Molecular and Genomic Characterization of the Pseudomonas syringae Phylogroup 4: An Emerging Pathogen of Arabidopsis thaliana and Nicotiana benthamiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Bacterial Culture and Plant Infections
2.3. Comparative Genomics, Gene Ontology Analysis, and Functional Annotation
2.4. Phylogenomics Analysis
2.5. Identification of Type III Secretion System and Other Virulence-Associated Genes
2.6. HopBJ1Psy Mutation and Nicotiana Transient Expression by Agroinfiltration
3. Results
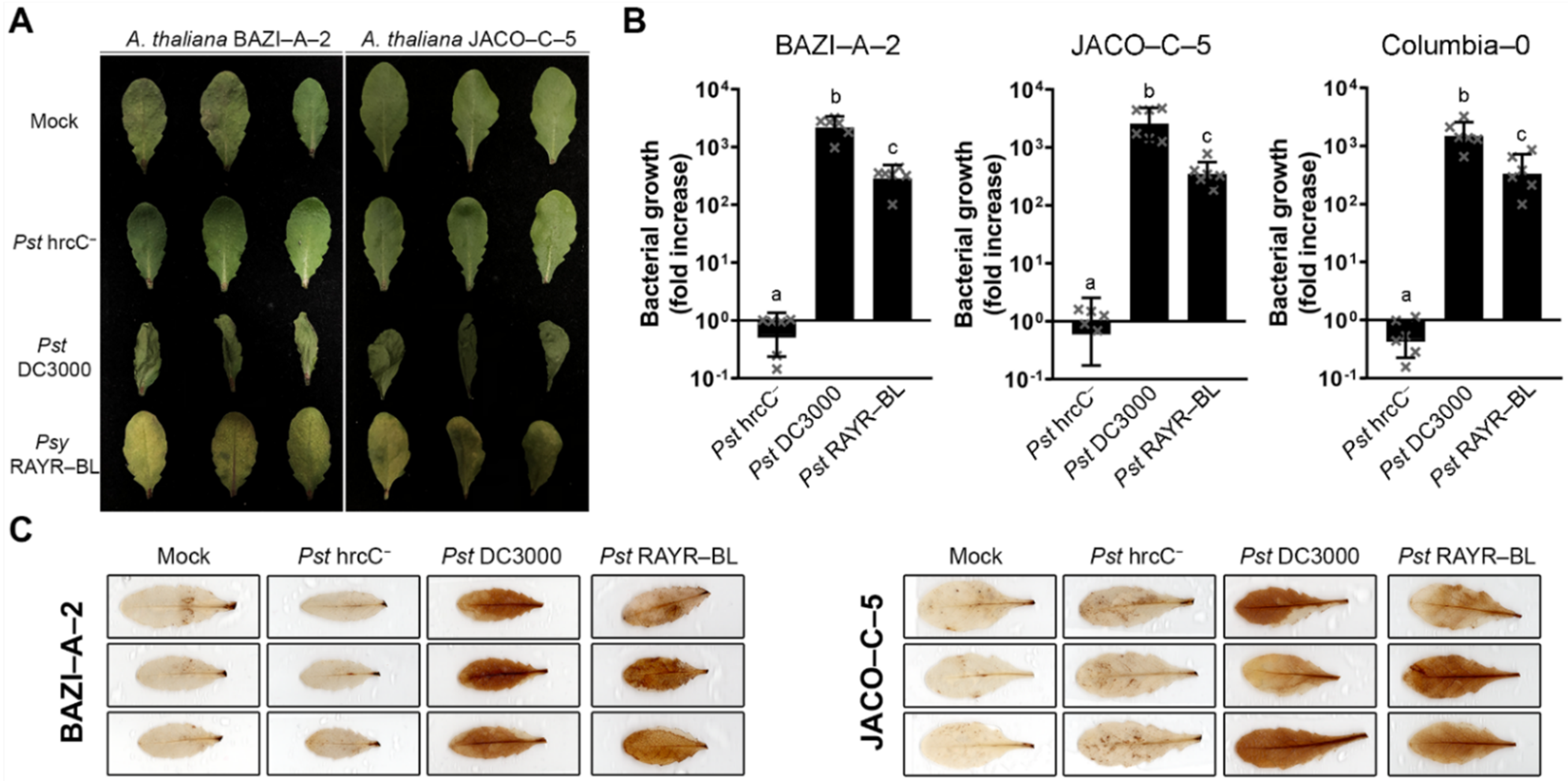
3.1. The Strain Pseudomonas syringae RAYR-BL Induces Severe Disease on A. thaliana
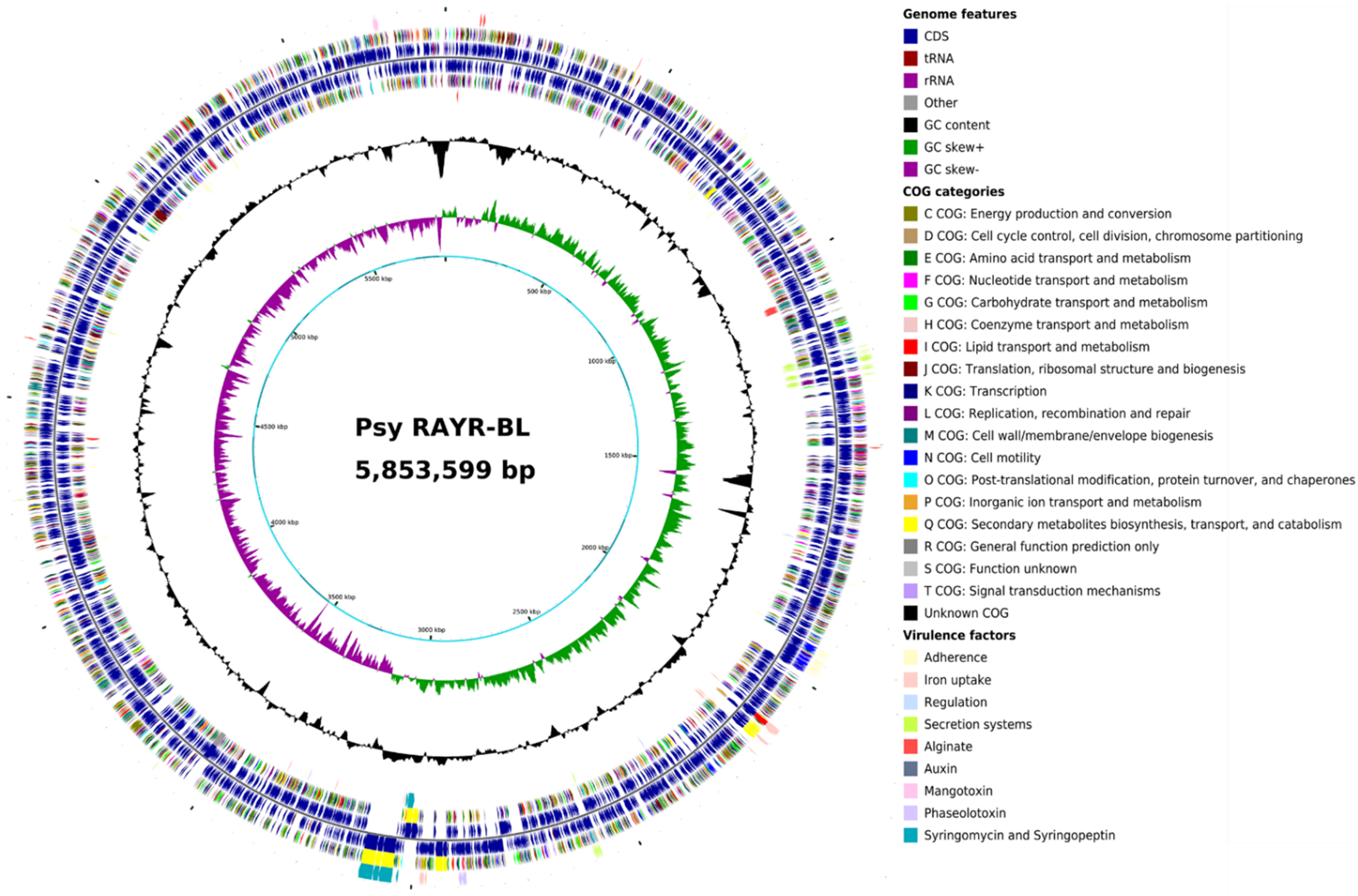
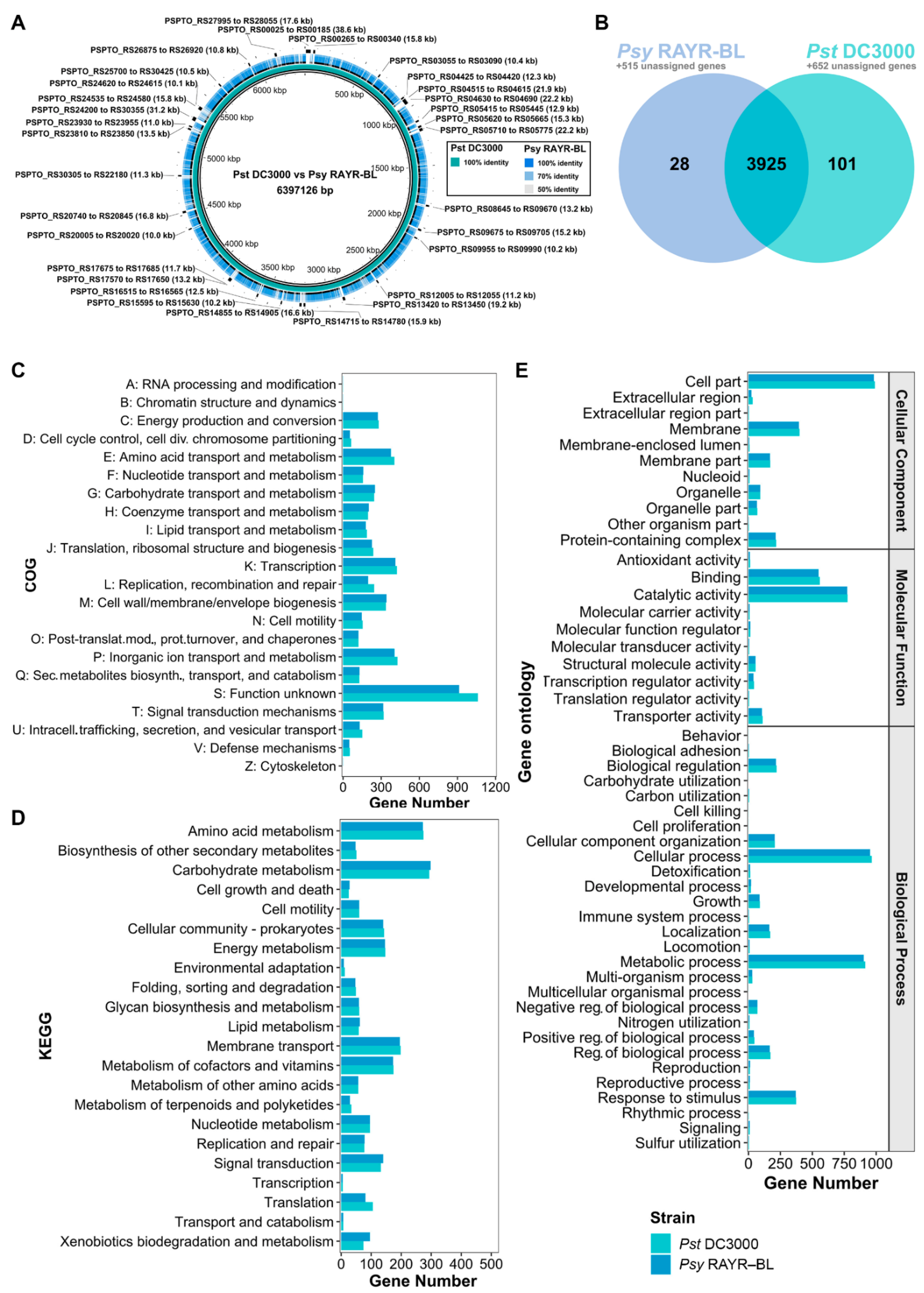
3.2. Comparative Genomics of Pseudomonas syringae DC3000 and Pseudomonas syringae RAYR-BL
