The Human Mycobiome in Chronic Respiratory Diseases: Current Situation and Future Perspectives
Abstract
1. Introduction
2. Prevalence and Clinical Significance of Fungi in Chronic Respiratory Disease
2.1. Chronic Obstructive Pulmonary Disease
2.2. Cystic Fibrosis
2.3. Non-Cystic Fibrosis Bronchiectasis
2.4. Asthma
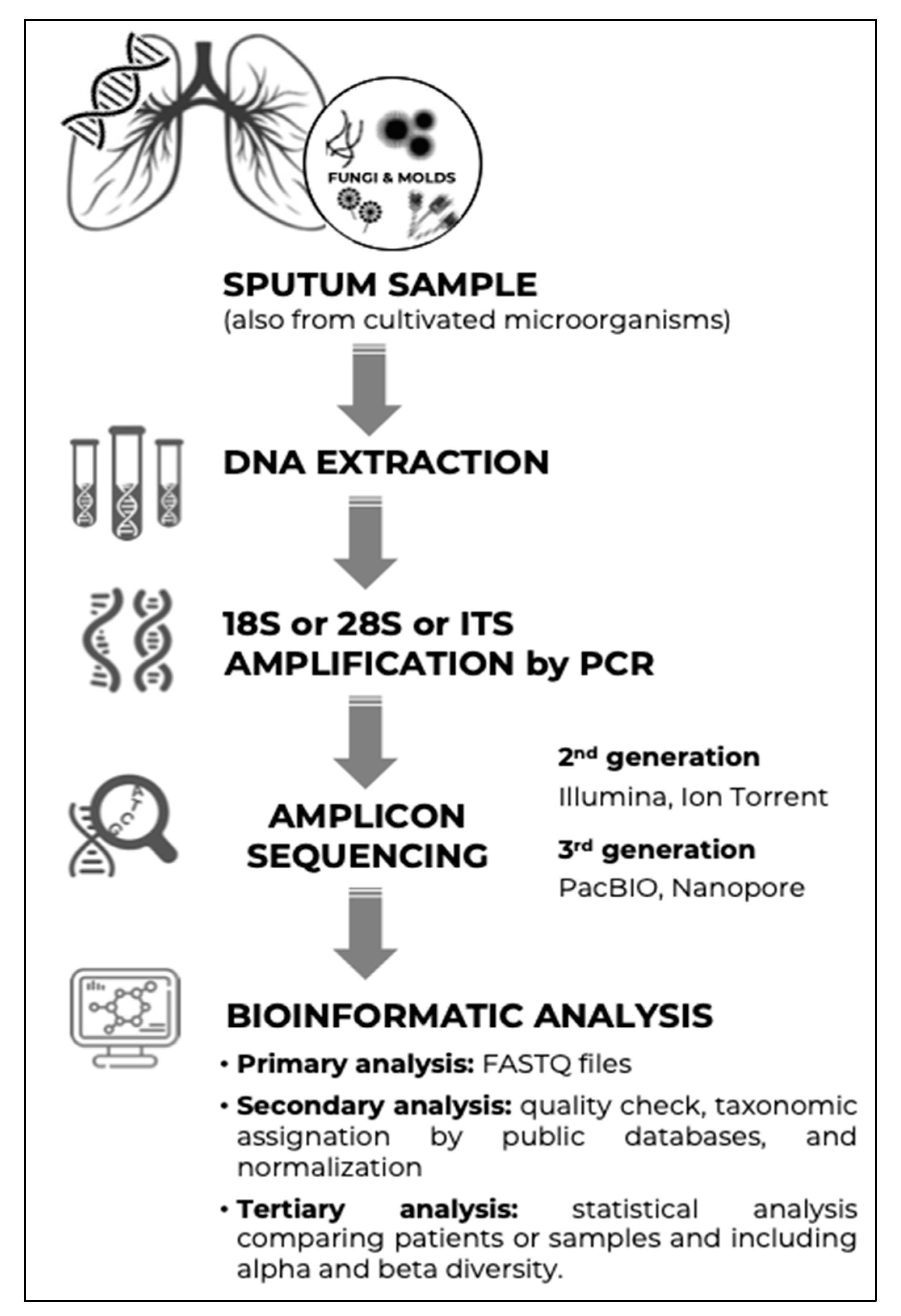
3. Studying the Human Mycobiome Using Next-Generation Sequencing
3.1. Introduction
3.2. Next-Generation Sequencing Strategies
3.3. Amplification Targets
3.4. Bioinformatics Analysis
- Primary analysis, which includes the transformation of the sequencer readings into base calls with their associated quality data. This is a closed process that is automatically performed by the sequencer itself and typically includes the demultiplexing process, whereby each read is assigned to its source sample (thanks to the unique barcode that is added to each library before sequencing). The outputs of this process in the case of Illumina platforms are FASTQ files.
- Secondary analysis, which refers to the curation of sequences and their counting and classification and typically includes the following:
- The trimming of adaptor and primer regions as well as conserved regions flanking the target region (ITS). The latter is a special feature that only applies when sequencing ITS. The size of this region varies according to the fungal species, requiring the use of tools that detect these ends for each particular sequence and cut at that point.
- Filtering out low-quality sequences (a factor given by the sequencing platform as a percentage of safety for each nucleotide).
- Filtering of sequences of lower-than-expected size.
- Merging of forward and reverse sequences (in the case of Illumina, paired-end protocols).
- The search and generation of representative sequences (amplicon sequence variants (ASVs)) and count tables per sample.
- The taxonomic assignment of ASVs. The main reference databases for the taxonomic assignment of fungi are UNITE, INSDC, SILVA, Warcup, and FindFungi.
- The elimination of ASVs not assigned to the kingdom Fungi.
- Data normalisation: Process that normalises the number of reads per sample for later comparison.
- Tertiary analysis, which begins once the processing of raw reads to achieve the ASVs is completed. This stage includes the statistical analysis of the sequencing data considering the clinical variables of the study samples. First, the alpha diversity (richness of each sample) is analysed, generally using the Shannon, Chao1, and Faith-PD indices. These parameters consider the number of species, their distribution, and, in the case of the latter, their phylogenetic relationships. The alignment of the ITS region is not useful for inferring the evolutionary distances between very distant species, although there are methods to partially overcome this limitation (https://github.com/JTFouquier/q2-ghost-tree, accessed on 10 April 2022). Moreover, beta-diversity analyses compare the composition between groups of samples (established according to the clinical variables collected) using indices that represent their similarity/dissimilarity, such as Bray Curtis, Jaccard, and UniFrac. These differences can be subjected to statistical analysis to assess their significance and/or represented graphically (e.g., via principal coordinate analysis). Differential abundance analyses can also be performed to identify which species primarily explain the differences between groups, using the linear discriminant analysis effect size tool based on linear discriminant analysis, as an example. Various platforms (either developed in a Linux environment or as web tools) facilitate the analysis, integrating the different tools and, above all, their statistical significance. These platforms include QIIME2 [82], mothur [83], CloVR-ITS [84], CONSTAX [85], and HumanMycobiomeScan [86].
