Influence of Different Vegetation Types on Soil Physicochemical Parameters and Fungal Communities
Abstract
1. Introduction
2. Material and Method
2.1. Site Description
2.2. Soil Sampling
2.3. Characterization of Soil Physicochemical Parameters
2.4. DNA Extraction, PCR Amplification, and MiSeq Sequencing
2.5. Analysis of Sequencing Data
2.6. Statistical Analyses
3. Results
3.1. Physicochemical Characteristics of Soil
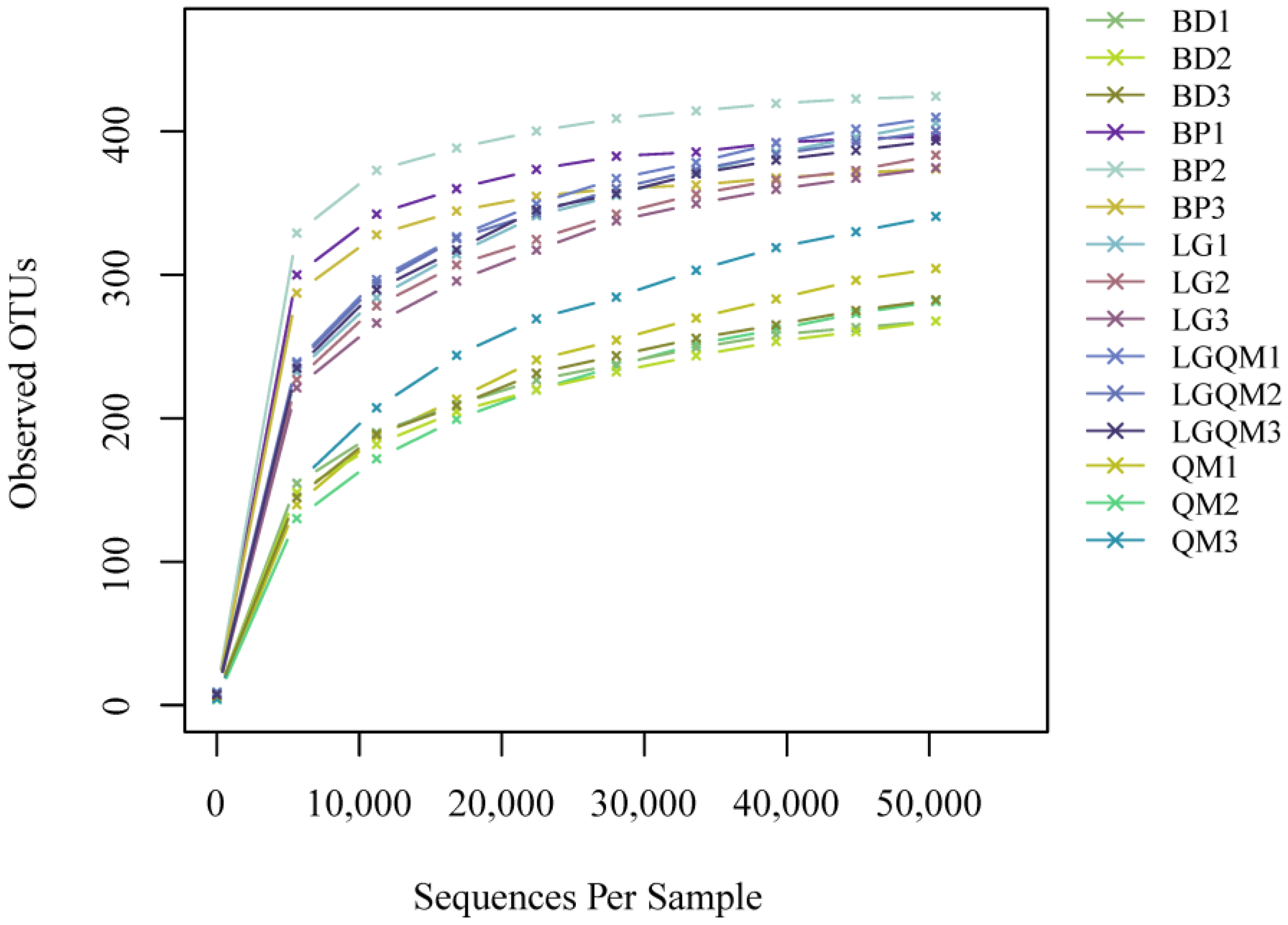
3.2. Effect of Different Vegetation Types on Fungal Diversity
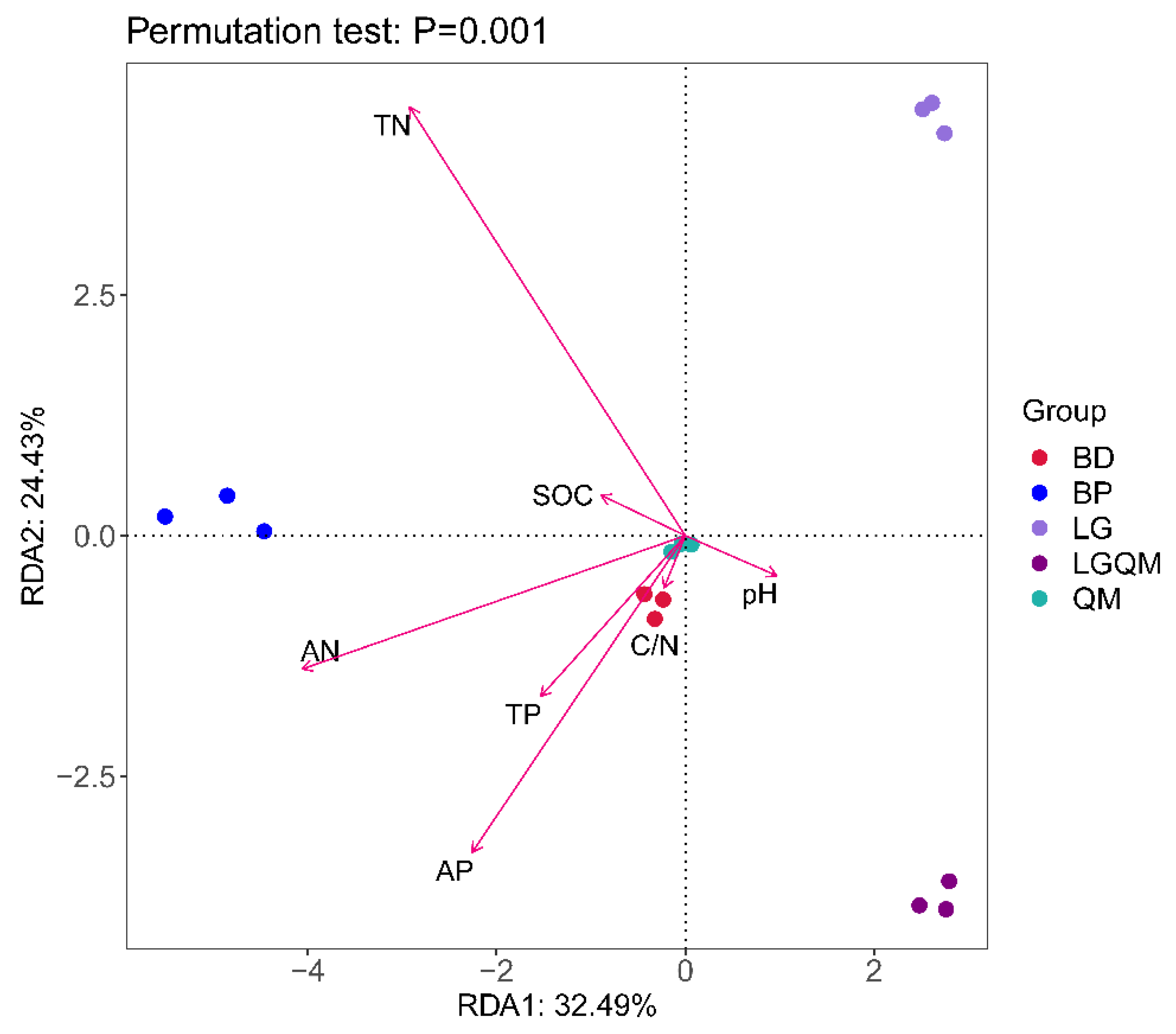
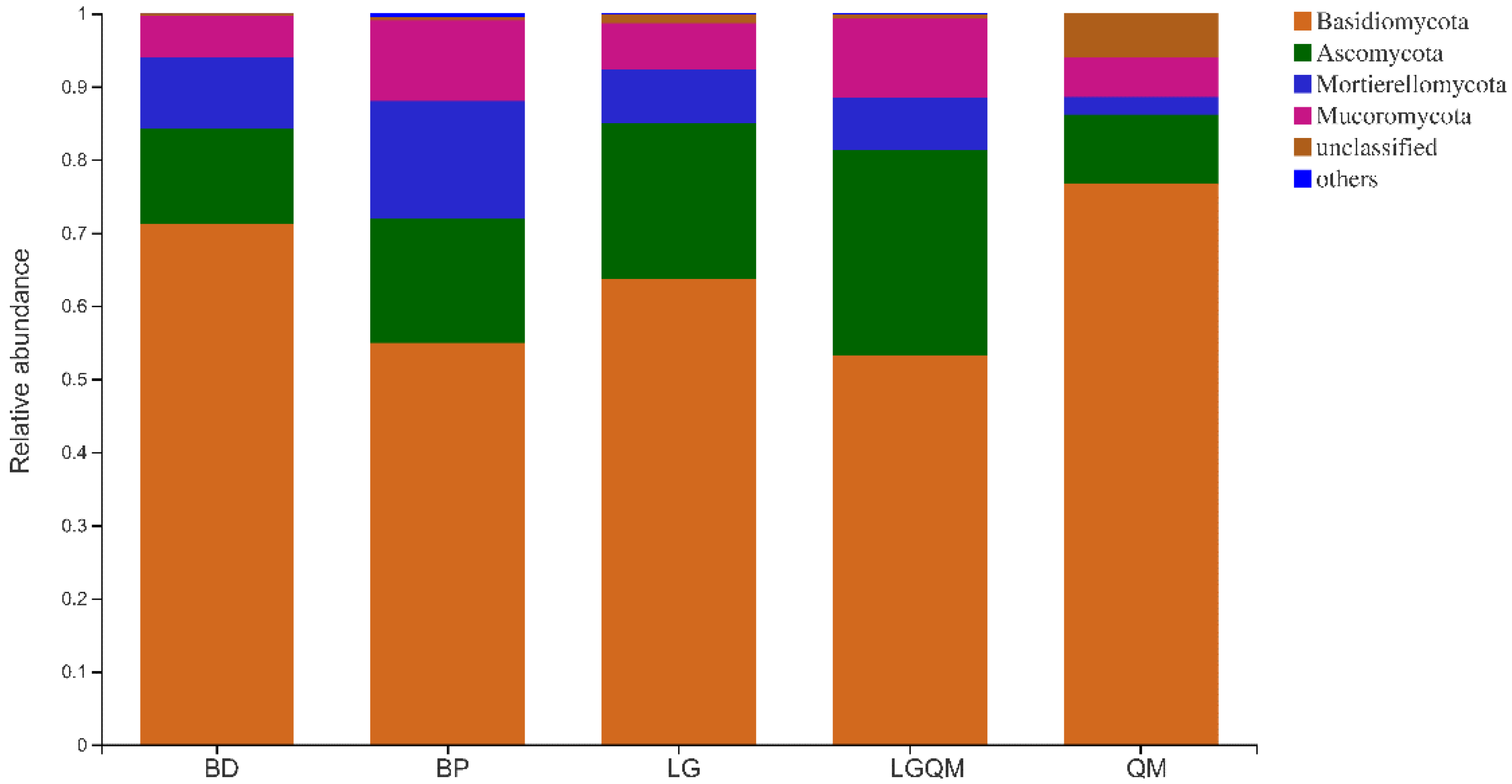
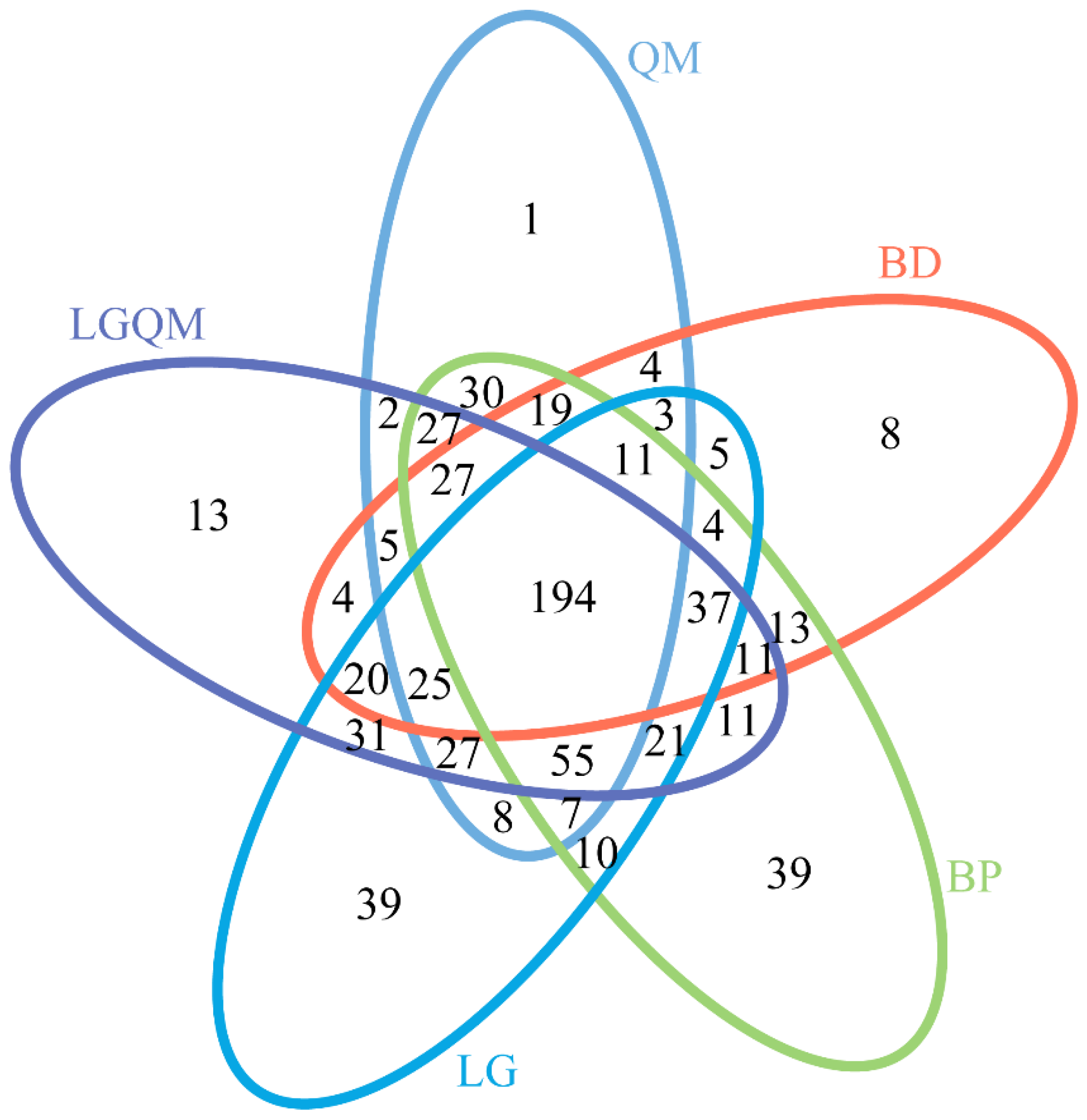
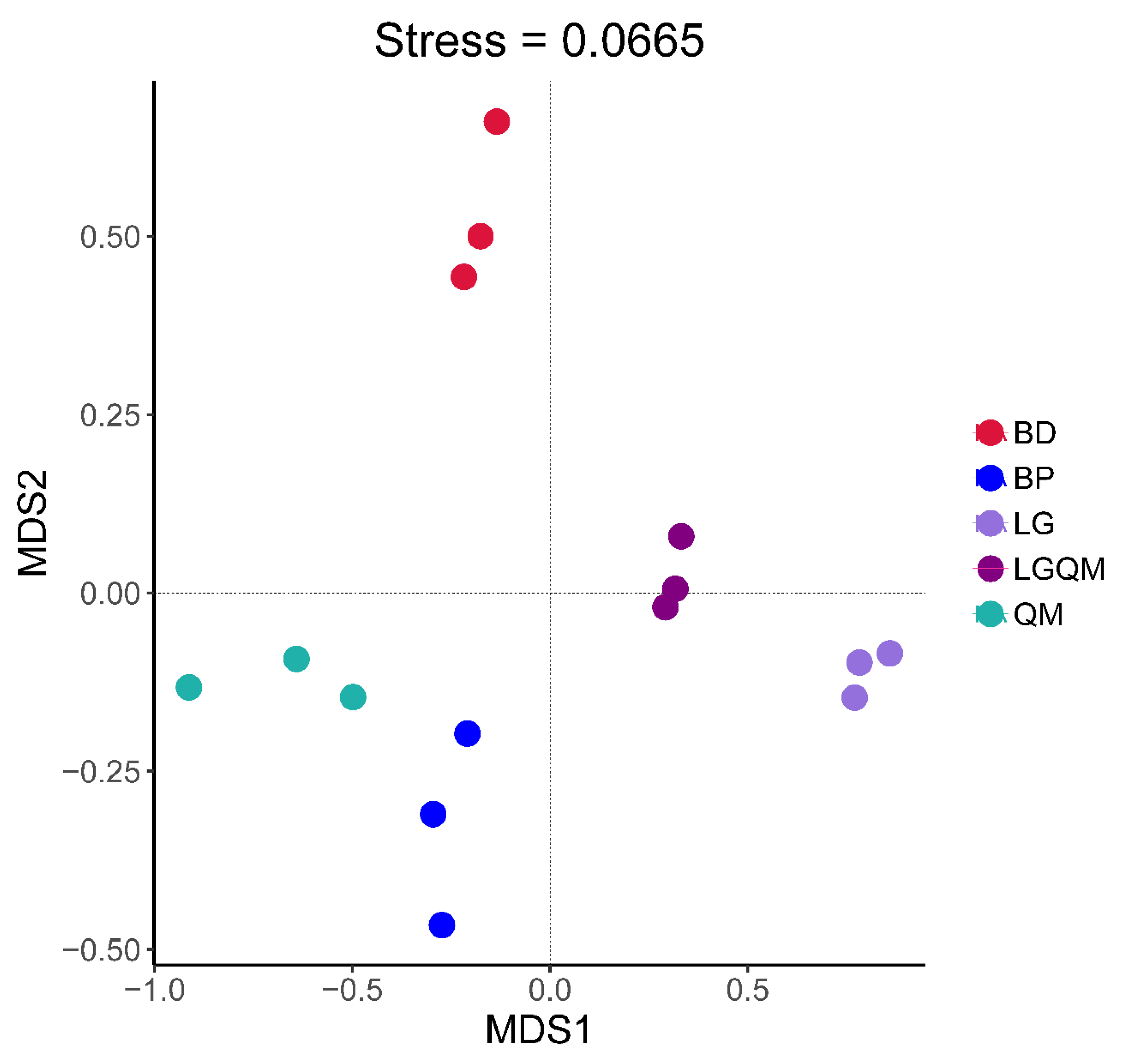
3.3. Effects of Different Vegetation Types on Fungal Community Structure
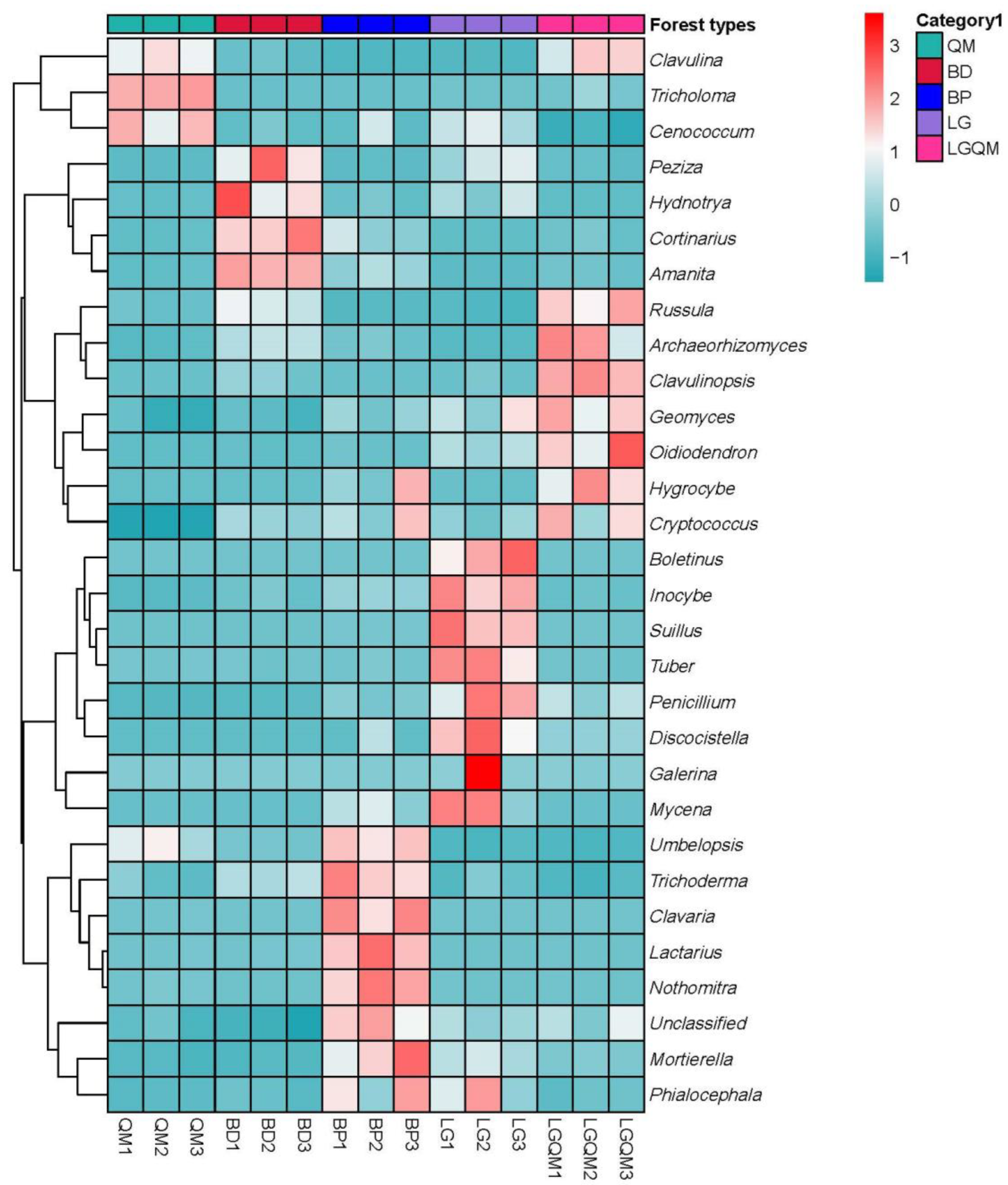
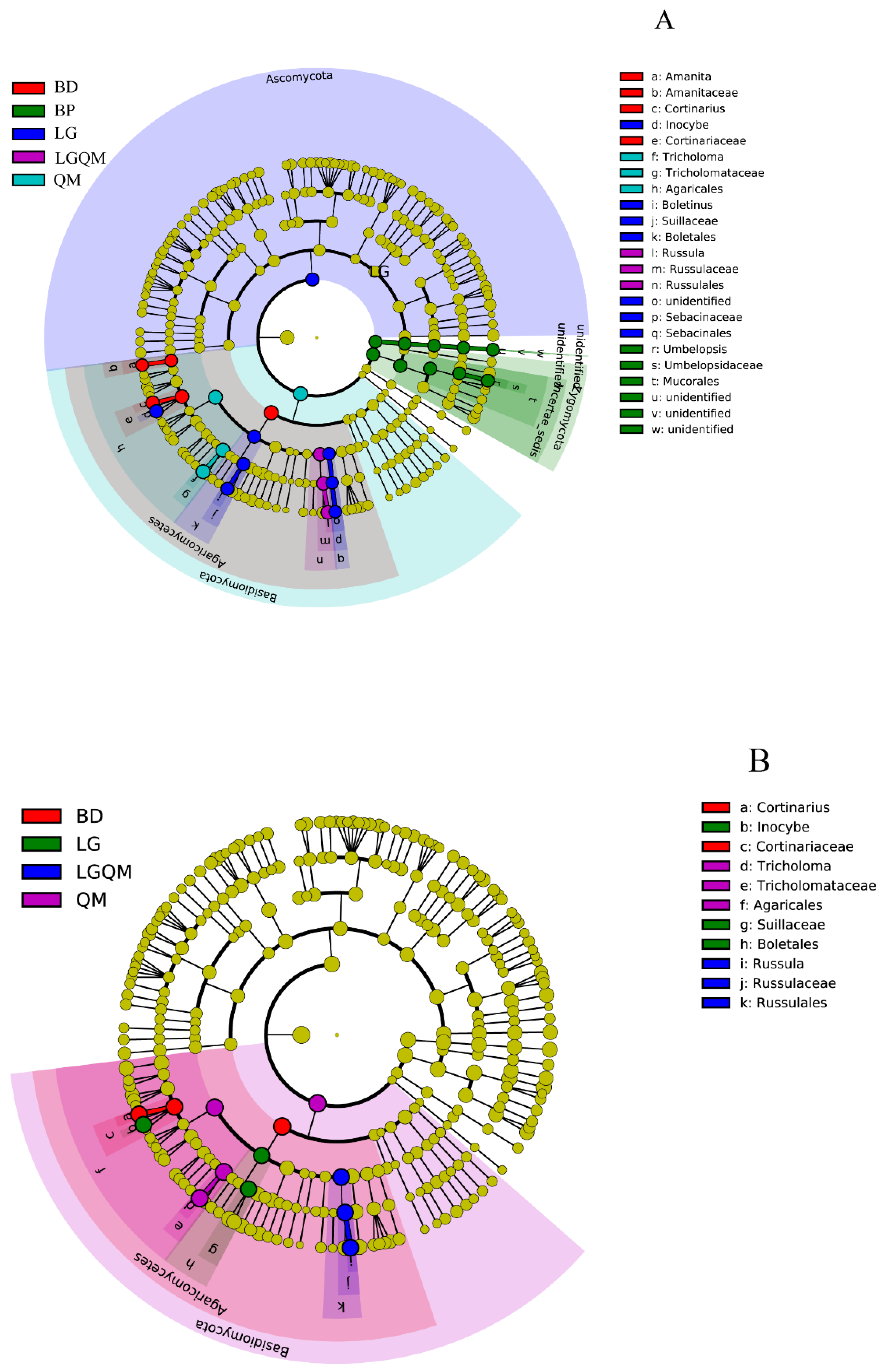
3.4. Indicator Species
3.5. Primary Shapers of Fungal Community Structure
4. Discussion
4.1. Characteristics of Soils in the Naturally Restored Forests
4.2. Effects of Vegetation Types on the Diversity and Structure of Fungal Communities
4.3. Effects of Soil Characteristics on Fungal Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Coomes, D.A.; Gibson, L.; Hu, G.; Liu, J.; Luo, Y.; Yu, M. Forest fragmentation in China and its effect on biodiversity. Biol. Rev. 2019, 94, 1636–1657. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Chen, A.; Fang, H.; Wu, J.; Liu, G. Effects of vegetation restoration types on soil quality in Yuanmou dry-hot valley, China. Soil Sci. Plant Nutr. 2013, 59, 347–360. [Google Scholar] [CrossRef]
- Nunezmir, G.C.; Iannone, B.V.; Curtis, K.; Fei, S. Evaluating the evolution of forest restoration research in a changing world: A “big literature” review. New Forest 2015, 46, 669–682. [Google Scholar] [CrossRef]
- Li, H.L.; Zhang, J.; Wang, L.; Yang, H.; Yan, H.E.; Lu, L.U. The Influence of Land Use Change on the Microbial Community in Two Acidic Soils. J. Chin. West Norm. Univ. 2017, 38, 373–381. [Google Scholar]
- Gallardo, B.; Cabezas, A.; Gonzalez, E.; Comín, F.A. Effectiveness of a newly created oxbow lake to mitigate habitat loss and increase biodiversity in a regulated floodplain. Restor Ecol. 2012, 20, 387–394. [Google Scholar] [CrossRef]
- Oh, W.S.; Lee, C. Recovery of Ecosystem Service Functions through Ecological Restoration Practice: A Case Study of Coal Mine Spoils, Samcheok, Central Eastern Korea. Environ. Biol. Res. 2014, 32, 102–111. [Google Scholar] [CrossRef]
- Zhang, Q.; Jia, X.; Zhao, C.; Shao, M.A. Revegetation with artificial plants improves topsoil hydrological properties but intensifies deep-soil drying in northern Loess Plateau, China. J. Arid Land 2018, 10, 335–346. [Google Scholar] [CrossRef]