3.3. Phylogenetic Position of Pseudomonas syringae RAYR-BL
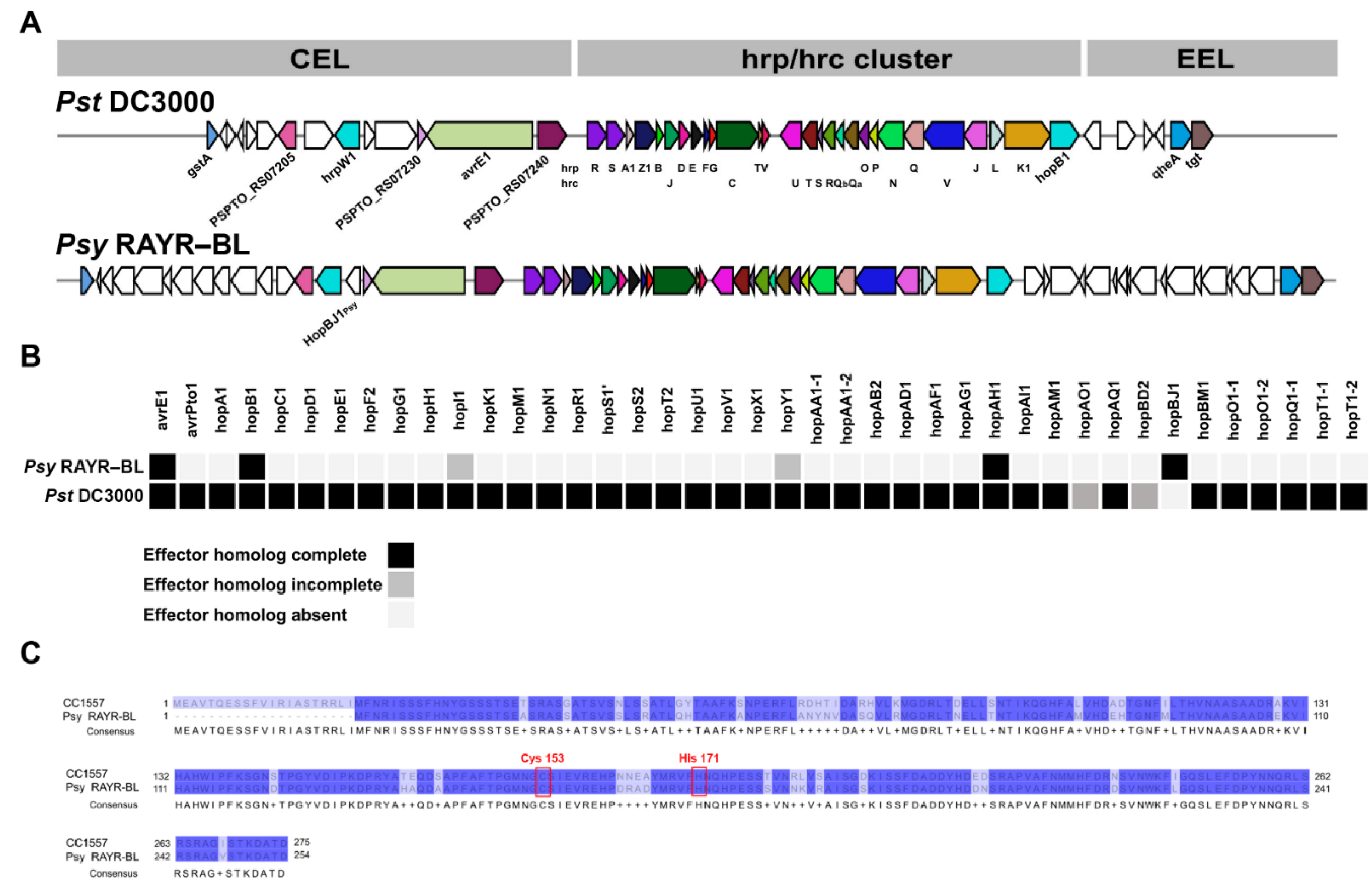
3.4. Analysis of Molecular Components That Participate in the Pseudomonas syringae Pathogenesis
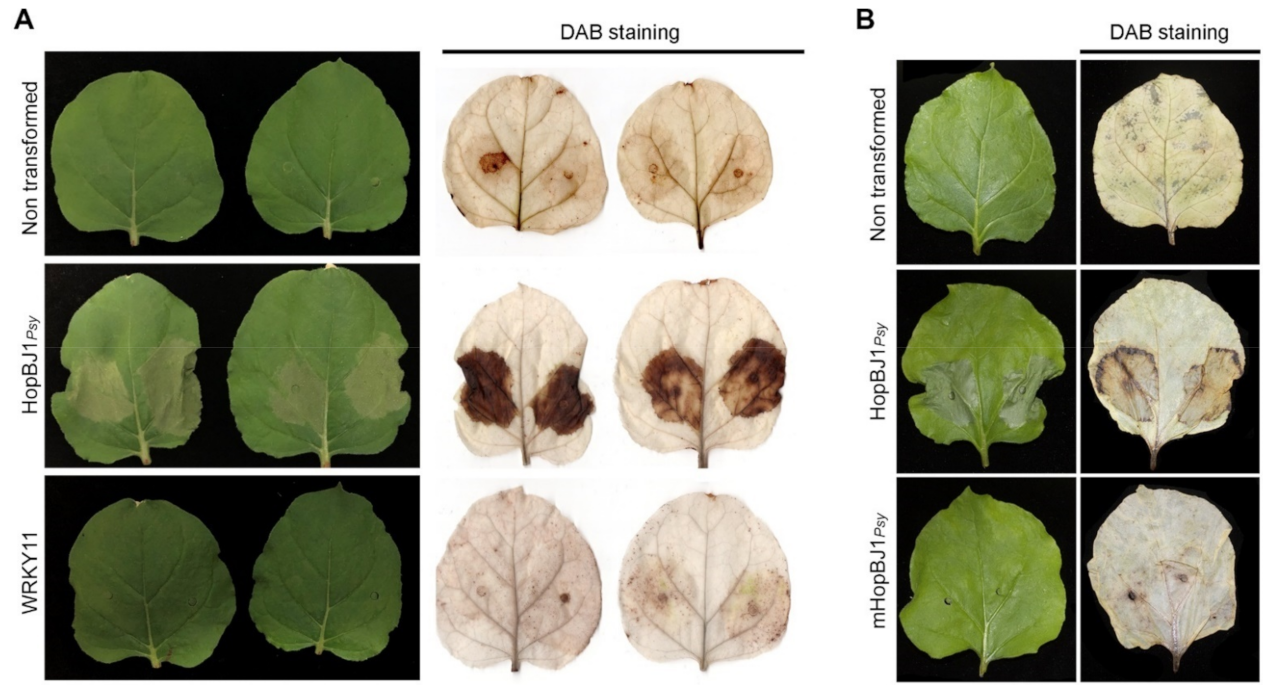
3.5. hopBJ1Psy Is a Virulence Factor Required for Psy RAYR-BL to Produce Plant Disease
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gullino, L.M.; Albajes, R.; Al-Jboory, I.; Angelotti, F.; Chakraborty, S.; Garrett, K.A.; Hurley, B.P.; Juroszek, P.; Makkouk, K.; Pan, X.; et al. Scientific review of the impact of climate change on plant pests, 2021. IPPC Secretariat Rome. Available online: https://www.infoteca.cnptia.embrapa.br/infoteca/bitstream/doc/1132870/1/Scientific-review-of-the-impact-of-climate-2021.pdf (accessed on 20 February 2022).