3.5. Limitations
4. Structure and Composition of the Lung Mycobiome
4.1. Conventional Culture versus Next-Generation Sequencing
4.2. Healthy Individuals
4.3. Patients with Chronic Obstructive Pulmonary Disease
4.4. Patients with Non-Cystic Fibrosis Bronchiectasis
4.5. Patients with Cystic Fibrosis
4.6. Patients with Asthma
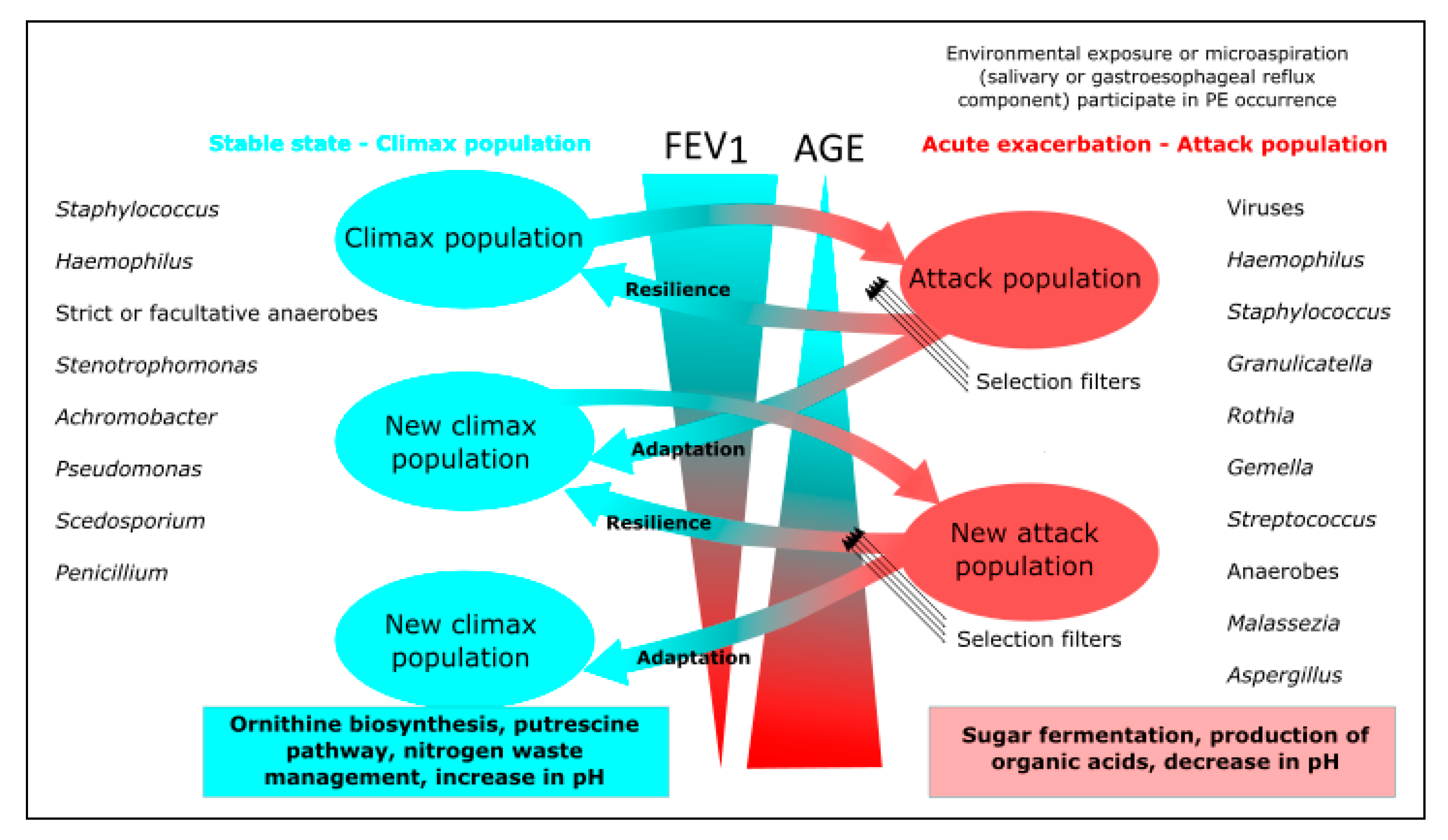
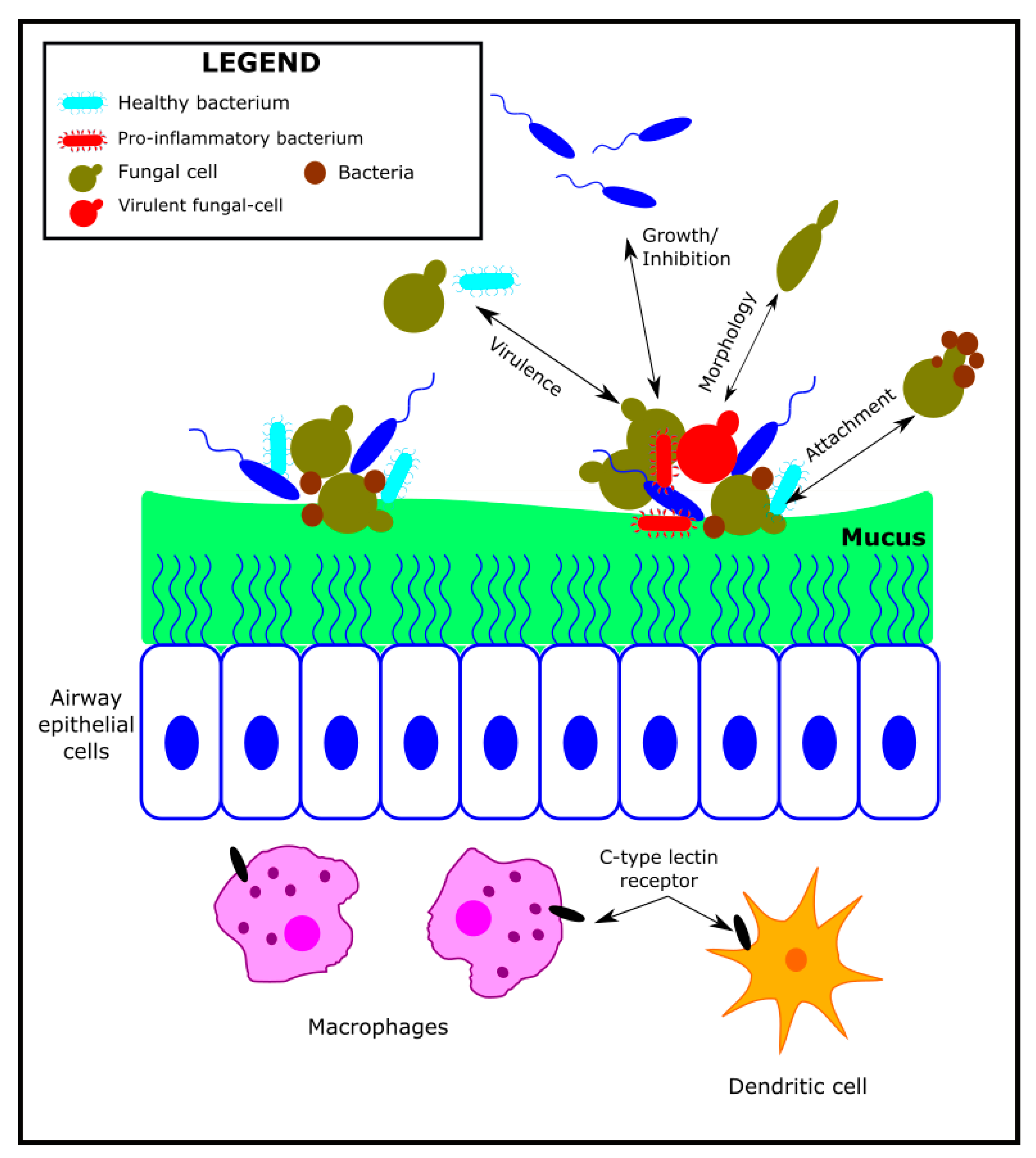
5. Role of the Mycobiome in Chronic Pulmonary Diseases
5.1. Direct Implications
5.2. Host and Bacterial Interactions with the Mycobiome
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paudel, K.R.; Jha, S.K.; Allam, V.S.R.R.; Prasher, P.; Gupta, P.K.; Bhattacharjee, R.; Jha, N.K.; Vishwas, S.; Singh, S.K.; Shrestha, J.; et al. Recent Advances in Chronotherapy Targeting Respiratory Diseases. Pharmaceutics 2021, 13, 2008. [Google Scholar] [CrossRef] [PubMed]
- Soler-Cataluña, J.J.; Novella, L.; Soler, C.; Nieto, M.L.; Esteban, V.; Sánchez-Toril, F.; Miravitlles, M. Clinical Characteristics and Risk of Exacerbations Associated With Different Diagnostic Criteria of Asthma-COPD Overlap. Arch. Bronconeumol. 2020, 56, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.B.; Alfageme, I.; Miravitlles, M.; de Lucas, P.; Soler-Cataluña, J.J.; García-Río, F.; Casanova, C.; González-Moro, J.M.R.; Cosío, B.G.; Sánchez, G.; et al. Prevalence and Determinants of COPD in Spain: EPISCAN II. Arch. Bronconeumol. 2021, 57, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Máiz, L.; Nieto, R.; Cantón, R.; de la Pedrosa, E.G.G.; Martinez-García, M.Á. Fungi in bronchiectasis: A concise review. Int. J. Mol. Sci. 2018, 19, 142. [Google Scholar] [CrossRef]
- Martinez-García, M.A.; Villa, C.; Dobarganes, Y.; Girón, R.; Máiz, L.; García-Clemente, M.; Sibila, O.; Golpe, R.; Rodríguez, J.; Barreiro, E.; et al. RIBRON: The spanish Online Bronchiectasis Registry. Characterization of the First 1912 Patients. Arch. Bronconeumol. 2021, 57, 28–35. [Google Scholar] [CrossRef]
- Olveira, C.; Padilla, A.; Martínez-García, M.Á.; Vendrell, M.; Máiz, L.; Borderías, L.; Polverino, E.; Martínez-Moragón, E.; Rajas, O.; Casas, F.; et al. Etiology of Bronchiectasis in a Cohort of 2047 Patients. An Analysis of the Spanish Historical Bronchiectasis Registry. Arch. Bronconeumol. 2017, 53, 366–374. [Google Scholar] [CrossRef]
- Martínez-García, M.A.; Perpiñá-Tordera, M.; Soler-Cataluña, J.J.; Román-Sánchez, P.; Lloris-Bayo, A.; González-Molina, A. Dissociation of lung function, dyspnea ratings and pulmonary extension in bronchiectasis. Respir. Med. 2007, 101, 2248–2253. [Google Scholar] [CrossRef]
- Martínez-Garćia, M.A.; Vendrell, M.; Girón, R.; Máiz-Carro, L.; de la Rosa Carrillo, D.; de Gracia, J.; Olveira, C. The Multiple Faces of Non-Cystic Fibrosis Bronchiectasis. A Cluster Analysis Approach. Ann. Am. Thorac. Soc. 2016, 13, 1468–1475. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.A.; Agustí, A. Heterogeneity and complexity in bronchiectasis: A pending challenge. Arch. Bronconeumol. 2019, 55, 187–188. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.A.; Miravitlles, M. Bronchiectasis in COPD patients: More than a comorbidity? Int. J. COPD 2017, 12, 1401–1411. [Google Scholar] [CrossRef]
- Padilla-Galo, A.; Olveira Fuster, C. Bronchiectasis in COPD and Asthma. More Than Just a Coincidence. Arch. Bronconeumol. 2019, 55, 181–182. [Google Scholar] [CrossRef] [PubMed]
- Stoltz, D.A.; Meyerholz, D.K.; Welsh, M.J. Origins of Cystic Fibrosis Lung Disease. N. Engl. J. Med. 2015, 372, 1574–1575. [Google Scholar] [CrossRef] [PubMed]
- LiPuma, J.J. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 2010, 23, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, S.A.; McGinniss, J.E.; Collman, R.G. The lung microbiome: Progress and promise. J. Clin. Investig. 2021, 131, e150473. [Google Scholar] [CrossRef]
- Nguyen, L.D.N.; Viscogliosi, E.; Delhaes, L. The lung mycobiome: An emerging field of the human respiratory microbiome. Front. Microbiol. 2015, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Waqas, S.; Dunne, K.; Talento, A.F.; Wilson, G.; Martín-Loeches, I.; Keane, J.; Rogers, T.R. Prospective observational study of respiratory Aspergillus colonization or disease in patients with various stages of chronic obstructive pulmonary disease utilizing culture versus nonculture techniques. Med. Mycol. 2021, 59, 557–563. [Google Scholar] [CrossRef]
- Hammond, E.E.; McDonald, C.S.; Vestbo, J.; Denning, D.W. The global impact of Aspergillus infection on COPD. BMC Pulm. Med. 2020, 20, 241. [Google Scholar] [CrossRef]
