Endophytic PGPR from Tomato Roots: Isolation, In Vitro Characterization and In Vivo Evaluation of Treated Tomatoes (Solanum lycopersicum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Endophytes from Roots
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Selection of Potentially Interesting PGPR Isolates
2.4. In Planta Tests
2.5. Statistical Analysis
2.6. Indole Acetic Acid Production
2.7. Phosphate Solubilization
3. Results
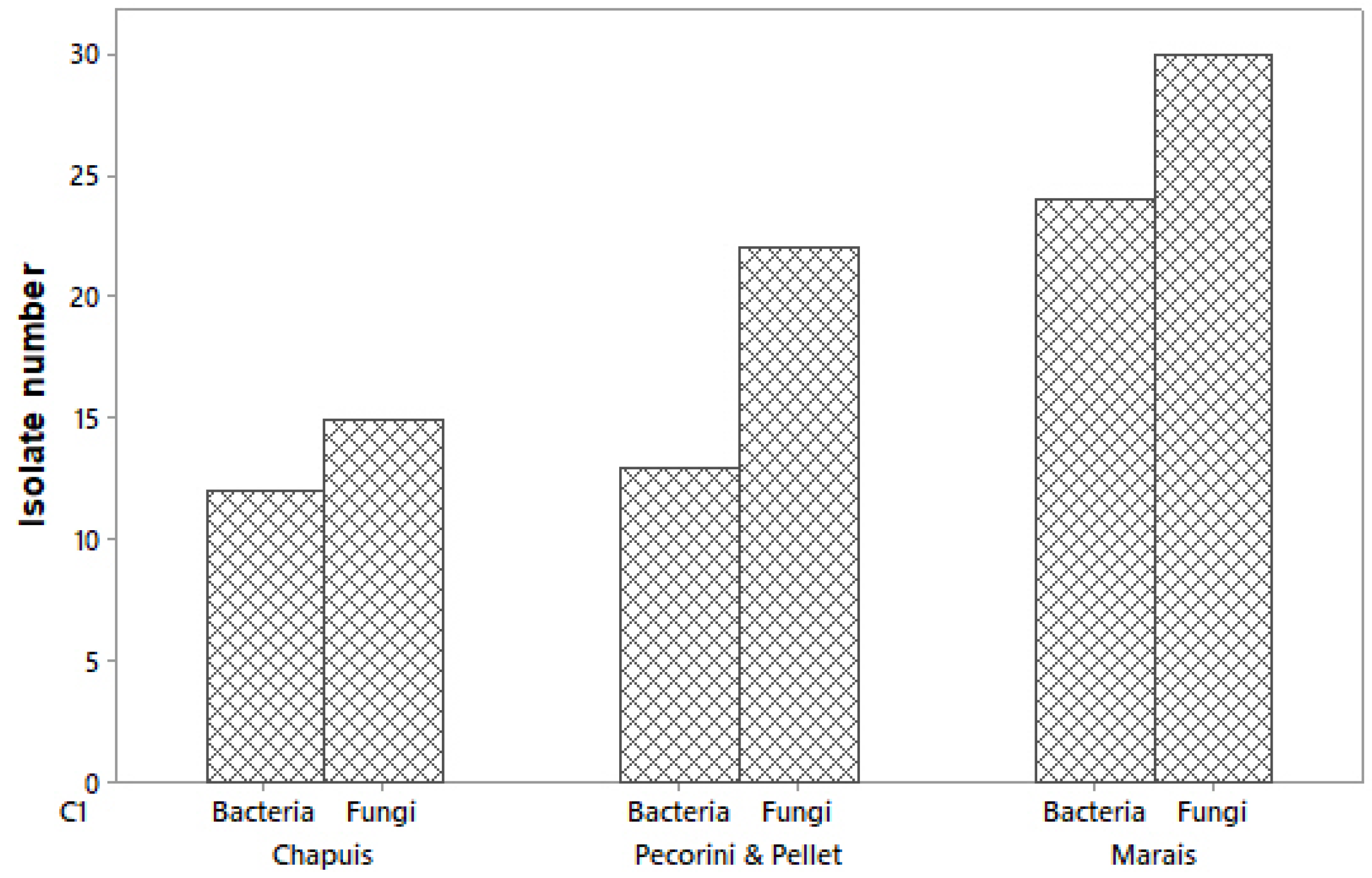
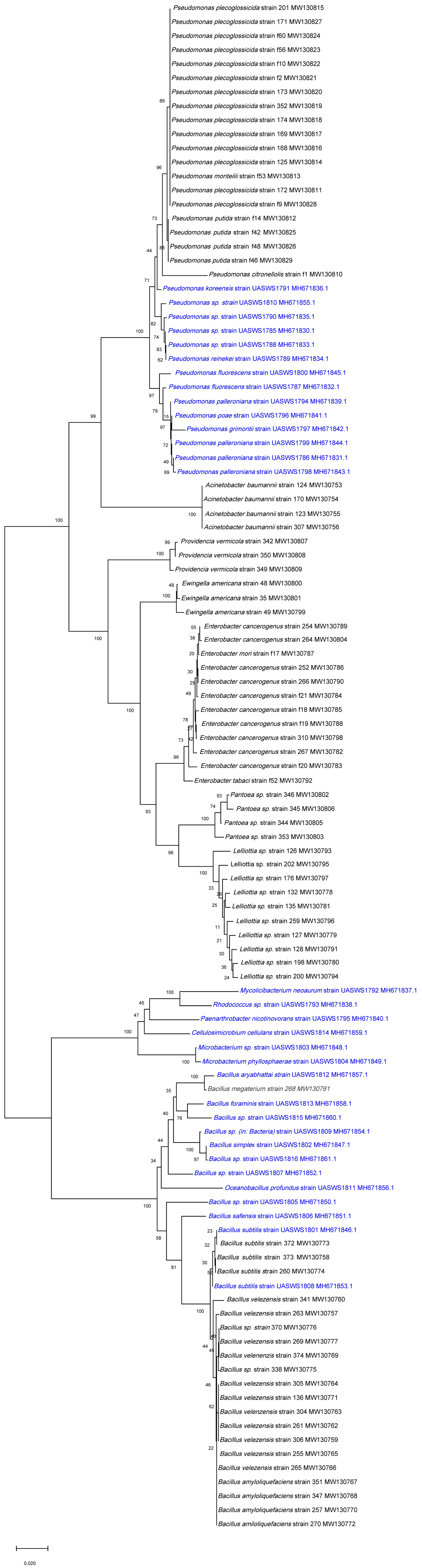
3.1. Isolation, Identification and Selection of Endophytes
3.2. Selected Microorganisms for Biochemical and in Planta Tests
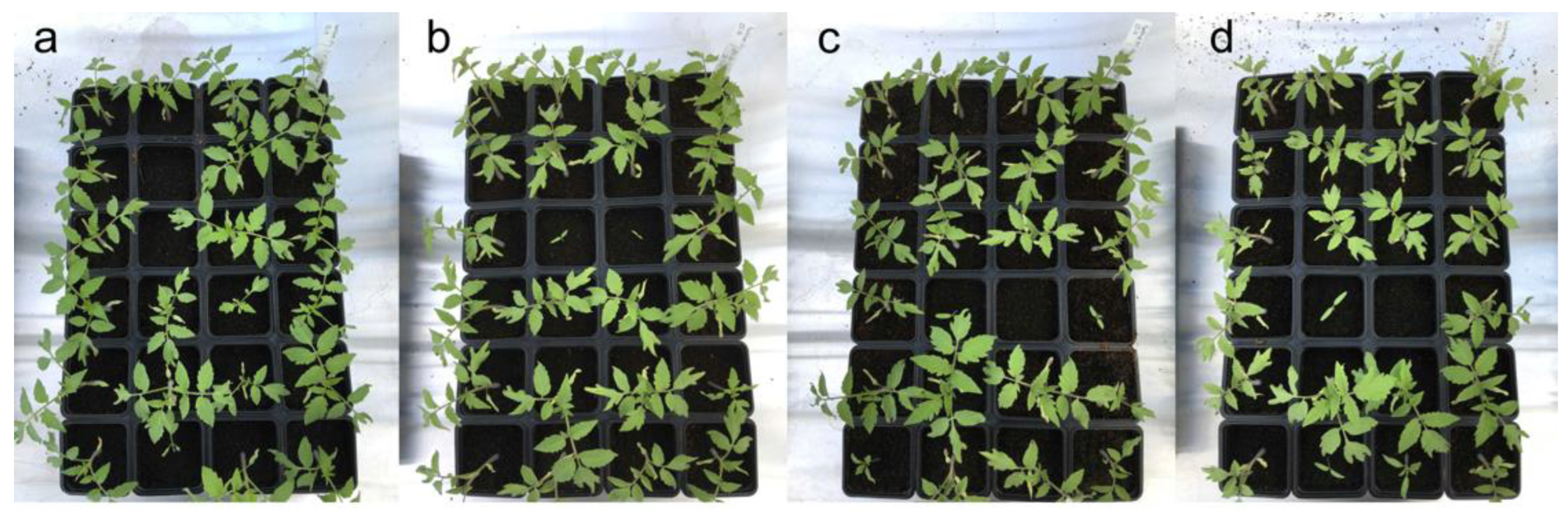
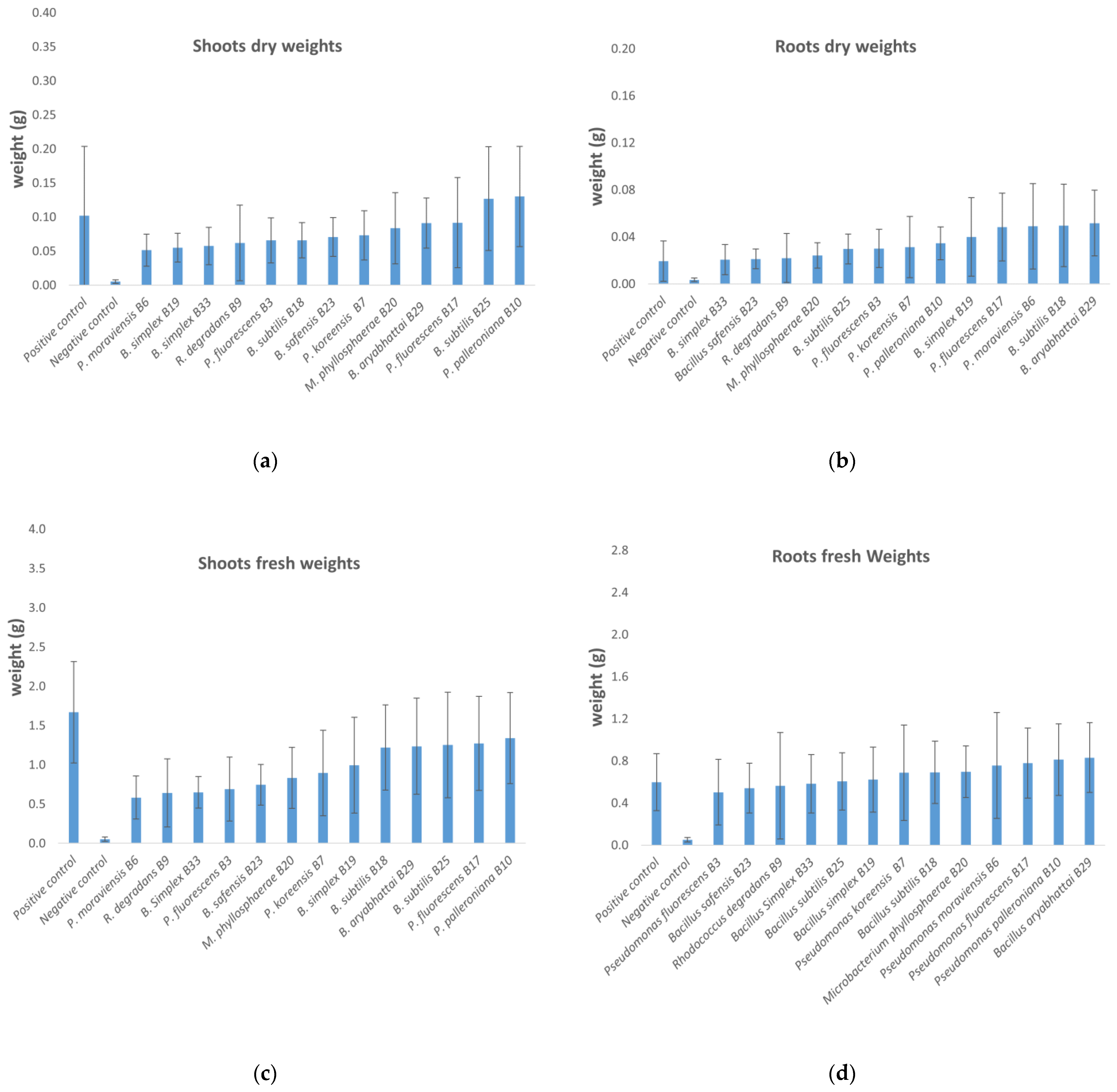
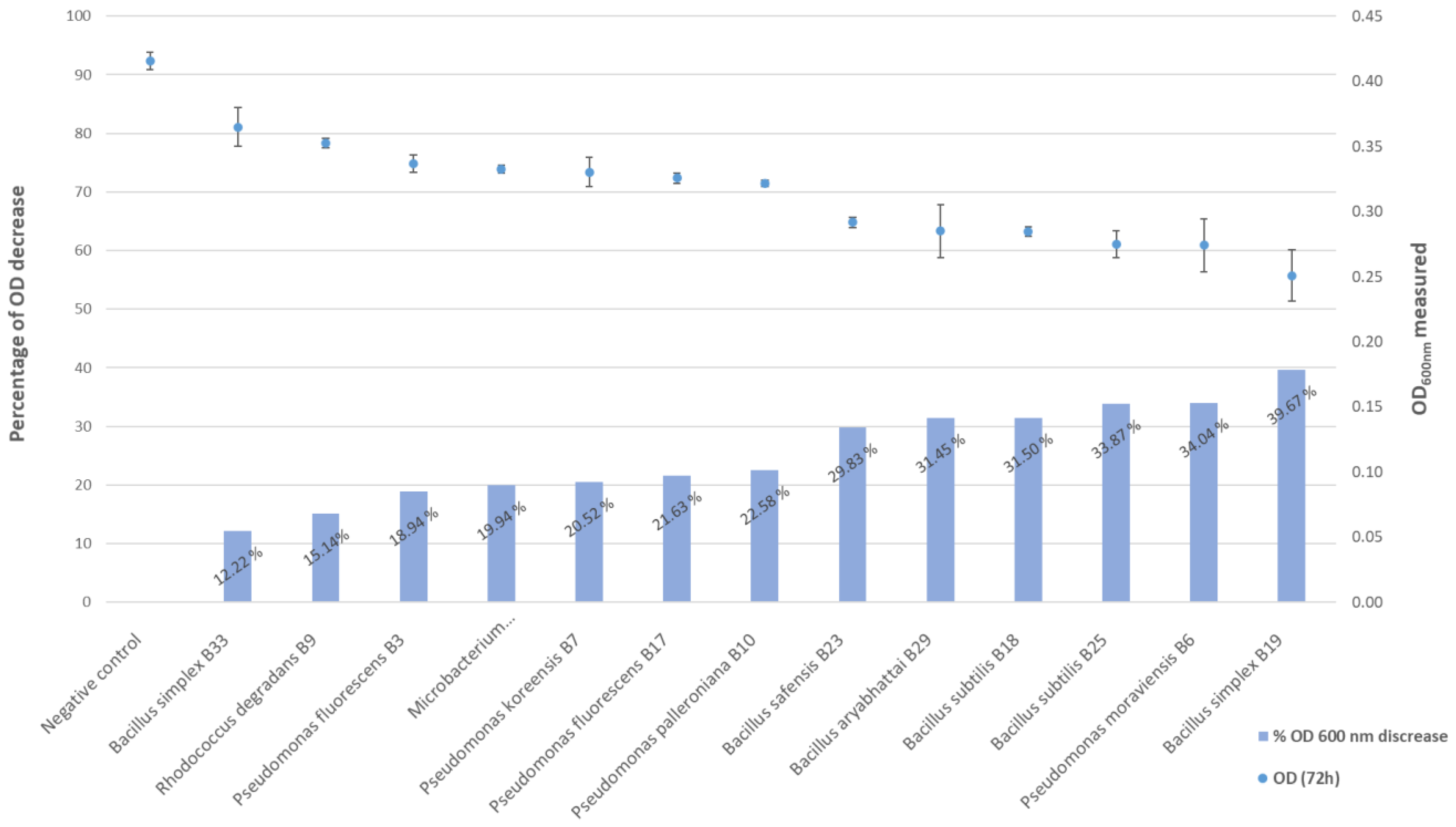
3.3. In Planta Tests
3.4. Phosphate Solubilization
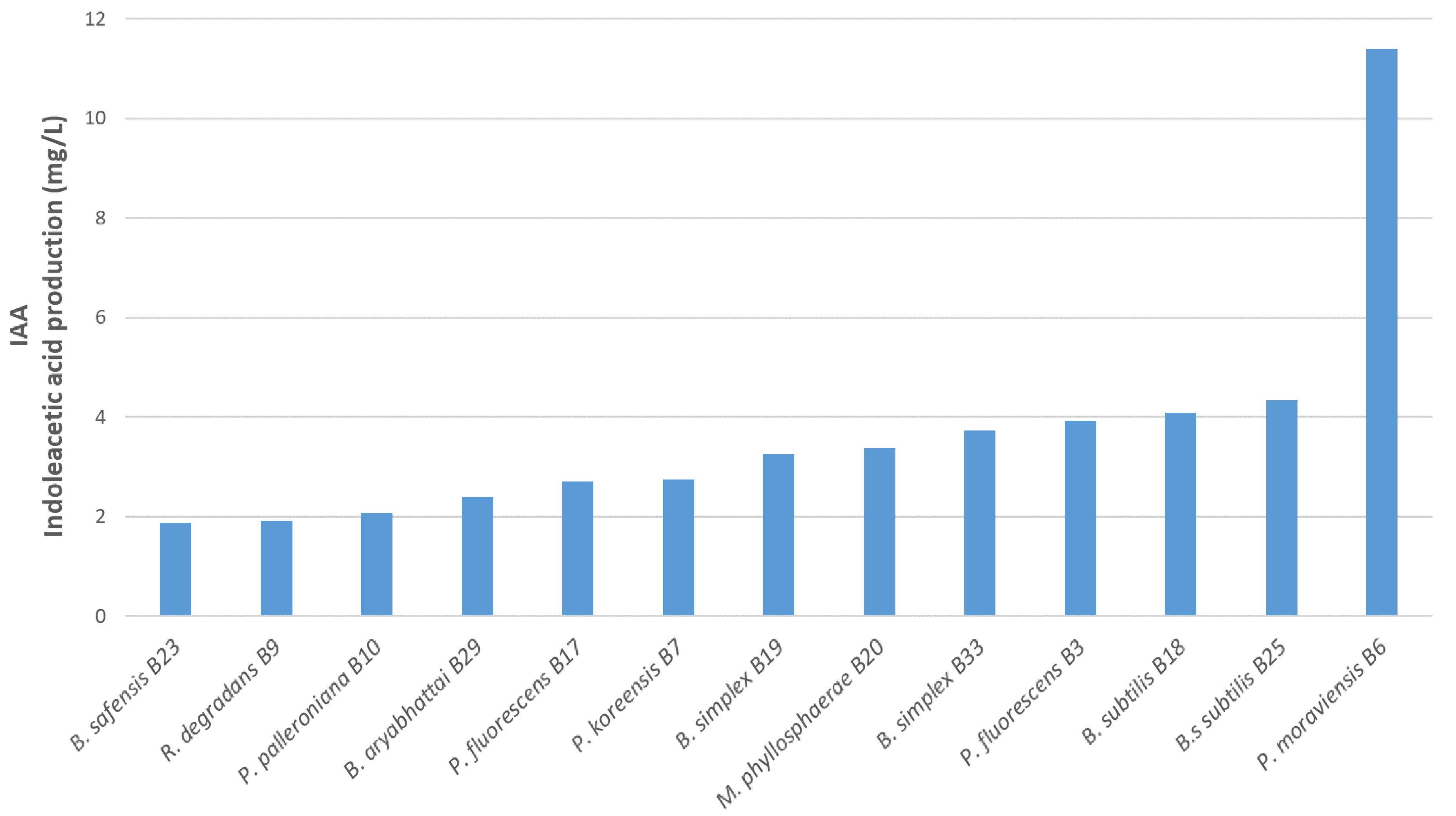
3.5. Indole Acetic Acid Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Computer Code and Software
References
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.K.; Fatima, T.; Mishra, I.; Verma, S. Microbe-based inoculants: Role in next green revolution. In Environmental Concerns and Sustainable Development; Volume 2: Biodiversity, Soil and Waste Management; Vertika, S., Narendra, K., Eds.; Springer: Singapore, 2019; pp. 191–246. [Google Scholar]
- Weerapol, Y.; Nimraksa, H.; Paradornuwat, A.; Sriamornsak, P. Development of ready-to-use products derived from Bacillus subtilis strain CMs026 for plant disease control. BioControl 2019, 64, 173–183. [Google Scholar] [CrossRef]