- Desaint, H.; Aoun, N.; Deslandes, L.; Vailleau, F.; Roux, F.; Berthomé, R. Fight hard or die trying: When plants face pathogens under heat stress. New Phytol. 2021, 229, 712–734. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Caballo-Ponce, E.; Pintado, A.; Moreno-Pérez, A.; Murillo, J.; Smalla, K.; Ramos, C. Pseudomonas savastanoi pv. mandevillae pv. nov., a Clonal Pathogen Causing an Emerging, Devastating Disease of the Ornamental Plant Mandevilla spp. Phytopathology 2021, 111, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.K.; Matiuzzo, M.; Bertani, I.; Bigirimana, V.D.P.; Ash, G.J.; Höfte, M.; Venturi, V. Identification of virulence associated loci in the emerging broad host range plant pathogen Pseudomonas fuscovaginae. BMC Microbiol. 2014, 14, 274. [Google Scholar] [CrossRef]
- Scortichini, M.; Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G. Pseudomonas syringae pv. actinidiae: A re-emerging, multi-faceted, pandemic pathogen. Mol. Plant Pathol. 2012, 13, 631–640. [Google Scholar] [CrossRef]
- Takikawa, Y.; Serizawa, S.; Ichikawa, T.; Tsuyumu, S.; Goto, M. Pseudomonas syringae pv. actinidiae pv. nov.: The causal bacterium of canker of kiwifruit in Japan. Jpn. J. Phytopathol. 1989, 55, 437–444. [Google Scholar] [CrossRef]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Genet. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Whalen, M.C.; Innes, R.W.; Bent, A.; Staskawicz, B.J. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 1991, 3, 49–59. [Google Scholar] [CrossRef]
- Wiebe, W.L. Characterization of Pseudomonas syringae pv. maculicola and Comparison with P. s. tomato. Plant Dis. 1993, 77, 414. [Google Scholar]
- Nomura, K.; Melotto, M.; He, S.-Y. Suppression of host defense in compatible plant–Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 2005, 8, 361–368. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive Oxygen Species Signaling in Response to Pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, M.; Anderson, J. Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals. Microorganisms 2021, 9, 1227. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.-L.; Preston, G.; Collmer, A.; Chang, C.-J.; Huang, H.-C. Characterization of the hrpC and hrpRS Operons of Pseudomonas syringae Pathovars Syringae, Tomato, and Glycinea and Analysis of the Ability of hrpF, hrpG, hrcC, hrpT, and hrpV Mutants to Elicit the Hypersensitive Response and Disease in Plants. J. Bacteriol. 1998, 180, 4523–4531. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; He, S.Y. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J. Bacteriol. 1996, 178, 6399–6402. [Google Scholar] [CrossRef]
- Wei, H.-L.; Collmer, A. Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors. Mol. Plant Pathol. 2018, 19, 1779–1794. [Google Scholar] [CrossRef]
- Cunnac, S.; Chakravarthy, S.; Kvitko, B.H.; Russell, A.B.; Martin, G.B.; Collmer, A. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 2011, 108, 2975–2980. [Google Scholar] [CrossRef]
- Wei, H.-L.; Chakravarthy, S.; Mathieu, J.; Helmann, T.; Stodghill, P.; Swingle, B.; Martin, G.B.; Collmer, A. Pseudomonas syringae pv. tomato DC3000 Type III Secretion Effector Polymutants Reveal an Interplay between HopAD1 and AvrPtoB. Cell Host Microbe 2015, 17, 752–762. [Google Scholar] [CrossRef]
- O’Brien, H.E.; Thakur, S.; Guttman, D.S. Evolution of Plant Pathogenesis in Pseudomonas syringae: A Genomics Perspective. Annu. Rev. Phytopathol. 2011, 49, 269–289. [Google Scholar] [CrossRef]
- Martínez-García, P.M.; Rodríguez-Palenzuela, P.; Arrebola, E.; Carrión, V.J.; Gutiérrez-Barranquero, J.A.; Pérez-García, A.; Ramos, C.; Cazorla, F.M.; de Vicente, A. Bioinformatics Analysis of the Complete Genome Sequence of the Mango Tree Pathogen Pseudomonas syringae pv. syringae UMAF0158 Reveals Traits Relevant to Virulence and Epiphytic Lifestyle. PLoS ONE 2015, 10, e0136101. [Google Scholar]
- Lo, T.; Koulena, N.; Seto, D.; Guttman, D.S.; Desveaux, D. The HopF family of Pseudomonas syringae type III secreted effectors. Mol. Plant Pathol. 2016, 18, 457–468. [Google Scholar] [CrossRef]
- Lee, J.; Teitzel, G.M.; Munkvold, K.; del Pozo, O.; Martin, G.; Michelmore, R.W.; Greenberg, J.T. Type III Secretion and Effectors Shape the Survival and Growth Pattern of Pseudomonas syringae on Leaf Surfaces. Plant Physiol. 2012, 158, 1803–1818. [Google Scholar] [CrossRef] [PubMed]