- Huerta, A.; Soler, N.; Esperatti, M.; Guerrero, M.; Menendez, R.; Gimeno, A.; Zalacaín, R.; Mir, N.; Aguado, J.M.; Torres, A. Importance of Aspergillus spp. isolation in acute exacerbations of severe COPD: Prevalence, factors and follow-up: The FUNGI-COPD study. Respir. Res. 2014, 15, 17. [Google Scholar] [CrossRef]
- Agarwal, R.; Hazarika, B.; Gupta, D.; Aggarwal, A.N.; Chakrabarti, A.; Jindal, S.K. Aspergillus hypersensitivity in patients with chronic obstructive pulmonary disease: COPD as a risk factor for ABPA? Med. Mycol. 2010, 48, 988–994. [Google Scholar] [CrossRef]
- Bafadhel, M.; Agbetile, J.; Fairs, A.; Desai, D.; Mistry, V.; Morley, J.P.; Pancholi, M.; Pavord, I.D.; Wardlaw, A.J.; Pashley, C.H.; et al. S91 Aspergillus fumigatus sensitisation in patients with chronic obstructive pulmonary disease. Thorax 2011, 66 (Suppl. S4), A43. [Google Scholar] [CrossRef]
- Connell, D.; Shah, A. The contribution of Aspergillus fumigatus to COPD exacerbations: A “sensitive” topic. Eur. Respir. J. 2020, 56, 2002223. [Google Scholar] [CrossRef] [PubMed]
- Tiew, P.Y.; San Ko, F.W.; Pang, S.L.; Matta, S.A.; Sio, Y.Y.; Poh, M.E.; Lau, K.J.X.; Mac Aogáin, M.; Jaggi, T.K.; Ivan, F.X.; et al. Environmental fungal sensitisation associates with poorer clinical outcomes in COPD. Eur. Respir. J. 2020, 56, 2000418. [Google Scholar] [CrossRef] [PubMed]
- Fraczek, M.G.; Chishimba, L.; Niven, R.M.; Bromley, M.; Simpson, A.; Smyth, L.; Denning, D.W.; Bowyer, P. Corticosteroid treatment is associated with increased filamentous fungal burden in allergic fungal disease. J. Allergy Clin. Immunol. 2018, 142, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Milla, C.E.; Wielinski, C.L.; Regelmann, W.E. Clinical significance of the recovery of Aspergillus species from the respiratory secretions of cystic fibrosis patients. Pediatr. Pulmonol. 1996, 21, 6–10. [Google Scholar] [CrossRef]
- Delhaes, L.; Touati, K.; Faure-Cognet, O.; Cornet, M.; Botterel, F.; Dannaoui, E.; Morio, F.; Le Pape, P.; Grenouillet, F.; Favennec, L.; et al. Prevalence, geographic risk factor, and development of a standardized protocol for fungal isolation in cystic fibrosis: Results from the international prospective study “MFIP”. J. Cyst. Fibrosis 2019, 18, 212–220. [Google Scholar] [CrossRef]
- Hedayati, M.T.; Tavakoli, M.; Maleki, M.; Heidari, S.; Mortezaee, V.; Gheisari, M.; Hassandaz, M.; Mirenayat, M.S.; Mahdaviani, S.A.; Pourabdollah, M.; et al. Fungal epidemiology in cystic fibrosis patients with a special focus on Scedosporium species complex. Microb. Pathog. 2019, 129, 168–175. [Google Scholar] [CrossRef]
- Máiz, L.; Cuevas, M.; Lamas, A.; Sousa, A.; Quirce, S.; Suárez, L. Aspergillus fumigatus and Candida albicans in cystic fibrosis: Clinical significance and specific immune response involving serum immunoglobulins G, A, and M. Arch. Bronconeumol. 2008, 44, 146–151. [Google Scholar] [CrossRef]
- Máiz, L.; Cuevas, M.; Quirce, S.; Cañón, J.F.; Pacheco, A.; Sousa, A.; Escobar, H. Serologic IgE immune responses against Aspergillus fumigatus and Candida albicans in patients with cystic fibrosis. Chest 2002, 121, 782–788. [Google Scholar] [CrossRef]
- Engel, T.G.P.; Slabbers, L.; de Jong, C.; Melchers, W.J.G.; Hagen, F.; Verweij, P.E.; Merkus, P.; Meis, J.F. Prevalence and diversity of filamentous fungi in the airways of cystic fibrosis patients—A Dutch, multicentre study. J. Cyst. Fibrosis 2019, 18, 221–226. [Google Scholar] [CrossRef]
- Schwarz, C.; Bouchara, J.P.; Buzina, W.; Chrenkova, V.; Dmeńska, H.; Gómez García de la Pedrosa, E.; Cantón, R.; Fiscarelli, E.; Le Govic, Y.; Kondori, N.; et al. Organization of Patient Management and Fungal Epidemiology in Cystic Fibrosis. Mycopathologia 2018, 183, 7–19. [Google Scholar] [CrossRef]
- Ramirez-Garcia, A.; Pellon, A.; Rementeria, A.; Buldain, I.; Barreto-Bertger, E.; Rollin-Pinheiro, R.; de Mireilles, J.V.; Xisto, M.I.D.S.; Ranque, S.; Havlicek, V.; et al. Scedosporium and Lomentospora: An updated overview of underrated opportunists. Med. Mycol. 2018, 56 (Suppl. S1), S102–S125. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.; Nachbaur, E.; Jaksch, P.; Dehlink, E. Update on Respiratory Fungal Infections in Cystic Fibrosis Lung Disease and after Lung Transplantation. J. Fungi 2020, 6, 381. [Google Scholar] [CrossRef] [PubMed]
- El-Dahr, J.M.; Fink, R.; Selden, R.; Arruda, L.K.; Platts-Mills, T.A.; Heymann, P.W. Development of immune responses to Aspergillus at an early age in children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 1994, 150 Pt 1, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Düesberg, U.; Wosniok, J.; Naehrlich, L.; Eschenhagen, P.; Schwarz, C. Risk factors for respiratory Aspergillus fumigatus in German Cystic Fibrosis patients and impact on lung function. Sci. Rep. 2020, 10, 18999. [Google Scholar] [CrossRef]