- Gravel, V.; Antoun, H.; Tweddell, R.J. Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: Possible role of indole acetic acid (IAA). Soil Biol. Biochem. 2007, 39, 1968–1977. [Google Scholar] [CrossRef]
- Ottesen, A.R.; González Peña, A.; White, J.R.; Pettengill, J.B.; Li, C.; Allard, S.; Rideout, S.; Allard, M.; Hill, T.; Evans, P.; et al. Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 2013, 13, 114. [Google Scholar] [CrossRef] [Green Version]
- Romero, F.M.; Marina, M.; Pieckenstain, F.L. The communities of tomato (Solanum lycopersicum L.) leaf endophytic bacteria, analyzed by 16S-ribosomal RNA gene pyrosequencing. FEMS Microbiol. Lett. 2014, 351, 187–194. [Google Scholar] [CrossRef] [Green Version]
- Bradáčová, K.; Florea, A.; Bar-Ta, A.; Minz, D.; Yermiyahu, U.; Shawahna, R.; Kraut-Cohen, J.; Zolti, A.; Erel, R.; Dietel, K.; et al. Microbial consortia versus single-strain inoculants: An advantage in PGPM-assisted tomato production? Agronomy 2019, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Durairaj, K.; Velmurugan, P.; Park, J.H.; Chang, W.S.; Park, Y.J.; Senthilkumar, P.; Choi, K.M.; Lee, J.H.; Oh, B.T. Characterization and assessment of two biocontrol bacteria against Pseudomonas syringae wilt in Solanum lycopersicum and its genetic responses. Microbiol. Res. 2018, 206, 43–49. [Google Scholar] [CrossRef]