- Wu, G.L.; Liu, Y.; Fang, N.F.; Deng, L.; Shi, Z.H. Soil physical properties response to grassland conversion from cropland on the semi-arid area. Ecohydrology 2016, 9, 1471–1479. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, J.; Liu, G.B.; Zhang, T. Effects of Revegetation Types on the Soil Carbon, Nitrogen and Phosphorus of the Gully Areas in Hilly Loess Plateau, China. Acta Agrectir. Sin. 2015, 23, 62–68. [Google Scholar]
- Schweitzer, J.A.; Bailey, J.K.; Fischer, D.G.; LeRoy, C.J.; Lonsdorf, E.V.; Whitham, T.G.; Hart, S.C. Plant–soil–microorganism interactions: Heritable relationship between plant genotype and associated soil microorganisms. Ecology 2008, 89, 773–781. [Google Scholar] [CrossRef]
- Cheng, F.; Peng, X.; Zhao, P.; Yuan, J.; Zhong, C.; Cheng, Y.; Zhang, S. Soil microbial biomass, basal respiration and enzyme activity of main forest types in the Qinling Mountains. PLoS ONE 2013, 8, e67353. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.L.; Zemunik, G.; Laliberté, E.; Drake, J.J.; Jones, F.A.; Saltonstall, K. Contrasting patterns of plant and microbial diversity during long-term ecosystem development. J. Ecol. 2019, 107, 606–621. [Google Scholar] [CrossRef]
- Fan, M.; Li, J.; Tang, Z.; Shangguan, Z. Soil bacterial community succession during desertification in a desert steppe ecosystem. Land. Degrad. Dev. 2020, 31, 1662–1674. [Google Scholar] [CrossRef]
- Paul, M.; Catterall, C.P.; Pollard, P.C.; Kanowski, J. Recovery of soil properties and functions in different rainforest restoration pathways. For. Ecol. Manag. 2010, 259, 2083–2092. [Google Scholar] [CrossRef]
- Yue, P.; Cui, X.; Zuo, X.; Li, K.; Wang, S.; Jia, Y.; Liu, X. The contribution of arbuscular mycorrhizal fungi to ecosystem respiration and methane flux in an ephemeral plants-dominated desert. Land Degrad. Dev. 2021, 32, 1844–1853. [Google Scholar] [CrossRef]
- Ding, T.; Yan, Z.; Zhang, W.; Duan, T. Green Manure Crops Affected Soil Chemical Properties and Fungal Diversity and Community of Apple Orchard in the Loess Plateau of China. J. Soil Sci. Plant Nutr. 2021, 21, 1089–1102. [Google Scholar] [CrossRef]
- Liu, D.; Liu, G.; Chen, L.; Wang, J.; Zhang, L. Soil pH determines fungal diversity along an elevation gradient in Southwestern China. Sci. China Life Sci. 2018, 61, 718–726. [Google Scholar] [CrossRef]
- Gil-Martínez, M.; López-García, A.; Domínguez, M.T.; Kjøller, R.; Navarro-Fernández, C.M.; Rosendahl, S.; Marañón, T. Soil fungal diversity and functionality are driven by plant species used in phytoremediation. Soil Biol. Biochem. 2021, 153, 108102. [Google Scholar] [CrossRef]