- Oguiza, J.A.; Asensio, A.C. The VirPphA/AvrPtoB family of type III effectors in Pseudomonas syringae. Res. Microbiol. 2005, 156, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; Joe, A.; Jeong, B.-R.; Korneli, C.; Boutrot, F.; Westedt, I.; Staiger, D.; Alfano, J.R.; Zipfel, C. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013, 32, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, Y.; Xue, L.; Chu, J.; Yan, C.; Fu, J.; Chen, M.; Innes, R.W.; Zhou, J. A Pseudomonas syringae protein perturbs Arabidopsis hormone signalling by activating MAP KINASE 4. Cell Host Microbe 2012, 7, 164–175. [Google Scholar] [CrossRef]
- Gangadharan, A.; Sreerekha, M.-V.; Whitehill, J.; Ham, J.H.; Mackey, D. The Pseudomonas syringae pv. tomato Type III Effector HopM1 Suppresses Arabidopsis Defenses Independent of Suppressing Salicylic Acid Signaling and of Targeting AtMIN7. PLoS ONE 2013, 8, e82032. [Google Scholar] [CrossRef]
- Jeleńska, J.; Lee, J.; Manning, A.J.; Wolfgeher, D.J.; Ahn, Y.; Walters-Marrah, G.; Lopez, I.E.; Garcia, L.; McClerklin, S.A.; Michelmore, R.W.; et al. Pseudomonas syringae effector HopZ3 suppresses the bacterial AvrPto1–tomato PTO immune complex via acetylation. PLoS Pathog. 2021, 17, e1010017. [Google Scholar] [CrossRef]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; DeBoy, R.T.; Durkin, A.S.; Kolonay, J.F.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar] [CrossRef]
- Geng, X.; Jin, L.; Shimada, M.; Kim, M.G.; Mackey, D. The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta 2014, 240, 1149–1165. [Google Scholar] [CrossRef]
- Arrebola, E.; Cazorla, F.M.; Pérez-García, A.; De Vicente, A. Chemical and Metabolic Aspects of Antimetabolite Toxins Produced by Pseudomonas syringae Pathovars. Toxins 2011, 3, 1089–1110. [Google Scholar] [CrossRef]
- Bender, C.L.; Alarcon-Chaidez, F.; Gross, D.C. Pseudomonas syringae Phytotoxins: Mode of Action, Regulation, and Biosynthesis by Peptide and Polyketide Synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar]
- Bartoli, C.; Frachon, L.; Barret, M.; Rigal, M.; Huard-Chauveau, C.; Mayjonade, B.; Zanchetta, C.; Bouchez, O.; Roby, D.; Carrère, S.; et al. In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 2018, 12, 2024–2038. [Google Scholar] [CrossRef]
- Berge, O.; Monteil, C.; Bartoli, C.; Chandeysson, C.; Guilbaud, C.; Sands, D.C.; Morris, C.E. A User’s Guide to a Data Base of the Diversity of Pseudomonas syringae and Its Application to Classifying Strains in This Phylogenetic Complex. PLoS ONE 2014, 9, e105547. [Google Scholar] [CrossRef]
- Daudi, A.; O’Brien, J.A. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio-Protocol 2012, 2, e263. [Google Scholar] [CrossRef] [PubMed]
- Fuenzalida-Valdivia, I.; Gangas, M.V.; Zavala, D.; Herrera-Vásquez, A.; Roux, F.; Meneses, C.; Blanco-Herrera, F. Draft Genome Sequence of Pseudomonas syringae RAYR-BL, a Strain Isolated from Natural Accessions of Arabidopsis thaliana Plants. Microbiol. Resour. Announc. 2022, 11, e01001-21. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.V.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35 (Suppl. S2), W182–W185. [Google Scholar] [CrossRef]
- Grin, I.; Linke, D. GCView: The genomic context viewer for protein homology searches. Nucleic Acids Res. 2011, 39, W353–W356. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Eichinger, V.; Nussbaumer, T.; Platzer, A.; Jehl, M.-A.; Arnold, R.; Rattei, T. EffectiveDB—updates and novel features for a better annotation of bacterial secreted proteins and Type III, IV, VI secretion systems. Nucleic Acids Res. 2016, 44, D669–D674. [Google Scholar] [CrossRef]
- Goldberg, T.; Rost, B.; Bromberg, Y. Computational prediction shines light on type III secretion origins. Sci. Rep. 2016, 6, srep34516. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Li, X. Infiltration of Nicotiana benthamiana Protocol for Transient Expression via Agrobacterium. BIO-PROTOCOL 2011, 1. [Google Scholar] [CrossRef]