- Ruzzi, M.; Aroca, R. Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 2015, 196, 124–134. [Google Scholar] [CrossRef]
- Schütz, L.; Gattinger, A.; Meier, M.; Müller, A.; Boller, T.; Mäder, P.; Mathimaran, N. Improving Crop yield and nutrient use efficiency via biofertilization—A global meta-analysis. Front. Plant Sci. 2018, 8, 2204. [Google Scholar] [CrossRef] [Green Version]
- Stamenković, S.; Beškoski, V.; Karabegović, I.; Lazić, M.; Nikolić, N. Microbial fertilizers: A comprehensive review of current findings and future perspectives. Span. J. Agric. Res. 2018, 16, e09R01-01. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.; Upreti, R. Evaluation of tomato seedling root-associated bacterial endophytes towards organic seedling production. Org. Agric. 2016, 6, 89–98. [Google Scholar] [CrossRef]
- Martínez-Viveros, O.; Jorquera, M.; Crowley, D.; Gajardo, G.; Mora, M. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 2010, 10, 293–319. [Google Scholar] [CrossRef] [Green Version]
- Gupta, G.; Pariha, S.S.; Ahirwar, N.K.; Snehi, S.K.; Singh, V. Plant Growth Promoting Rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 2015, 7, 96–102. [Google Scholar]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.S.; Patra, J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, Z.A. PGPR: Prospective biocontrol agents of plant pathogens. In PGPR: Biocontrol and Biofertilization; Siddiqui, Z.A., Ed.; Springer: Dordrecht, NL, USA, 2006; pp. 111–142. [Google Scholar]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2013, 2, 587. [Google Scholar] [CrossRef] [Green Version]