- Frey, S.D.; Knorr, M.; Parrent, J.L.; Simpson, R.T. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. Forest Ecol. Manag. 2004, 196, 159–171. [Google Scholar] [CrossRef]
- Chaudhary, V.B.; Cuenca, G.; Johnson, N.C. Tropical—Temperate comparison of landscape-scale arbuscular mycorrhizal fungal species distributions. Divers. Distrib. 2018, 24, 116–128. [Google Scholar] [CrossRef]
- Kang, E.; Li, Y.; Zhang, X.; Yan, Z.; Wu, H.; Li, M.; Kang, X. Soil pH and nutrients shape the vertical distribution of microbial communities in an alpine wetland. Sci. Total Environ. 2021, 774, 145780. [Google Scholar] [CrossRef]
- Wang, J.; Shi, X.; Zheng, C.; Suter, H.; Huang, Z. Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 2021, 755, 142449. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Mills, J.G.; Gellie, N.J.; Bissett, A.; Lowe, A.J.; Breed, M.F. High-throughput eDNA monitoring of fungi to track functional recovery in ecological restoration. Biol. Conserv. 2018, 217, 113–120. [Google Scholar] [CrossRef]
- Harrison, K.A.; Bardgett, R.D. Influence of plant species and soil conditions on plant–soil feedback in mixed grassland communities. J. Ecol. 2010, 98, 384–395. [Google Scholar] [CrossRef]
- Deng, J.; Yin, Y.; Luo, J.; Zhu, W.; Zhou, Y. Different revegetation types alter soil physical-chemical characteristics and fungal community in the Baishilazi Nature Reserve. Peer J. 2019, 6, e6251. [Google Scholar] [CrossRef]
- Chai, Y.F.; Cao, Y.; Yue, M.; Tian, T.T.; Yin, Q.L.; Dang, H.; Quan, J.X.; Zhang RCWang, M. Soil abiotic properties and plant functional traits mediate associations between soil microbial and plant communities during a secondary forest succession on the Loess Plateau. Front. Microbiol. 2019, 10, 895. [Google Scholar] [CrossRef]
- Lin, Q.; Baldrian, P.; Li, L.J.; Novotny, V.; Heděnec, P.; Kukla, J.; Umari, R.; Meszárošová, L.; Frouz, J. Dynamics of soil bacterial and fungal communities during the secondary succession following swidden agriculture in lowland forests. Front. Microbiol. 2021, 12, 1421. [Google Scholar] [CrossRef]
- Weng, X.; Li, J.; Sui, X.; Li, M.; Yin, W.; Ma, W.; Yang, L.; Mu, L. Soil Microbial Functional Diversity Responses to Different Revegetation Types in Heilongjiang Zhongyangzhan Black-billed Capercaillie Nature Reserve. Ann. Microbiol. 2021, 71, 26. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Caporaso, J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, 259–264. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: http://www.R-project.org/ (accessed on 10 April 2021).