- Guilbaud, C.; Morris, C.E.; Barakat, M.; Ortet, P.; Berge, O. Isolation and identification ofPseudomonas syringaefacilitated by a PCR targeting the wholeP. syringaegroup. FEMS Microbiol. Ecol. 2015, 92, fiv146. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Varvaro, L. A new medium for the detection of fluorescent pigment production by pseudomonads. Plant Pathol. 2013, 62, 624–632. [Google Scholar] [CrossRef]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Cuppels, D.A. Generation and Characterization of Tn 5 Insertion Mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 1986, 51, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, M.; Mossialos, D.; Oliver, S.G.; Amoutzias, G.D. Comparative Analysis of the Core Proteomes among the Pseudomonas Major Evolutionary Groups Reveals Species-Specific Adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis. Diversity 2020, 12, 289. [Google Scholar] [CrossRef]
- Ling, J.; Pan, H.; Gao, Q.; Xiong, L.; Zhou, Y.; Zhang, D.; Gao, S.; Liu, X. Aerobactin Synthesis Genes iucA and iucC Contribute to the Pathogenicity of Avian Pathogenic Escherichia coli O2 Strain E058. PLoS ONE 2013, 8, e57794. [Google Scholar] [CrossRef]
- Ji, X.; Sun, Y.; Liu, J.; Zhu, L.; Guo, X.; Lang, X.; Feng, S. A novel virulence-associated protein, vapE, in Streptococcus suis serotype 2. Mol. Med. Rep. 2016, 13, 2871–2877. [Google Scholar] [CrossRef][Green Version]
- Hesse, C.; Schulz, F.; Bull, C.T.; Shaffer, B.T.; Yan, Q.; Shapiro, N.; Hassan, K.A.; Varghese, N.; Elbourne, L.; Paulsen, I.T.; et al. Genome-based evolutionary history of Pseudomonas spp. Environ. Microbiol. 2018, 20, 2142–2159. [Google Scholar] [CrossRef]
- Baltrus, D.; Nishimura, M.; Romanchuk, A.; Chang, J.; Mukhtar, S.; Cherkis, K.; Roach, J.; Grant, S.R.; Jones, C.D.; Dangl, J.L. Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 Pseudomonas syringae Isolates. PLoS Pathog. 2011, 7, e1002132. [Google Scholar] [CrossRef]
- Gomila, M.; Busquets, A.; Mulet, M.; García-Valdés, E.; Lalucat, J. Clarification of Taxonomic Status within the Pseudomonas syringae Species Group Based on a Phylogenomic Analysis. Front. Microbiol. 2017, 8, 2422. [Google Scholar] [CrossRef]
- Dillon, M.M.; Thakur, S.; Almeida, R.N.D.; Wang, P.W.; Weir, B.S.; Guttman, D.S. Recombination of ecologically and evolutionarily significant loci maintains genetic cohesion in the Pseudomonas syringae species complex. Genome Biol. 2019, 20, 1–28. [Google Scholar] [CrossRef]
- Hockett, K.; Nishimura, M.; Karlsrud, E.; Dougherty, K.; Baltrus, D. Pseudomonas syringae CC1557: A Highly Virulent Strain With an Unusually Small Type III Effector Repertoire That Includes a Novel Effector. Mol. Plant-Microbe Interact. 2014, 27, 923–932. [Google Scholar] [CrossRef]
- Rico, A.; McCraw, S.L.; Preston, G.M. The metabolic interface between Pseudomonas syringae and plant cells. Curr. Opin. Microbiol. 2011, 14, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Rico, A.; Preston, G.M. Pseudomonas syringae pv. tomato DC3000 Uses Constitutive and Apoplast-Induced Nutrient Assimilation Pathways to Catabolize Nutrients That Are Abundant in the Tomato Apoplast. Mol. Plant-Microbe Interact. 2008, 21, 269–282. [Google Scholar] [CrossRef]
- Lemonnier, M.; Landraud, L.; Lemichez, E. Rho GTPase-activating bacterial toxins: From bacterial virulence regulation to eukaryotic cell biology. FEMS Microbiol. Rev. 2007, 31, 515–534. [Google Scholar] [CrossRef]
- Jakob, K.; Goss, E.M.; Araki, H.; Van, T.; Kreitman, M.; Bergelson, J. Pseudomonas viridiflava and P. syringae—Natural Pathogens of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2002, 15, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Baltrus, D.A.; Yourstone, S.; Lind, A.; Guilbaud, C.; Sands, D.C.; Jones, C.D.; Morris, C.E.; Dangl, J.L. Draft Genome Sequences of a Phylogenetically Diverse Suite of Pseudomonas syringae Strains from Multiple Source Populations. Genome Announc. 2014, 2, e01195-13. [Google Scholar] [CrossRef] [PubMed]
- Karasov, T.L.; Almario, J.; Friedemann, C.; Ding, W.; Giolai, M.; Heavens, D.; Kersten, S.; Lundberg, D.S.; Neumann, M.; Regalado, J.; et al. Arabidopsis thaliana and Pseudomonas Pathogens Exhibit Stable Associations over Evolutionary Timescales. Cell Host Microbe 2018, 24, 168–179.e4. [Google Scholar] [CrossRef]
- Mucyn, T.S.; Yourstone, S.; Lind, A.L.; Biswas, S.; Nishimura, M.; Baltrus, D.; Cumbie, J.S.; Chang, J.; Jones, C.D.; Dangl, J.L.; et al. Variable Suites of Non-effector Genes Are Co-regulated in the Type III Secretion Virulence Regulon across the Pseudomonas syringae Phylogeny. PLoS Pathog. 2014, 10, e1003807. [Google Scholar] [CrossRef]