- Mena-Violante, H.G.; Olalde-Portugal, V. Alteration of tomato fruit quality by root inoculation with plant growth-promoting rhizobacteria (PGPR): Bacillus subtilis BEB-13bs. Sci. Hortic. 2007, 113, 103–106. [Google Scholar] [CrossRef]
- Stinson, M.; Ezra, D.; Hess, W.M.; Sears, J.; Strobel, G. An endophytic Gliocladium sp. of Eucryphia cordifolia producing selective volatile antimicrobial compounds. Plant Sci. 2003, 165, 913–922. [Google Scholar] [CrossRef]
- Kai, M.; Effmert, U.; Berg, G.; Piechulla, B. Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 2007, 187, 351–360. [Google Scholar] [CrossRef]
- Yang, J.; Kloepper, J.W.; Ryu, C. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2008, 14, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kegge, W.; Pierik, R. Biogenic Volatile organic compounds and plant competition. Trends Plant Sci. 2010, 15, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Senthilraja, G.; Anand, T.; Kennedy, J.S.; Raguchander, T.; Samiyappan, R. Plant Growth Promoting Rhizobacteria (PGPR) and entomopathogenic fungus bioformulation enhance the expression of defense enzymes and pathogenesis-related proteins in groundnut plants against leafminer insect and collar rot pathogen. Physiol. Mol. Plant Pathol. 2013, 82, 10–19. [Google Scholar] [CrossRef]