- Jubin, V.; Ranque, S.; Le Bel, N.S.; Sarles, J.; Dubus, J.C. Risk factors for Aspergillus colonization and allergic bronchopulmonary aspergillosis in children with cystic fibrosis. Pediatr. Pulmonol. 2010, 45, 764–771. [Google Scholar] [CrossRef]
- Bargon, J.; Dauletbaev, N.; Köhler, B.; Wolf, M.; Posselt, H.G.; Wagner, T.O. Prophylactic antibiotic therapy is associated with an increased prevalence of Aspergillus colonization in adult cystic fibrosis patients. Respir. Med. 1999, 93, 835–838. [Google Scholar] [CrossRef]
- Sudfeld, C.R.; Dasenbrook, E.C.; Merz, W.G.; Carroll, K.C.; Boyle, M.P. Prevalence and risk factors for recovery of filamentous fungi in individuals with cystic fibrosis. J. Cyst. Fibrosis 2010, 9, 110–116. [Google Scholar] [CrossRef]
- Laufer, P.; Fink, J.N.; Bruns, W.T.; Unger, G.F.; Kalbfleisch, J.H.; Greenberger, P.A.; Patterson, R. Allergic bronchopulmonary aspergillosis in cystic fibrosis. J. Allergy Clin. Immunol. 1984, 73 Pt 1, 44–48. [Google Scholar] [CrossRef]
- Maturu, V.N.; Agarwal, R. Prevalence of Aspergillus sensitization and allergic bronchopulmonary aspergillosis in cystic fibrosis: Systematic review and meta-analysis. Clin. Exp. Allergy 2015, 45, 1765–1778. [Google Scholar] [CrossRef]
- Amin, R.; Dupuis, A.; Aaron, S.D.; Ratjen, F. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 2010, 137, 171–176. [Google Scholar] [CrossRef]
- Baxter, C.G.; Moore, C.B.; Jones, A.M.; Webb, A.K.; Denning, D.W. IgE-mediated immune responses and airway detection of Aspergillus and Candida in adult cystic fibrosis. Chest 2013, 143, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Breuer, O.; Schultz, A.; Garratt, L.W.; Turkovic, L.; Rosenow, T.; Murray, C.P.; Karpievitch, Y.V.; Akesson, L.; Dalton, S.; Sly, P.D.; et al. Aspergillus infections and progression of structural lung disease in children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2020, 201, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Middleton, P.G.; Chen, S.C.A.; Meyer, W. Fungal infections and treatment in cystic fibrosis. Curr. Opin. Pulm. Med. 2013, 19, 670–675. [Google Scholar] [CrossRef] [PubMed]
- de Vrankrijker, A.M.M.; van der Ent, C.K.; van Berkhout, F.T.; Stellato, R.K.; Willems, R.J.L.; Bonten, M.J.M.; Wolfs, T.F.W. Aspergillus fumigatus colonization in cystic fibrosis: Implications for lung function? Clin. Microbiol. Infect. 2011, 17, 1381–1386. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; O’Donoghue, E.; Bennett, K.; Gunaratnam, C.; O’Neill, S.J.; McElvaney, N.G. Sputum Candida albicans presages FEV₁ decline and hospital-treated exacerbations in cystic fibrosis. Chest 2010, 138, 1186–1195. [Google Scholar] [CrossRef]
- Gileles-Hillel, A.; Shoseyov, D.; Polacheck, I.; Korem, M.; Kerem, E.; Cohen-Cymberknoh, M. Association of chronic Candida albicans respiratory infection with a more severe lung disease in patients with cystic fibrosis. Pediatr. Pulmonol. 2015, 50, 1082–1089. [Google Scholar] [CrossRef]
- Singh, A.; Ralhan, A.; Schwarz, C.; Hartl, D.; Hector, A. Fungal Pathogens in CF Airways: Leave or Treat? Mycopathologia 2018, 183, 119–137. [Google Scholar] [CrossRef]
- Muthig, M.; Hebestreit, A.; Ziegler, U.; Seidler, M.; Müller, F.M.C. Persistence of Candida species in the respiratory tract of cystic fibrosis patients. Med. Mycol. 2010, 48, 56–63. [Google Scholar] [CrossRef][Green Version]
- Esther, C.R.J.; Plongla, R.; Kerr, A.; Lin, F.-C.; Gilligan, P. Clinical outcomes in cystic fibrosis patients with Trichosporon respiratory infection. J. Cyst. Fibrosis 2016, 15, e45–e49. [Google Scholar] [CrossRef]
- Kroner, C.; Kappler, M.; Grimmelt, A.-C.; Laniado, G.; Wurstl, B.; Griese, M. The basidiomycetous yeast Trichosporon may cause severe lung exacerbation in cystic fibrosis patients—Clinical analysis of Trichosporon positive patients in a Munich cohort. BMC Pulm. Med. 2013, 13, 61. [Google Scholar] [CrossRef]
- de Almeida, J.N.; Hennequin, C. Invasive Trichosporon Infection: A Systematic Review on a Re-emerging Fungal Pathogen. Front. Microbiol. 2016, 7, 1629. [Google Scholar] [CrossRef] [PubMed]
- Tiew, P.Y.; Jaggi, T.K.; Chan, L.L.Y.; Chotirmall, S.H. The airway microbiome in COPD, bronchiectasis and bronchiectasis-COPD overlap. Clin. Respir. J. 2021, 15, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, R.; Mac Aogáin, M.; Chalmers, J.D.; Elborn, S.J.; Chotirmall, S.H. Geographic variation in the aetiology, epidemiology and microbiology of bronchiectasis. BMC Pulm. Med. 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Felton, I.; James, P.; Cox, M.J.; Bilton, D.; Schelenz, S.; Loebinger, M.R.; Cookson, W.O.C.; Simmonds, N.J.; Moffatt, M.F. The fungal airway microbiome in cystic fibrosis and non-cystic fibrosis bronchiectasis. J. Cyst. Fibrosis 2021, 20, 295–302. [Google Scholar] [CrossRef]
