, V. Biodegradation pattern of hydrocarbons from a fuel oil-type complex residue by an emulsifier-producing microbial consortium. J. Hazard. Mater. 2008, 154, 96–104. [Google Scholar] [CrossRef]
- Hmidet, N.; Ayed, H.; Jacques, P.; Nasri, M. Enhancement of Surfactin and Fengycin Production by Bacillus mojavensis A21: Application for Diesel Biodegradation. BioMed Res. Int. 2017, 2017, 5893123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supaphol, S.; Jenkins, S.N.; Intomo, P.; Waite, I.S.; O’Donnell, A.G. Microbial community dynamics in mesophilic anaerobic co-digestion of mixed waste. Bioresour. Technol. 2011, 102, 4021–4027. [Google Scholar] [CrossRef] [PubMed]
- Maimona, S.; Noshin, I.; Muhammad, A.; Muhammad, S.; Iftikhar, A.; Arghya, B. Development of a plant microbiome bioremediation system for crude oil contamination. J. Environ. Chem. Eng. 2021, 9, 105401. [Google Scholar] [CrossRef]
- Xing, S.; Min, D.; Jiawei, Y.; Lin, S.; Xiao, T.; Changsheng, P.; Imran, A. A critical review on the phytoremediation of heavy metals from environment: Performance and challenges. Chemosphere 2022, 291, 132979. [Google Scholar] [CrossRef]
- Langenbach, T.; Nascimento, A.; Sarpa, M. Influence of heavy metals on nitrogen fixation and growth of Azospirillum strains. Rev. Latinoam. Microbiol. 1988, 30, 139–142. [Google Scholar]
- Belimov, A.A.; Dietz, K.J. Effect of associative bacteria on element composition of barley seedlings grown in solution culture at toxic cadmium concentrations. Microbiol. Res. 2000, 155, 113–121. [Google Scholar] [CrossRef]
- Belimov, A.A.; Kunakova, A.M.; Safronova, V.I.; Stepanok, V.V.; Yudkin, L.Y.; Alekseev, Y.V.; Kozhemyakov, A.P. Employment of rhizobacteria for the inoculation of barley plants cultivated in soil contaminated with lead and cadmium. Microbiology 2004, 73, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Akond, M.A.; Khan, Z.U. Role of carbon sources and heavy metals on growth and nitrogen fixing potential of Azospirillum and the effect of Azospirillum strains on vegetative growth of rice. J. Environ. Sci. Stud. (Dhaka) 2005, 3, 1–8. [Google Scholar]
- Kamnev Alexander, A.; Anna, V.T.; Lyudmila, P.A.; Petros, A.T.; Moschos, G.P.; Philip, H.E. Effects of heavy metals on plant-associated rhizobacteria: Comparison of endophytic and non-endophytic strains of Azospirillum brasilense. J. Trace Elem. Med. Biol. 2005, 19, 91–95. [Google Scholar] [CrossRef]
- Lyubun, Y.V.; Fritzsche, A.; Chernyshova, M.P.; Dudel, E.G.; Fedorov, E.E. Arsenic transformation by Azospirillum brasilense Sp245 in association with wheat (Triticum aestivum L.) roots. Plant Soil 2006, 286, 219–227. [Google Scholar] [CrossRef]
- Vezza, M.E.; Olmos Nicotra, M.F.; Agostini, E.; Talano, M.A. Biochemical and molecular characterization of arsenic response from Azospirillum brasilense Cd, a bacterial strain used as plant inoculant. Environ. Sci. Pollut. Res. Int. 2020, 27, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Ogar, A.; Sobczyk, Ł.; Turnau, K. Effect of combined microbes on plant tolerance to Zn-Pb contaminations. Environ. Sci. Pollut. Res. Int. 2015, 22, 19142–19156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesh, K.S.; Sundaramoorthy, P.; Nagarajan, M. Organic soil amendments: Potential source for heavy metal accumulation. World Sci. News 2015, 16, 28–39. [Google Scholar]
- Rojas Aparicio, A.; Vázquez, J.; Marbella, J.; Romero, G.N.; Rodríguez Barrera, M.A.; Toribio, J.J.; Romero, R.Y. Evaluación de compost con presencia de metales pesados en el crecimiento de Azospirillum brasilense y Glomus intraradices. Rev. Mex. Cienc. Agric. 2016, 7, 2047–2054. [Google Scholar]
- Arora, K.; Sharma, S.; Monti, A. Bio-remediation of Pb and Cd polluted soils by switchgrass: A case study in India. Int. J. Phytoremediation 2016, 18, 704–709. [Google Scholar] [CrossRef]
- Balakrishnan, B.; Sahu, B.K.; Kothilmozhian Ranishree, J.; Lourduraj, A.V.; Nithyanandam, M.; Packiriswamy, N.; Panchatcharam, P. Assessment of heavy metal concentrations and associated resistant bacterial communities in bulk and rhizosphere soil of Avicennia marina of Pichavaram mangrove, India. Environ. Earth Sci. 2017, 76, 58. [Google Scholar] [CrossRef]