- Correa, O.S.; Romero, A.M.; Montecchia, M.S.; Soria, M.A. Tomato genotype and Azospirillum inoculation modulate the changes in bacterial communities associated with roots and leaves. J. Appl. Microbiol. 2007, 102, 781–786. [Google Scholar] [CrossRef]
- Enya, J.; Shinohara, H.; Yoshida, S.; Tsukiboshi, T.; Negishi, H.; Suyama, K.; Tsushima, S. Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microb. Ecol. 2007, 53, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S. In vitro characterization of bacterial endophytes from tomato (Solanum lycopersicum Mill.) for phytobeneficial traits. Appl. Ecol. Environ. Res. 2018, 16, 1037–1051. [Google Scholar] [CrossRef]
- Jangir, M.; Pathak, R.; Sharma, S.; Sharma, S. Biocontrol mechanisms of Bacillus sp., isolated from tomato rhizosphere, against Fusarium oxysporum f. sp. lycopersici. Biol. Control. 2018, 123, 60–70. [Google Scholar] [CrossRef]
- Anzalone, A.; Di Guardo, M.; Bella, P.; Ghadamgahi, F.; Dimaria, G.; Zago, R.; Cirvilleri, G.; Catara, V. Bioprospecting of beneficial bacteria traits associated with tomato root in greenhouse environment reveals that sampling sites impact more than the root compartment. Front. Plant Sci. 2021, 12, 637582. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, J.; Bon, M.C.; Jones, W. Optimalisation de L’extraction d’ADN Génomique de la Morelle Jaune (Solanum elaeagnifolium Cav.), une Plante Invasive des Milieux Cultivés en Région Méditerranéenne. BASE 2011, 1, 95–100. Available online: https://popups.uliege.be/1780-4507/index.php?id=7020 (accessed on 18 February 2022).