- Vitali, F.; Mastromei, G.; Senatore, G.; Caroppo, C.; Casalone, E. Long lasting effects of the conversion from natural forest to poplar plantation on soil microbial communities. Microbiol. Res. 2016, 182, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, D.; Wang, X.; Settles, M.L.; Wang, J.; Hao, Z.; Ma, Z.S. Soil bacterial communities of different natural forest types in Northeast China. Plant Soil 2014, 383, 203–216. [Google Scholar] [CrossRef]
- Gao, F.; Jiang, H.; Cui, X.Y. Soil organic carbon pools and their turnover under two different types of forest in Xiaoxing’an Mountains, northeast China. J. Appl. Ecol. 2015, 26, 1913–1920. [Google Scholar]
- Ge, X.G.; Huang, Z.L.; Cheng, R.M.; Zeng, L.X.; Xiao, W.F.; Tan, B.W. Effects of litterfall and root input on soil physical and chemical properties in Pinus massoniana plantations in Three Gorges Reservoir Area, China. J. Appl. Ecol. 2012, 23, 3301–3308. [Google Scholar]
- Anderson, J.A.; Hooper, M.J.; Zak, J.C.; Cox, S.B. Characterization of the structural and functional diversity of indigenous soil microbial communities in smelter-impacted and nonimpacted soils. Environ. Toxicol. Chem. 2009, 28, 534–541. [Google Scholar] [CrossRef]
- Degrune, F.; Dufrêne, M.; Colinet, G.; Massart, S.; Taminiau, B.; Bodson, B.; Vandenbol, M. A novel sub-phylum method discriminates better the impact of crop management on soil microbial community. Agron. Sustain. Dev. 2015, 35, 1157–1166. [Google Scholar] [CrossRef]
- Gorfer, M.; Mayer, M.; Berger, H.; Rewald, B.; Tallian, C.; Matthews, B.; Godbold, D.L. High fungal diversity but low seasonal dynamics and ectomycorrhizal abundance in a Mountain Beech forest. Microb Ecol. 2021, 82, 243–256. [Google Scholar] [CrossRef]
- Waring, B.G.; Adams, R.; Branco, S.; Powers, J.S. Scale—Dependent variation in nitrogen cycling and soil fungal communities along gradients of forest composition and age in regenerating tropical dry forests. New Phytol. 2016, 209, 845–854. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.; Frey, B.; Yang, L.; Liu, Y.; Ni, H.; Li, M.H. Soil physicochemical properties drive the variation in soil microbial communities along a forest successional series in a degraded wetland in northeastern China. Ecol. Evol. 2021, 11, 2194–2208. [Google Scholar] [CrossRef]
- Alfaro, M.; Castanera, R.; Lavín, J.L.; Grigoriev, I.V.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative and transcriptional analysis of the predicted secretome in the lignocellulose—Degrading basidiomycete fungus Pleurotus ostreatus. Environ. Microbiol. 2016, 18, 4710–4726. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, C.B.; Waldrop, M.P.; Zak, D.R.; Sinsabaugh, R.L. Molecular analysis of fungal communities and laccase genes in decomposing litter reveals differences among forest types but no impact of nitrogen deposition. Environ. Microbiol. 2007, 9, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.W.; Kang, Y.; Lu, H.; Wang, Q.Y. Basic Soil Physicochemical Properties and Soil Fungal Diversity under Different Forest Types of Urban Forest. J. North For. Univ. 2016, 44, 89–100. [Google Scholar]