- Maiz, L.; Vendrell, M.; Olveira, C.; Giron, R.; Nieto, R.; Martinez-Garcia, M.A. Prevalence and factors associated with isolation of Aspergillus and Candida from sputum in patients with non-cystic fibrosis bronchiectasis. Respiration 2015, 89, 396–403. [Google Scholar] [CrossRef]
- Pashley, C.H. Fungal culture and sensitisation in asthma, cystic fibrosis and chronic obstructive pulmonary disorder: What does it tell us? Mycopathologia 2014, 178, 457–463. [Google Scholar] [CrossRef]
- Mac Aogáin, M.; Chandrasekaran, R.; Hou Lim, A.Y.; Low, T.B.; Tan, G.L.; Hassan, T.; Ong, T.H.; Ng, A.H.Q.; Bertrand, D.; Koh, J.Y.; et al. Immunological corollary of the pulmonary mycobiome in bronchiectasis: The CAMEB study. Eur. Respir. J. 2018, 52, 1800766. [Google Scholar] [CrossRef]
- Mac Aogáin, M.; Tiew, P.Y.; Lim, A.Y.H.; Low, T.B.; Tan, G.L.; Hassan, T.; Ong, T.H.; Pang, S.L.; Lee, Z.Y.; Gwee, X.W.; et al. Distinct “immunoallertypes” of disease and high frequencies of sensitization in non-cystic fibrosis bronchiectasis. Am. J. Respir. Crit. Care Med. 2019, 199, 842–853. [Google Scholar] [CrossRef]
- García-Rivero, J.L. The Microbiome and Asthma. Arch. Bronconeumol. 2020, 56, 1–2. [Google Scholar] [CrossRef]
- van Tilburg Bernardes, E.; Gutierrez, M.W.; Arrieta, M.C. The Fungal Microbiome and Asthma. Front. Cell. Infect. Microbiol. 2020, 10, 583418. [Google Scholar] [CrossRef]
- Monsó, E. The respiratory microbiome: Beyond the culture. Arch. Bronconeumol. 2017, 53, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Páez Codeso, F.M.; Bermúdez Ruiz, M.P.; Dorado Galindo, A. Radiology, Bronchoscopy and Microbiology in Bronchopulmonary Aspergillosis. Arch. Bronconeumol. 2018, 54, 625. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, R.A.; Bearman, N.; Thornton, C.R.; Husk, K.; Osborne, N.J. Indoor fungal diversity and asthma: A meta-analysis and systematic review of risk factors. J. Allergy Clin. Immunol. 2015, 135, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Morris, A.; Ghedin, E. The human mycobiome in health and disease. Genome Med. 2013, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Delhaes, L.; Monchy, S.; Fréalle, E.; Hubans, C.; Salleron, J.; Leroy, S.; Prevotat, A.; Wallet, F.; Wallaert, B.; Dei-Cas, E.; et al. The airway microbiota in cystic fibrosis: A complex fungal and bacterial community-implications for therapeutic management. PLoS ONE 2012, 7, e36313. [Google Scholar] [CrossRef]
- Větrovský, T.; Morais, D.; Kohout, P.; Lepinay, C.; Algora, C.; Hollá, S.A.; Bahnmann, B.D.; Bílohnêdá, K.; Brabcová, V.; D'Aló, F.; et al. GlobalFungi, a global database of fungal occurrences from high-throughput-sequencing metabarcoding studies. Sci. Data 2020, 7, 228. [Google Scholar] [CrossRef]
- Donovan, P.D.; Gonzalez, G.; Higgins, D.G.; Butler, G.; Ito, K. Identification of fungi in shotgun metagenomics datasets. PLoS ONE 2018, 13, e0192898. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gessell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Sun, Y.; Zuo, T.; Cheung, C.P.; Gu, W.; Wan, Y.; Zhang, F.; Chen, N.; Zhan, H.; Yeoh, Y.K.; Niu, J.; et al. Population-Level Configurations of Gut Mycobiome Across 6 Ethnicities in Urban and Rural China. Gastroenterology 2021, 160, 272–286.e11. [Google Scholar] [CrossRef]
- McTaggart, L.R.; Copeland, J.K.; Surendra, A.; Wang, P.W.; Husain, S.; Coburn, B.; Guttman, D.S.; Kus, J.V. Mycobiome Sequencing and Analysis Applied to Fungal Community Profiling of the Lower Respiratory Tract During Fungal Pathogenesis. Front. Microbiol. 2019, 10, 512. [Google Scholar] [CrossRef] [PubMed]
- Bello, S.; Vengoechea, J.J.; Ponce-Alonso, M.; Figueredo, A.L.; Mincholé, E.; Rezusta, A.; Gambó, P.; Pastor, J.M.; Galeano, J.; del Campo, R. Core Microbiota in Central Lung Cancer With Streptococcal Enrichment as a Possible Diagnostic Marker. Arch. Bronconeumol. 2020, 57, 681–689. [Google Scholar] [CrossRef]
- Soret, P.; Vandenborght, L.E.; Francis, F.; Coron, N.; Enaud, R.; Avalos, M.; Schaeverbeke, T.; Berger, P.; Fayon, M.; Thiebaut, R.; et al. Respiratory mycobiome and suggestion of inter-kingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci. Rep. 2020, 10, 3589. [Google Scholar] [CrossRef] [PubMed]
- Huseyin, C.E.; Rubio, R.C.; O’Sullivan, O.; Cotter, P.D.; Scanlan, P.D. The Fungal Frontier: A Comparative Analysis of Methods Used in the Study of the Human Gut Mycobiome. Front. Microbiol. 2017, 8, 1432. [Google Scholar] [CrossRef] [PubMed]