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Matsuda, R.; Handayani, M.L.; Sasaki, H.; Takechi, K.; Takano, H.; Takio, S. Production of indoleacetic acid by strains of the epiphytic bacteria Neptunomonas spp. isolated from the red alga Pyropia yezoensis and the seagrass Zostera marina. Arch. Microbiol. 2018, 200, 255–265. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Bona, E.; Todeschini, V.; Cantamessa, S.; Cesaro, P.; Copetta, A.; Lingua, G.; Gamalero, E.; Berta, G.; Massa, N. Combined bacterial and mycorrhizal inocula improve tomato quality at reduced fertilization. Sci. Hortic. 2018, 234, 160–165. [Google Scholar] [CrossRef]
- Kandan, A.; Ramiah, M.; Vasanthi, V.J.; Radjacommare, R.; Nandakumar, R.; Ramanathan, A.; Samiyappan, R. Use of Pseudomonas fluorescens–based formulations for management of tomato spotted wilt virus (TSWV) and enhanced yield in tomato. Biocontrol Sci. Technol. 2005, 15, 553–569. [Google Scholar] [CrossRef]
- Hassan, T.U.; Bano, A. Biofertilizer: A novel formulation for improving wheat growth, physiology and yield. Pak. J. Bot. 2016, 48, 2233–2241. [Google Scholar]
- Hultberg, M.; Alsberg, T.; Khalil, S.; Alsanius, B. Suppression of disease in tomato infected by Pythium ultimum with a biosurfactant produced by Pseudomonas koreensis. BioControl 2010, 55, 435–444. [Google Scholar] [CrossRef]
- Kasotia, A.; Choudhary, D.K. Induced Inorganic Phosphate Solubilization Through N-Methyl-N′-Nitro-N-Nitrosoguanidine Treated Mutants of Pseudomonas koreensis Strain AK-1 (MTCC Number 12058) Under Polyethylene Glycol. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2016, 86, 115–123. [Google Scholar] [CrossRef]
- Oh, S.-Y.; Kim, M.; Eimes, J.A.; Lim, Y.W. Effect of fruiting body bacteria on the growth of Tricholoma matsutake and its related molds. PLoS ONE 2018, 13, e0190948. [Google Scholar] [CrossRef] [Green Version]
- Bell, K.S.; Philp, J.C.; Aw, D.W.J.; Christofi, N. A review: The genus Rhodococcus. J. Appl. Microbiol. 1998, 85, 195–210. [Google Scholar] [CrossRef]
- Francis, I.M.; Stes, E.; Zhang, Y.; Rangel, D.; Audenaert, K.; Vereecke, D. Mining the genome of Rhodococcus fascians, a plant growth-promoting bacterium gone astray. New Biotechnol. 2016, 33, 706–717. [Google Scholar] [CrossRef]
- Tomer, S.; Suyal, D.C.; Shukla, A.; Rajwar, J.; Yadav, A.; Shouche, Y.; Goel, R. Isolation and characterization of phosphate solubilizing bacteria from Western Indian Himalayan soils. 3 Biotech 2017, 7, 95. [Google Scholar] [CrossRef]
- Cabra Cendales, T.; Rodríguez González, C.A.; Villota Cuásquer, C.P.; Tapasco Alzate, O.A.; Hernández Rodríguez, A. Bacillus effect on the germination and growth of tomato seedlings (Solanum lycopersicum L.). Acta Biol. Colomb. 2017, 22, 37. [Google Scholar] [CrossRef]
- Amaresan, N.; Jayakumar, V.; Kumar, K.; Thajuddin, N. Isolation and characterization of plant growth promoting endophytic bacteria and their effect on tomato (Lycopersicon esculentum) and chilli (Capsicum annuum) seedling growth. Ann. Microbiol. 2012, 62, 805–810. [Google Scholar] [CrossRef]
- Valenzuela-Soto, J.H.; Estrada-Hernández, M.G.; Ibarra-Laclette, E.; Délano-Frier, J.P. Inoculation of tomato plants (Solanum lycopersicum) with growth-promoting Bacillus subtilis retards whitefly Bemisia tabaci development. Planta 2010, 231, 397–410. [Google Scholar] [CrossRef]
- Schwartz, A.; Ortiz, I.; Maymon, M.; Herbold, C.; Fujishige, N.; Vijanderan, J.; Villella, W.; Hanamoto, K.; Diener, A.; Sanders, E.; et al. Bacillus simplex—A little known PGPB with anti-fungal activity—Alters pea legume root architecture and nodule morphology when co-inoculated with Rhizobium leguminosarum bv. viciae. Agronomy 2013, 3, 595–620. [Google Scholar] [CrossRef] [Green Version]
- Hassen, A.I.; Labuschagne, N. Root colonization and growth enhancement in wheat and tomato by rhizobacteria isolated from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 2010, 26, 1837–1846. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Iqrar, I. Selective isolation and characterization of agriculturally beneficial endophytic bacteria from wild hemp using canola. Pak. J. Bot. 2015, 47, 1999–2008. [Google Scholar]