- Lundell, T.K.; Mäkelä, M.R.; Hildén, K. Lignig—Modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J. Basic Microb. 2010, 50, 5–20. [Google Scholar] [CrossRef]
- Meng, M.; Wang, B.; Zhang, Q.; Tian, Y. Driving force of soil microbial community structure in a burned area of Daxing’anling, China. J. For. Res. 2021, 32, 1723–1738. [Google Scholar] [CrossRef]
- Frey, B.; Carnol, M.; Dharmarajah, A.; Brunner, I.; Schleppi, P. Only minor changes in the soil microbiome of a sub-alpine forest after 20 years of moderately increased nitrogen loads. Front. Forest Glob. Chan. 2020, 3, 77. [Google Scholar] [CrossRef]
- Frey, B.; Walthert, L.; Perez-Mon, C.; Stierli, B.; Köchli, R.; Dharmarajah, A.; Brunner, I. Deep soil layers of drought-exposed forests harbor poorly known bacterial and fungal communities. Front. Microb. 2021, 12, 1061. [Google Scholar] [CrossRef]
- Yang, L.B.; Sui, X.; Zhu, D.G.; Cui, F.X.; Li, J.B.; Song, R.Q.; Ni, H.W. Study on fungal communities characteristics of different Larix gmelini forest typesin cold temperate zone. J. Cent. South Univ. Technol. 2017, 37, 76–84. [Google Scholar]
- Harrower, E.; Bougher, N.L.; Henkel, T.W.; Horak, E.; Matheny, P.B. Long-distance dispersal and speciation of Australasian and American species of Cortinarius sect. Cortinarius. Mycologia 2015, 107, 697–709. [Google Scholar] [CrossRef]
- Wagai, R.; Kitayama, K.; Satomura, T.; Fujinuma, R.; Balser, T. Interactive influences of climate and parent material on soil microbial community structure in Bornean tropical forest ecosystems. Ecol. Res. 2011, 26, 627–636. [Google Scholar] [CrossRef]
- Sun, H.; Terhonen, E.; Kovalchuk, A.; Tuovila, H.; Chen, H.; Oghenekaro, A.O.; Asiegbu, F.O. Dominant tree species and soil type affect the fungal community structure in a boreal peatland forest. Appl. Environ. Microb. 2016, 82, 2632–2643. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Niklaus, P.A.; Zimmermann, S.; Schmutz, S.; Kremer, J.; Abarenkov, K.; Lüscher, P.; Widmer, F.; Frey, B. Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J. 2014, 8, 226–244. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Li, Z.W.; Wang, Z.Y.; Wang, W.; Jia, Y.; Tian, S. Impacts of artificially planted vegetation on the ecological restoration of movable sand dunes in the Mugetan Desert, northeastern Qinghai-Tibet Plateau. Int. J. Sediment Res. 2017, 32, 277–287. [Google Scholar] [CrossRef]