- Tiew, P.Y.; Mac Aogain, M.; Ali, N.A.B.M.; Thng, K.X.; Goh, K.; Lau, K.J.X.; Chotirmall, S.H. The Mycobiome in Health and Disease: Emerging Concepts, Methodologies and Challenges. Mycopathologia 2020, 185, 207–231. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 2020, 367, eaba1223. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Ali, N.A.B.M.; Mac Aogáin, M.; Morales, R.F.; Tiew, P.Y.; Chotirmall, S.H. Optimisation and Benchmarking of Targeted Amplicon Sequencing for Mycobiome Analysis of Respiratory Specimens. Int. J. Mol. Sci. 2019, 20, 4991. [Google Scholar] [CrossRef]
- Frau, A.; Kenny, J.G.; Lenzi, L.; Campbell, B.J.; Ijaz, U.; Duckworth, C.A.; Burkitt, M.D.; Hall, N.; Anson, J.; Darby, A.C.; et al. DNA extraction and amplicon production strategies deeply influence the outcome of gut mycobiome studies. Sci. Rep. 2019, 9, 9328. [Google Scholar] [CrossRef]
- Li, S.; Deng, Y.; Wang, Z.; Zhang, Z.; Kong, X.; Zhou, W.; Yi, Y.; Qu, Y. Exploring the accuracy of amplicon-based internal transcribed spacer markers for a fungal community. Mol. Ecol. Resour. 2020, 20, 170–184. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; González Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D. Reintroducing mothur: 10 Years Later. Appl. Environ. Microbiol. 2020, 86, e02343. [Google Scholar] [CrossRef] [PubMed]
- White, J.R.; Maddox, C.; White, O.; Angiuoli, S.V.; Fricke, W.F. CloVR-ITS: Automated internal transcribed spacer amplicon sequence analysis pipeline for the characterization of fungal microbiota. Microbiome 2013, 1, 6. [Google Scholar] [CrossRef]
- Gdanetz, K.; Benucci, G.M.N.; Vande Pol, N.; Bonito, G. CONSTAX: A tool for improved taxonomic resolution of environmental fungal ITS sequences. BMC Bioinform. 2017, 18, 538. [Google Scholar] [CrossRef]
- Soverini, M.; Turroni, S.; Biagi, E.; Brigidi, P.; Candela, M.; Rampelli, S. HumanMycobiomeScan: A new bioinformatics tool for the characterization of the fungal fraction in metagenomic samples. BMC Genom. 2019, 20, 496. [Google Scholar] [CrossRef]
- Vesty, A.; Biswas, K.; Taylor, M.W.; Gear, K.; Douglas, R.G. Evaluating the Impact of DNA Extraction Method on the Representation of Human Oral Bacterial and Fungal Communities. PLoS ONE 2017, 12, e0169877. [Google Scholar] [CrossRef]
- Rosenbaum, J.; Usyk, M.; Chen, Z.; Zolnik, C.P.; Jones, H.E.; Waldron, L.; Dowd, J.B.; Thorpe, L.E.; Burk, R.D. Evaluation of Oral Cavity DNA Extraction Methods on Bacterial and Fungal Microbiota. Sci. Rep. 2019, 9, 1531. [Google Scholar] [CrossRef]
- Nguyen, L.D.N.; Deschaght, P.; Merlin, S.; Loywick, A.; Audebert, C.; Van Daele, S.; Viscogliosi, E.; Vaneechoutte, M.; Delhaes, L. Effects of Propidium Monoazide (PMA) Treatment on Mycobiome and Bacteriome Analysis of Cystic Fibrosis Airways during Exacerbation. PLoS ONE 2016, 11, e0168860. [Google Scholar] [CrossRef]
- Mirzayi, C.; Renson, A.; Genomic Standards Consortium; Massive Analysis and Quality Control Society; Zohra, F.; Elsafoury, S.; Geistlinger, L.; Kasselman, L.J.; Eckenrode, K.; van de Wijgert, J.; et al. Reporting guidelines for human microbiome research: The STORMS checklist. Nat. Med. 2021, 27, 1885–1892. [Google Scholar] [CrossRef]
- Pashley, C.H.; Fairs, A.; Morley, J.P.; Tailor, S.; Agbetile, J.; Bafadhel, M.; Brightling, C.E.; Wardlaw, A.J. Routine processing procedures for isolating filamentous fungi from respiratory sputum samples may underestimate fungal prevalence. Med. Mycol. 2012, 50, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.; Sauer-Heilborn, A.; Welte, T.; Guzman, C.A.; Abraham, W.-R.; Höfle, M.G. Cohort Study of Airway Mycobiome in Adult Cystic Fibrosis Patients: Differences in Community Structure between Fungi and Bacteria Reveal Predominance of Transient Fungal Elements. J. Clin. Microbiol. 2015, 53, 2900–2907. [Google Scholar] [CrossRef] [PubMed]
- Masoud-Landgraf, L.; Badura, A.; Eber, E.; Feierl, G.; Marth, E.; Buzina, W. Modified culture method detects a high diversity of fungal species in cystic fibrosis patients. Med. Mycol. 2014, 52, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Engel, T.G.P.; Tehupeiory-Kooreman, M.; Melchers, W.J.G.; Reijers, M.H.; Merkus, P.; Verweij, P.E. Evaluation of a New Culture Protocol for Enhancing Fungal Detection Rates in Respiratory Samples of Cystic Fibrosis Patients. J. Fungi 2020, 6, 82. [Google Scholar] [CrossRef]
- Willger, S.D.; Grim, S.L.; Dolben, E.L.; Shipunova, A.; Hampton, T.H.; Morrison, H.G.; Filkins, L.M.; O'Toole, G.A.; Moulton, L.A.; Ashare, A.; et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2014, 2, 40. [Google Scholar] [CrossRef]
- van Woerden, H.C.; Gregory, C.; Brown, R.; Marchesi, J.R.; Hoogendoorn, B.; Matthews, I.P. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: A community based case control study. BMC Infect. Dis. 2013, 13, 69. [Google Scholar] [CrossRef]