- De Oliveira Costa, L.E.; de Queiroz, M.V.; Borges, A.C.; de Moraes, C.A.; de Araújo, E.F. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Braz. J. Microbiol. 2012, 43, 1562–1575. [Google Scholar] [CrossRef] [PubMed]
- Akinrinlola, R.J.; Yuen, G.Y.; Drijber, R.A.; Adesemoye, A.O. Evaluation of Bacillus strains for plant growth promotion and predictability of efficacy by in vitro physiological traits. Int. J. Microbiol. 2018, 2018, 5686874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.L.; Halo, B.A.; Elyassi, A.; Ali, S.; Al-Hosni, K.; Hussain, J.; Al-Harrasi, A.; Lee, I.J. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron. J. Biotechnol. 2016, 21, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, C.; Bakshi, U.; Mallick, I.; Mukherji, S.; Bera, B.; Ghosh, A. Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile Bacillus aryabhattai strain AB211. Front. Microbiol. 2017, 8, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.G.; Mun, B.G.; Kang, S.M.; Hussain, A.; Shahzad, R.; Seo, C.W.; Yun, B.W. Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS ONE 2017, 12, e0173203. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.Y.; Huang, H.B.; Wang, X.H.; Li, S.; Feng, N.X.; Zhao, H.M.; Huang, X.P.; Li, Y.W.; Li, H.; Cai, Q.Y.; et al. Novel phosphate-solubilising bacteria isolated from sewage sludge and the mechanism of phosphate solubilisation. Sci. Total Environ. 2019, 658, 474–484. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [Green Version]
- Joly, P.; Calteau, A.; Wauquier, A.; Dumas, R.; Beuvin, M.; Vallenet, D.; Crovadore, J.; Cochard, B.; Lefort, F.; Berthon, J.Y. From strain characterization to field authorization: Highlights on Bacillus velezensis strain B25 beneficial properties for plants and its activities on phytopathogenic fungi. Microorganisms 2021, 9, 1924. [Google Scholar] [CrossRef] [PubMed]
- Lemanceau, P.; Corberand, T.; Gardan, L.; Latour, X.; Laguerre, G.; Boeufgras, J.M.; Alabouvette, C. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent Pseudomonas. Appl. Environ. Microbiol. 1995, 61, 1004–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friman, J.; Pineda, A.; van Loon, J.J.; Dicke, M. Bidirectional plant-mediated interactions between rhizobacteria and shoot-feeding herbivorous insects: A community ecology perspective. Ecol Entomol. 2021, 46, 1–10. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, C.; Ye, Y.; Wen, J.; Wu, Y.; Wang, H.; Li, H.; Cai, S.; Cai, W.; Cheng, Z.; et al. Beneficial traits of bacterial endophytes belonging to the core communities of the tomato root microbiome. Agric. Ecosyst. Environ. 2017, 247, 149–156. [Google Scholar] [CrossRef]
- Domenech, J.; Reddy, M.S.; Kloepper, J.W.; Ramos, B.; Gutierrez-Mañero, J. Combined application of the biological product LS213 with Bacillus, Pseudomonas or Chryseobacterium for growth promotion and biological control of soil-borne diseases in pepper and tomato. Biocontrol 2006, 51, 245–258. [Google Scholar] [CrossRef]
- Rocha, F.Y.O.; de Oliveira, C.M.; da Silva, P.R.A.; de Melo, L.H.V.; do Carmo, M.G.F.; Baldani, J.I. Taxonomical and functional characterization of Bacillus strains isolated from tomato plants and their biocontrol activity against races 1, 2 and 3 of Fusarium oxysporum f. sp. Lycopersici. Appl. Soil. Ecol. 2017, 120, 8–19. [Google Scholar] [CrossRef]
- Stamenov, D.R.; Djuric, S.; Hajnal-Jafari, T.; Andjelkovic, S. Influence of Pseudomonas and Bacillus strains isolated from Lolium perenne rhizospheric soil in Vojvodina (Serbia) on planth growth and soil microbial communities. Pol. J. Microbiol. 2017, 66, 269–272. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Pantigoso, H.A.; Wu, Z.; Vivanco, J.M. Co-inoculation of Bacillus sp. and Pseudomonas putida at different development stages acts as a biostimulant to promote growth, yield and nutrient uptake of tomato. J. Appl. Microbiol. 2019, 127, 196–207. [Google Scholar] [CrossRef]