- Liu, M.; Liu, J.; Chen, X.; Jiang, C.; Wu, M.; Li, Z.P. Shifts in bacterial and fungal diversity in a paddy soil faced with phosphorus surplus. Biol. Fertil. Soils 2018, 54, 259–267. [Google Scholar] [CrossRef]
- Huang, F.; Liu, Z.; Mou, H.; Zhang, P.; Jia, Z. Effects of different long-term farmland mulching practices on the loessial soil fungal community in a semiarid region of China. Appl. Soil Ecol. 2019, 137, 111–119. [Google Scholar] [CrossRef]
- Wang, J.T.; Zheng, Y.M.; Hu, H.W.; Zhang, L.M.; Li, J.; He, J.Z. Soil pH determines the alpha diversity but not beta diversity of soil fungal community along altitude in a typical Tibetan forest ecosystem. J. Soil Sediment 2015, 15, 1224–1232. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, X.; Liu, C.; Bai, L.; Zhao, M.; Li, L. Diversity and characteristics of colonization of root-associated fungi of Vaccinium uliginosum. Sci. Rep. 2018, 8, 15283. [Google Scholar] [CrossRef]
- Yao, Q.; Liu, J.; Yu, Z.; Li, Y.; Jin, J.; Liu, X.; Wang, G. Three years of biochar amendment alters soil physiochemical properties and fungal community composition in a black soil of northeast China. Soil Biol. Biochem. 2017, 110, 56–67. [Google Scholar] [CrossRef]
- Treseder, K.K.; Maltz, M.R.; Hawkins, B.A.; Fierer, N.; Stajich, J.E.; McGuire, K.L. Evolutionary histories of soil fungi are reflected in their large-scale biogeography. Ecol. Lett. 2014, 17, 1086–1093. [Google Scholar] [CrossRef]
- Tian, Q.; Taniguchi, T.; Shi, W.Y.; Li, G.; Yamanaka, N.; Du, S. Land-use types and soil chemical properties influence soil microbial communities in the semiarid Loess Plateau region in China. Sci. Rep. 2017, 7, 45289. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.; Xia, H.; Zou, B.; Li, N.; Liu, J.; Zhu, W. Effects of nitrogen-fixing and non-nitrogen-fixing tree species on soil properties and nitrogen transformation during forest restoration in southern China. Soil Sci. Plant Nutr. 2010, 56, 297–306. [Google Scholar] [CrossRef]
- Dang, P.; Yu, X.; Le, H.; Liu, J.; Shen, Z.; Zhao, Z. Effects of stand age and soil properties on soil bacterial and fungal community composition in Chinese pine plantations on the Loess Plateau. PLoS ONE 2017, 12, e0186501. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.N.; Liu, Y.X.; Zhou, X.; Wang, T.; Gao, Q.; Gao, Y.; Liu, S. Effects of long-term different fertilization on soil fungal communities in black soil based on the Illumina MiSeq platform. Acta Microbiol. Sin. 2018, 58, 1658–1671. [Google Scholar]
- Vaario, L.M.; Fritze, H.; Spetz, P.; Heinonsalo, J.; Hanajík, P.; Pennanen, T. Tricholoma matsutake dominates diverse microbial communities in different forest soils. Appl. Environ. Microbiol. 2011, 77, 8523–8531. [Google Scholar] [CrossRef]