- Gaitanis, G.; Magiatis, P.; Hantschke, M.; Bassukas, I.D.; Velegraki, A. The Malassezia genus in skin and systemic diseases. Clin. Microbiol. Rev. 2012, 25, 106–141. [Google Scholar] [CrossRef]
- Huang, C.; Yu, Y.; Du, W.; Liu, Y.; Dai, R.; Tang, W.; Wang, P.; Zhang, C.; Shi, G. Fungal and bacterial microbiome dysbiosis and imbalance of trans-kingdom network in asthma. Clin. Transl. Allergy 2020, 10, 42. [Google Scholar] [CrossRef]
- Sharma, A.; Laxman, B.; Naureckas, E.T.; Hogarth, D.K.; Sperling, A.I.; Solway, J.; Ober, C.; Gilbert, J.A.; White, S.R. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J. Allergy Clin. Immunol. 2019, 144, 1214–1227.e7. [Google Scholar] [CrossRef]
- Charlson, E.S.; Diamond, J.M.; Bittinger, K.; Fitzgerald, A.S.; Yadav, A.; Haas, A.R.; Bushman, F.D.; Collman, R.G. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am. J. Respir. Crit. Care Med. 2012, 186, 536–545. [Google Scholar] [CrossRef]
- Segal, L.N.; Clemente, J.C.; Tsay, J.C.J.; Koralov, S.B.; Keller, B.C.; Wu, B.G.; Li, Y.; Shen, N.; Ghedin, E.; Morris, A.; et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016, 1, 16031. [Google Scholar] [CrossRef] [PubMed]
- Gollwitzer, E.S.; Saglani, S.; Trompette, A.; Yadava, K.; Sherburn, R.; McCoy, K.D.; Nicod, L.P.; Lloyd, C.M.; Marsland, B.J. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 2014, 20, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Herbst, T.; Sichelstiel, A.; Schär, C.; Yadava, K.; Bürki, K.; Cahenzli, J.; McCoy, K.; Marsland, B.J.; Harris, N.L. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 2011, 184, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.G.; Sulaiman, I.; Tsay, J.C.J.; Pérez, L.; Franca, B.; Li, Y.; Wang, J.; González, A.N.; El-Ashmawy, M.; Carpenito, J.; et al. Episodic Aspiration with Oral Commensals Induces a MyD88-dependent, Pulmonary T-Helper Cell Type 17 Response that Mitigates Susceptibility to Streptococcus pneumoniae. Am. J. Respir. Crit. Care Med. 2021, 203, 1099–1111. [Google Scholar] [CrossRef]
- Brown, R.L.; Sequeira, R.P.; Clarke, T.B. The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 2017, 8, 1512. [Google Scholar] [CrossRef]
- Cui, L.; Lucht, L.; Tipton, L.; Rogers, M.B.; Fitch, A.; Kessinger, C.; Camp, D.; Kingsley, L.; Leo, N.; Greenblatt, R.M.; et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am. J. Respir. Crit. Care Med. 2015, 191, 932–942. [Google Scholar] [CrossRef]
- Su, J.; Liu, H.; Tan, X.; Ji, Y.; Jiang, Y.X.; Prabhakar, M.; Rong, Z.H.; Zhou, H.W.; Zhang, G.X. Sputum Bacterial and Fungal Dynamics during Exacerbations of Severe COPD. PLoS ONE 2015, 10, e0130736. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Dicker, A.J.; Keir, H.R.; Poh, M.E.; Pang, S.L.; Mac Aogáin, M.; Chua, B.Q.Y.; Tan, J.L.; Xu, H.; Koh, M.S.; et al. A high-risk airway mycobiome is associated with frequent exacerbation and mortality in COPD. Eur. Respir. J. 2021, 57, 2002050. [Google Scholar] [CrossRef]
- Martinsen, E.M.H.; Eagan, T.M.L.; Leiten, E.O.; Haaland, I.; Husebø, G.R.; Knudsen, K.S.; Drengenes, C.; Sanseverino, W.; Paytuví-Gallart, A.; Nielsen, R. The pulmonary mycobiome—A study of subjects with and without chronic obstructive pulmonary disease. PLoS ONE 2021, 16, e0248967. [Google Scholar] [CrossRef]
- Martínez-García, M.Á.; Máiz, L.; Olveira, C.; Girón, R.M.; de la Rosa, D.; Blanco, M.; Cantón, R.; Vendrell, M.; Polverino, E.; de Gracia, J.; et al. Spanish Guidelines on Treatment of Bronchiectasis in Adults. Arch. Bronconeumol. 2018, 54, 88–98. [Google Scholar] [CrossRef]
- Sanders, D.B.; Bittner, R.C.L.; Rosenfeld, M.; Hoffman, L.R.; Redding, G.J.; Goss, C.H. Failure to recover to baseline pulmonary function after cystic fibrosis pulmonary exacerbation. Am. J. Respir. Crit. Care Med. 2010, 182, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Conrad, D.; Haynes, M.; Salamon, P.; Rainey, P.B.; Youle, M.; Rohwer, F. Cystic fibrosis therapy: A community ecology perspective. Am. J. Respir. Cell. Mol. Biol. 2013, 48, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, I.; Pletcher, S.D.; Goldberg, A.N.; Barker, B.M.; Cope, E.K. Fungal Microbiota in Chronic Airway Inflammatory Disease and Emerging Relationships with the Host Immune Response. Front. Microbiol. 2017, 8, 2477. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, A. The role of the house dust mite-induced innate immunity in development of allergic response. Int. Arch. Allergy Immunol. 2011, 155, 95–105. [Google Scholar] [CrossRef]