- Xu, Q.; Pan, W.; Zhang, R.; Lu, Q.; Xue, W.; Wu, C.; Song, B.; Du, S. Inoculation with Bacillus subtilis and Azospirillum brasilense Produces Abscisic Acid That Reduces Irt1-Mediated Cadmium Uptake of Roots. J. Agric. Food Chem. 2018, 66, 5229–5236. [Google Scholar] [CrossRef]
- Armendariz, A.L.; Talano, M.A.; Olmos, N.M.F.; Escudero, L.; Breser, M.L.; Porporatto, C.; Agostini, E. Impact of double inoculation with Bradyrhizobium japonicum E109 and Azospirillum brasilense Az39 on soybean plants grown under arsenic stress. Plant Physiol. Biochem. 2019, 138, 26–35. [Google Scholar] [CrossRef]
- Pan, W.; Lu, Q.; Xu, Q.R.; Zhang, R.R.; Li, H.Y.; Yang, Y.H.; Liu, H.J.; Du, S.T. Abscisic acid-generating bacteria can reduce Cd concentration in pakchoi grown in Cd-contaminated soil. Ecotoxicol. Environ. Saf. 2019, 177, 100–107. [Google Scholar] [CrossRef]
- Vázquez, A.; Zawoznik, M.; Benavides, M.P.; Groppa, M.D. Azospirillum brasilense Az39 restricts cadmium entrance into wheat plants and mitigates cadmium stress. Plant Sci. 2021, 312, 111056. [Google Scholar] [CrossRef]
| Name | Country | Source | Reference |
|---|---|---|---|
| Azospirillum largimobile (A. largimogile) | Senegal | Grass | [37] |
| Azospirillum orizae (A. orizae) | Japan | Rice | [38] |
| Azospirillum lipoferum (A. lipoferum) | Brasil | Wheat | [39] |
| Azospirillum irakense (A. irakense) | Iraq | Rice | [40] |
| Azospirillum formosense (A. formosense) | Taiwan | Rice | [41] |
| Azospirillum thiophilum (A.tiophilum) | Russia | Water | [42] |
| Azospirillum griseum (A. griseum) | China | Agua | [43] |
| Azospirillum oleicastium (A. oleicastium) | China | Oil | [44] |
| Azospirillum rugosum (A. rugosum) | Taiwan | Contaminated soil | [45] |
| Azospirillum picis (A. picis) | Taiwan | Tar | [46] |
| Azospirillum fermentarium (A. fermentarium) | Taiwan | Fermenter | [47] |
| Azospirillum humicireducens (A. humicireducens) | China | Microbial fuel cell | [48] |
| Azospirillum brasilense (A. brasilense) | Brazil | Grass | [49] |
| Azospirillum halopraeferens (A. halopraeferens) | Pakistan | Grass | [50] |
| Azospirillum doebereinerae (A. doebereinerae) | Germany | Grass | [51] |
| Azospirillum melinis (A. melinis) | China | Grass | [52] |
| Azospirillum canadense (A. canadense) | Canada | Corn | [53] |
| Azospirillum zeae (A. zeae) | Canada | Corn | [54] |
| Azospirillum palatum (A. palatum) | China | Soil | [55] |
| Azospirillum soli (A. soli) | Taiwan | Agricultural soil | [56] |
| Azospirillum agricola (A. agricola) | Taiwan | Agricultural soil | [57] |
| Azospirillum palustre (A. palustre) | Russia | Soil | [58] |
| Azospirillum ramasamyi (A. ramasamyi) | Korea | Fermented bovine products | [59] |
| Azospirillum baldaniorum (A. baldoniorum) | Brazil | Rhizosphere | [60] |
| Azospirillum thermophilum (A. thermophilum) | China | Hot spring | [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Hernández, M.A.; Mendoza-Herrera, A.; Bocanegra-García, V.; Rivera, G. Azospirillum spp. from Plant Growth-Promoting Bacteria to Their Use in Bioremediation. Microorganisms 2022, 10, 1057. https://doi.org/10.3390/microorganisms10051057
Cruz-Hernández MA, Mendoza-Herrera A, Bocanegra-García V, Rivera G. Azospirillum spp. from Plant Growth-Promoting Bacteria to Their Use in Bioremediation. Microorganisms. 2022; 10(5):1057. https://doi.org/10.3390/microorganisms10051057
Chicago/Turabian StyleCruz-Hernández, María Antonia, Alberto Mendoza-Herrera, Virgilio Bocanegra-García, and Gildardo Rivera. 2022. "Azospirillum spp. from Plant Growth-Promoting Bacteria to Their Use in Bioremediation" Microorganisms 10, no. 5: 1057. https://doi.org/10.3390/microorganisms10051057
APA StyleCruz-Hernández, M. A., Mendoza-Herrera, A., Bocanegra-García, V., & Rivera, G. (2022). Azospirillum spp. from Plant Growth-Promoting Bacteria to Their Use in Bioremediation. Microorganisms, 10(5), 1057. https://doi.org/10.3390/microorganisms10051057