- Tedersoo, L.; Kõljalg, U.; Hallenberg, N.; Larsson, K.H. Fine scale distribution of ectomycorrhizal fungi and roots across substrate layers including coarse woody debris in a mixed forest. New Phytol. 2003, 159, 153–165. [Google Scholar] [CrossRef]


| Vegetation Type | Primary Tree Species | Elevation (m) | Forest Type |
|---|---|---|---|
| BD | Populus tremula, Betula dahurica | 552.1 | Natural secondary forest |
| BP | Betula platyphylla, Populus tremula | 535.5 | Natural secondary forest |
| QM | Quercus mongolica, Tilia amurensis | 520.4 | Natural secondary forest |
| LGQM | Betula platyphylla, Populus tremula, Quercus mongolica, Larix gmelinii | 565.4 | Natural secondary forest |
| LG | Larix gmelinii | 585.7 | Natural climax forest |
| Soil Physicochemical Parameters | LG | BD | LGQM | BP | QM | F-Value | p-Value |
|---|---|---|---|---|---|---|---|
| pH | 5.5 ± 0.1 a | 4.6 ± 0.4 b | 5.9 ± 0.3 a | 5.3 ± 0.1 a | 5.4 ± 0.2 a | 10.233 | p < 0.001 |
| SOC (g/kg) | 60.3 ± 3.5 c | 54.0 ± 3.53 d | 49.9 ± 3.95 d | 76.2 ± 2.04 b | 107.4 ± 22.4 a | 191.954 | p < 0.001 |
| TN (g/kg) | 4.1 ± 0.1 a | 3.47 ± 0.34 b | 2.76 ± 0.13 c | 4.27 ± 0.15 a | 4.20 ± 0.10 a | 33.856 | p < 0.001 |
| C/N | 14.7 ± 1.3 c | 15.59 ± 0.76 c | 18.07 ± 1.58 b | 17.86 ± 0.35 b | 25.82 ± 0.61 a | 55.253 | p < 0.001 |
| TP (g/kg) | 1.77 ± 0.15 c | 2.07 ± 0.06 bc | 2.17 ± 0.29 bc | 2.33 ± 0.15 ab | 2.60 ± 0.20 a | 8.374 | p < 0.001 |
| AN (mg/kg) | 24.63 ± 2.60 d | 78.67 ± 2.96 b | 38.59 ± 2.18 c | 83.75 ± 2.22 a | 87.34 ± 1.59 a | 450.879 | p < 0.001 |
| AP (mg/kg) | 27.43 ± 0.81 c | 36.17 ± 1.23 b | 35.86 ± 1.03 b | 37.28 ± 0.42 b | 41.25 ± 1.40 a | 88.517 | p < 0.001 |
| Types | Richness | Shannon Index | ACE Index | Chao1 Index | Simpson Index |
|---|---|---|---|---|---|
| BD | 278 ±9 c | 4.21 ± 0.31 c | 326.57 ±16.90 b | 333.84 ±29.21 b | 0.86 ± 0.04 b |
| BP | 400 ± 25 a | 6.30 ± 0.18 a | 410.24 ± 23.94 a | 419.49 ± 20.88 a | 0.97 ± 0.00 a |
| LG | 395 ± 17 a | 5.28 ± 0.14 b | 453.18 ± 29.73 a | 447.63 ± 28.58 a | 0.94 ± 0.01 a |
| LGQM | 408 ± 9 a | 5.36 ± 0.04 b | 450.89 ± 15.21 a | 451.24 ± 18.81 a | 0.94 ± 0.00 a |
| QM | 318 ± 30 b | 2.81 ±0.07 d | 396.44 ± 36.84 a | 401.18 ± 43.13 a | 0.64 ±0.03 c |
| pH | SOC | TN | C/N | TP | AN | AP | |
|---|---|---|---|---|---|---|---|
| Richness | 0.52 * | 0.54 * | 0.12 | 0.79 ** | 0.53 * | 0.20 | 0.64 * |
| ACE | 0.21 | 0.58 * | 0.04 | 0.75 ** | 0.45 | 0.37 | 0.66 ** |
| Chao1 | 0.24 | 0.61 * | 0.03 | 0.78 ** | 0.50 | 0.37 | 0.67 ** |
| Shannon | 0.81 ** | 0.06 | 0.48 | 0.40 | 0.27 | 0.29 | 0.24 |
| Simpson | 0.70 ** | −0.21 | 0.71 ** | 0.21 | −0.15 | −0.36 | 0.16 |
| Fungal Taxa | pH | SOC | TN | C/N | TP | AN | AP |
|---|---|---|---|---|---|---|---|
| Phylum | - | - | - | - | - | - | - |