- Carpagnano, G.E.; Malerba, M.; Lacedonia, D.; Susca, A.; Logrieco, A.; Carone, M.; Cotugno, G.; Palmiotti, G.A.; Foschino-Barbaro, M.P. Analysis of the fungal microbiome in exhaled breath condensate of patients with asthma. Allergy Asthma Proc. 2016, 37, e41–e46. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Mac Aogáin, M.; Chotirmall, S.H. The current understanding and future directions for sputum microbiome profiling in chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 2022, 28, 121–133. [Google Scholar] [CrossRef]
- Filho, F.S.L.; Alotaibi, N.M.; Ngan, D.; Tam, S.; Yang, J.; Hollander, Z.; Chen, V.; Fitzgerald, J.M.; Nislow, C.; Leung, J.M.; et al. Sputum Microbiome Is Associated with 1-Year Mortality after Chronic Obstructive Pulmonary Disease Hospitalizations. Am. J. Respir. Crit. Care Med. 2019, 199, 1205–1213. [Google Scholar] [CrossRef]
- Guinea, J.; Torres-Narbona, M.; Gijón, P.; Muñoz, P.; Pozo, F.; Peláez, T.; de Miguel, J.; Bouza, E. Pulmonary aspergillosis in patients with chronic obstructive pulmonary disease: Incidence, risk factors, and outcome. Clin. Microbiol. Infect. 2010, 16, 870–877. [Google Scholar] [CrossRef]
- Bafadhel, M.; McKenna, S.; Agbetile, J.; Fairs, A.; Desai, D.; Mistry, V.; Morley, J.P.; Pancholi, M.; Pavord, I.D.; Wardlaw, A.J.; et al. Aspergillus fumigatus during stable state and exacerbations of COPD. Eur. Respir. J. 2014, 43, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Krüger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.D.; Niemiec, M.J. Fungal-bacterial interactions in health and disease. Pathogens 2019, 8, 70. [Google Scholar] [CrossRef]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell. Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Rall, G.; Knoll, L.J. Development of Complex Models to Study Co- and Polymicrobial Infections and Diseases. PLoS Pathog. 2016, 12, e1005858. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.R.; Taylor, G.W.; Rutman, A.; Høiby, N.; Cole, P.J.; Wilson, R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 1999, 52, 385–387. [Google Scholar] [CrossRef]
- Reece, E.; Doyle, S.; Greally, P.; Renwick, J.; McClean, S. Aspergillus fumigatus Inhibits Pseudomonas aeruginosa in Co-culture: Implications of a Mutually Antagonistic Relationship on Virulence and Inflammation in the CF Airway. Front. Microbiol. 2018, 9, 1205. [Google Scholar] [CrossRef] [PubMed]
- Kolwijck, E.; van de Veerdonk, F.L. The potential impact of the pulmonary microbiome on immunopathogenesis of Aspergillus-related lung disease. Eur. J. Immunol. 2014, 44, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Udayanga, K.G.S.; Totsuka, N.; Weinberg, J.B.; Núñez, G.; Shibuya, A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 2014, 15, 95–102. [Google Scholar] [CrossRef]
- Hartl, D. Immunological mechanisms behind the cystic fibrosis-ABPA link. Med. Mycol. 2009, 47 (Suppl. S1), S183–S191. [Google Scholar] [CrossRef]
| Yeasts | Filamentous Fungi |
|---|---|
| Candida albicans | Aspergillus fumigatus |
| Candida glabrata | Aspergillus niger |
| Candida parapsilosis | Aspergillus terreus |
| Saccharomyces cerevisiae | Scedosporium apiospermum |
| Trichosporon beigelii | Penicillium spp. |
| Exophiala dermatitidis | Fusarium spp. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Dios Caballero, J.; Cantón, R.; Ponce-Alonso, M.; García-Clemente, M.M.; Gómez G. de la Pedrosa, E.; López-Campos, J.L.; Máiz, L.; del Campo, R.; Martínez-García, M.Á. The Human Mycobiome in Chronic Respiratory Diseases: Current Situation and Future Perspectives. Microorganisms 2022, 10, 810. https://doi.org/10.3390/microorganisms10040810
de Dios Caballero J, Cantón R, Ponce-Alonso M, García-Clemente MM, Gómez G. de la Pedrosa E, López-Campos JL, Máiz L, del Campo R, Martínez-García MÁ. The Human Mycobiome in Chronic Respiratory Diseases: Current Situation and Future Perspectives. Microorganisms. 2022; 10(4):810. https://doi.org/10.3390/microorganisms10040810
Chicago/Turabian Stylede Dios Caballero, Juan, Rafael Cantón, Manuel Ponce-Alonso, Marta María García-Clemente, Elia Gómez G. de la Pedrosa, José Luis López-Campos, Luis Máiz, Rosa del Campo, and Miguel Ángel Martínez-García. 2022. "The Human Mycobiome in Chronic Respiratory Diseases: Current Situation and Future Perspectives" Microorganisms 10, no. 4: 810. https://doi.org/10.3390/microorganisms10040810
APA Stylede Dios Caballero, J., Cantón, R., Ponce-Alonso, M., García-Clemente, M. M., Gómez G. de la Pedrosa, E., López-Campos, J. L., Máiz, L., del Campo, R., & Martínez-García, M. Á. (2022). The Human Mycobiome in Chronic Respiratory Diseases: Current Situation and Future Perspectives. Microorganisms, 10(4), 810. https://doi.org/10.3390/microorganisms10040810