- Molla, A.H.; Manjurul, H.; Amdadul, H.; Ilias, G.N.M. Trichoderma-Enriched biofertilizer enhances production and nutritional quality of tomato (Lycopersicon esculentum Mill.) and minimizes NPK fertilizer use. Agric. Res. 2012, 1, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Lei, S.; Li, Y.; Huang, W.; Ma, R.; Xiong, J.; Zhang, T.; Jin, L.; Haq, H.u.; Xu, X.; et al. Revealing the variation and stability of bacterial communities in tomato rhizosphere microbiota. Microorganisms 2020, 8, 170. [Google Scholar] [CrossRef] [Green Version]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]


| Genus | Specie | Isolate Code | NCBI GenBank Accession | UASWS Code | Provenance |
|---|---|---|---|---|---|
| Pseudomonas | reinekei | B1 | MH671830.1 | UASWS1785 | 1 |
| Pseudomonas | palleroniana | B2 | MH671831.1 | UASWS1786 | 1 |
| Pseudomonas | fluorescens | B3 | MH671832.1 | UASWS1787 | 1 |
| Pseudomonas | reinekei | B4 | MH671833.1 | UASWS1788 | 1 |
| Pseudomonas | reinekei | B5 | MH671834.1 | UASWS1789 | 1 |
| Pseudomonas | moraviensis | B6 | MH671835.1 | UASWS1790 | 1 |
| Pseudomonas | koreensis | B7 | MH671836.1 | UASWS1791 | 1 |
| Mycolicibacterium | neoaurum | B8 | MH671837.1 | UASWS1792 | 1 |
| Rhodococcus | degradans | B9 | MH671838.1 | UASWS1793 | 1 |
| Pseudomonas | palleroniana | B10 | MH671839.1 | UASWS1794 | 1 |
| Paenarthrobacter | nicotinovorans | B11 | MH671840.1 | UASWS1795 | 1 |
| Pseudomonas | poae | B12 | MH671841.1 | UASWS1796 | 1 |
| Pseudomonas | grimontii | B13 | MH671842.1 | UASWS1797 | 1 |
| Pseudomonas | palleroniana | B15 | MH671843.1 | UASWS1798 | 1 |
| Pseudomonas | palleroniana | B16 | MH671844.1 | UASWS1799 | 1 |
| Pseudomonas | fluorescens | B17 | MH671845.1 | UASWS1800 | 2 |
| Bacillus | subtilis | B18 | MH671846.1 | UASWS1801 | 2 |
| Bacillus | simplex | B19 | MH671847.1 | UASWS1802 | 2 |
| Microbacterium | phyllosphaerae | B20 | MH671848.1 | UASWS1803 | 2 |
| Microbacterium | phyllosphaerae | B21 | MH671849.1 | UASWS1804 | 2 |
| Bacillus | wiedmannii | B22 | MH671850.1 | UASWS1805 | 2 |
| Bacillus | safensis | B23 | MH671851.1 | UASWS1806 | 3 |
| Bacillus | subterraneus | B24 | MH671852.1 | UASWS1807 | 3 |
| Bacillus | subtilis | B25 | MH671853.1 | UASWS1808 | 3 |
| Bacillus | simplex | B26 | MH671854.1 | UASWS1809 | 3 |
| Pseudomonas | sp. | B27 | MH671855.1 | UASWS1810 | 3 |
| Oceanobacillus | profundus | B28 | MH671856.1 | UASWS1811 | 3 |
| Bacillus | aryabhattai | B29 | MH671857.1 | UASWS1812 | 3 |
| Bacillus | foraminis | B30 | MH671859.1 | UASWS1813 | 3 |
| Cellulosimicrobium | cellulans | B31 | MH671859.1 | UASWS1814 | 3 |
| Bacillus | solani | B32 | MH671860.1 | UASWS1815 | 3 |
| Bacillus | simplex | B33 | MH671861.1 | UASWS1816 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cochard, B.; Giroud, B.; Crovadore, J.; Chablais, R.; Arminjon, L.; Lefort, F. Endophytic PGPR from Tomato Roots: Isolation, In Vitro Characterization and In Vivo Evaluation of Treated Tomatoes (Solanum lycopersicum L.). Microorganisms 2022, 10, 765. https://doi.org/10.3390/microorganisms10040765
Cochard B, Giroud B, Crovadore J, Chablais R, Arminjon L, Lefort F. Endophytic PGPR from Tomato Roots: Isolation, In Vitro Characterization and In Vivo Evaluation of Treated Tomatoes (Solanum lycopersicum L.). Microorganisms. 2022; 10(4):765. https://doi.org/10.3390/microorganisms10040765
Chicago/Turabian StyleCochard, Bastien, Basile Giroud, Julien Crovadore, Romain Chablais, Lucas Arminjon, and François Lefort. 2022. "Endophytic PGPR from Tomato Roots: Isolation, In Vitro Characterization and In Vivo Evaluation of Treated Tomatoes (Solanum lycopersicum L.)" Microorganisms 10, no. 4: 765. https://doi.org/10.3390/microorganisms10040765
APA StyleCochard, B., Giroud, B., Crovadore, J., Chablais, R., Arminjon, L., & Lefort, F. (2022). Endophytic PGPR from Tomato Roots: Isolation, In Vitro Characterization and In Vivo Evaluation of Treated Tomatoes (Solanum lycopersicum L.). Microorganisms, 10(4), 765. https://doi.org/10.3390/microorganisms10040765