| Basidiomycota | −0.294 | 0.230 | −0.024 | 0.283 | 0.097 | 0.205 | 0.192 |
| Ascomycota | 0.474 | −0.487 | −0.178 | −0.480 | −0.389 | −0.708 ** | −0.613 * |
| Mortierellomycota | −0.210 | −0.134 | 0.332 | −0.378 | −0.121 | 0.228 | −0.056 |
| Mucoromycota | 0.170 | −0.026 | 0.093 | −0.068 | 0.099 | 0.072 | 0.066 |
| Glomeromycota | 0.152 | −0.016 | 0.119 | −0.090 | 0.210 | 0.253 | 0.213 |
| Genus | - | - | - | - | - | - | - |
| Tricholoma | 0.220 | 0.827 ** | 0.279 | 0.880 ** | 0.625 * | 0.411 | 0.607 * |
| Russula | 0.036 | −0.564 * | −0.948 ** | −0.116 | −0.001 | −0.180 | 0.145 |
| Cortinarius | −0.727 ** | −0.351 | −0.152 | −0.385 | −0.127 | 0.402 | 0.128 |
| Inocybe | 0.100 | −0.276 | 0.344 | −0.565 * | −0.669 ** | −0.676 ** | −0.904 ** |
| Umbelopsis | −0.164 | 0.707 ** | 0.617 * | 0.515 * | 0.648 ** | 0.763 ** | 0.607 * |
| Cenococcum | 0.025 | 0.722 ** | 0.669 ** | 0.501 | 0.295 | 0.235 | 0.141 |
| Boletus | 0.193 | −0.194 | 0.266 | −0.412 | −0.689 ** | −0.698 ** | −0.844 ** |
| Amanita | −0.804 ** | −0.363 | −0.193 | −0.374 | −0.137 | 0.427 | 0.134 |
| Mortierella | 0.130 | −0.030 | 0.433 | −0.297 | −0.147 | −0.048 | −0.250 |
| Archaeorhizomyces | 0.203 | −0.609 * | −0.869 ** | −0.239 | −0.187 | −0.265 | 0.092 |
| Clavulina | 0.518 * | 0.264 | −0.479 | 0.651 ** | 0.499 | 0.006 | 0.520 * |
| Lactarius | −0.109 | 0.161 | 0.425 | −0.056 | 0.230 | 0.440 | 0.213 |
| Suillus | 0.161 | −0.222 | 0.313 | −0.475 | −0.636 * | −0.725 ** | −0.890 ** |
| Thelephora | 0.332 | −0.524 * | −0.458 | −0.355 | −0.045 | −0.249 | −0.096 |
| Hygrocybe | 0.552 * | −0.329 | −0.520 * | −0.079 | 0.015 | −0.232 | 0.095 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sui, X.; Zeng, X.; Li, M.; Weng, X.; Frey, B.; Yang, L.; Li, M. Influence of Different Vegetation Types on Soil Physicochemical Parameters and Fungal Communities. Microorganisms 2022, 10, 829. https://doi.org/10.3390/microorganisms10040829
Sui X, Zeng X, Li M, Weng X, Frey B, Yang L, Li M. Influence of Different Vegetation Types on Soil Physicochemical Parameters and Fungal Communities. Microorganisms. 2022; 10(4):829. https://doi.org/10.3390/microorganisms10040829
Chicago/Turabian StyleSui, Xin, Xiannan Zeng, Mengsha Li, Xiaohong Weng, Beat Frey, Libin Yang, and Maihe Li. 2022. "Influence of Different Vegetation Types on Soil Physicochemical Parameters and Fungal Communities" Microorganisms 10, no. 4: 829. https://doi.org/10.3390/microorganisms10040829
APA StyleSui, X., Zeng, X., Li, M., Weng, X., Frey, B., Yang, L., & Li, M. (2022). Influence of Different Vegetation Types on Soil Physicochemical Parameters and Fungal Communities. Microorganisms, 10(4), 829. https://doi.org/10.3390/microorganisms10040829