- Dillon, M.M.; Almeida, R.N.; Laflamme, B.; Martel, A.; Weir, B.S.; Desveaux, D.; Guttman, D.S. Molecular Evolution of Pseudomonas syringae Type III Secreted Effector Proteins. Front. Plant Sci. 2019, 10, 418. [Google Scholar] [CrossRef]
- Bartoli, C.; Berge, O.; Monteil, C.; Guilbaud, C.; Balestra, G.; Varvaro, L.; Jones, C.; Dangl, J.L.; Baltrus, D.; Sands, D.C.; et al. ThePseudomonas viridiflavaphylogroups in theP. syringaespecies complex are characterized by genetic variability and phenotypic plasticity of pathogenicity-related traits. Environ. Microbiol. 2014, 16, 2301–2315. [Google Scholar] [CrossRef]
- Smertenko, A.; E Franklin-Tong, V. Organisation and regulation of the cytoskeleton in plant programmed cell death. Cell Death Differ. 2011, 18, 1263–1270. [Google Scholar] [CrossRef]
- Staiger, C.J.; Franklin-Tong, V.E. The actin cytoskeleton is a target of the self-incompatibility response in Papaver rhoeas. J. Exp. Bot. 2003, 54, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Gourlay, C.; Ayscough, K.R. The actin cytoskeleton: A key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 2005, 6, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Shimono, M.; Lu, Y.-J.; Porter, K.; Kvitko, B.H.; Henty-Ridilla, J.; Creason, A.; He, S.Y.; Chang, J.H.; Staiger, C.J.; Day, B. The Pseudomonas syringae Type III Effector HopG1 Induces Actin Remodeling to Promote Symptom Development and Susceptibility during Infection. Plant Physiol. 2016, 171, 2239–2255. [Google Scholar] [PubMed]
- Kang, Y.; Jelenska, J.; Cecchini, N.M.; Li, Y.; Lee, M.W.; Kovar, D.R.; Greenberg, J.T. HopW1 from Pseudomonas syringae Disrupts the Actin Cytoskeleton to Promote Virulence in Arabidopsis. PLoS Pathog. 2014, 10, e1004232. [Google Scholar] [CrossRef]
- Jelenska, J.; Kang, Y.; Greenberg, J. Plant pathogenic bacteria target the actin microfilament network involved in the trafficking of disease defense components. BioArchitecture 2014, 4, 149–153. [Google Scholar] [CrossRef] [PubMed][Green Version]



| Feature | Psy RAYR-BL | Pst DC3000 a | ||
|---|---|---|---|---|
| Molecule | Draft genome | Chromosome | pDC300A | pDC3000B |
| Size | 5,871,397 | 6,397,126 | 73,661 | 67,473 |
| G+C content (%) | 58.98 | 58.4 | 55.1 | 56.1 |
| Number of contigs (≥ 200) | 110 | 1 | 1 | 1 |
| Largest contig | 1,064,144 | 6,397,126 | 73,661 | 67,473 |
| N50 | 193,008 | 6,397,126 | 73,661 | 67,473 |
| L50 | 8 | 1 | 1 | 1 |
| Genes | 5188 | 5765 | 77 | 77 |
| CDSs (coding) | 5053 | 5466 | 74 | 68 |
| rRNA | 6 | 16 | - | - |
| tRNA | 57 | 63 | - | - |
| Number of CDSs with assigned function | 4453 (88.1%) | 4556 (83.4%) | 54 (72.9%) | 47 (69.1%) |
| Number of CDSs without assigned function | 600 (11.9%) | 910 (16.6%) | 20 (27.0%) | 21 (30.9%) |
| Phytotoxin | Reference Organism | Genes ID | Number of Genes in Psy RAYR-BL | ||
|---|---|---|---|---|---|
| Reference Genes | Psy RAYR-BL | Total | Percentage (%) | ||
| Coronatine | Pseudomonas syringae pv tomato DC3000 | cfl, cfa1, cfa2, cfa3, cfa4, cfa5, cfa6, cfa7, cfa8, cfa9, corR, corS, cmaD, cmaE, cmaA, cmaB, cmaC, cmaT | - | - | - |
| Mangotoxin | Pseudomonas syringae pv. syringae UMAF0158 | mgoD, mgoA, mgoC, mgoB, ORF2, ORF1 | K0038_05071, K0038_05070,K0038_05069, K0038_05068, K0038_05067, K0038_05066, | 6/6 | 100 |
| Phaseolotoxin | Pseudomonas savatanoi pv phaseolicola 1448A | argK, phtA, phtB, phtC, phtD, phtE, phtF, phtG, phtH, desO, phtJ, phtK, phtL, phtM, phtN, phtP, amtA, phtQ, phtS, phtT, phtU, phtV | K0038_02673, K0038_02672, K0038_02670, K0038_02671, K0038_02666, K0038_02665, K0038_02664, K0038_02663, K0038_02662, K0038_02661, K0038_02660, K0038_02659, K0038_02668 | 13/22 | 59.1 |
| Syringomycin and syringopeptin | Pseudomonas syringae pv syringae B728a | YP_235685.1, YP_235686.1, YP_235687.1, YP_235688.1, YP_235689.1, YP_235690.1, YP_235691.1, YP_235692.1, YP_235693.1, | K0038_02743, K0038_02745, K0038_02746, K0038_02747, K0038_02748, K0038_02749, K0038_02756, K0038_02757, K0038_02758 | 9/9 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zavala, D.; Fuenzalida, I.; Gangas, M.V.; Peppino Margutti, M.; Bartoli, C.; Roux, F.; Meneses, C.; Herrera-Vásquez, A.; Blanco-Herrera, F. Molecular and Genomic Characterization of the Pseudomonas syringae Phylogroup 4: An Emerging Pathogen of Arabidopsis thaliana and Nicotiana benthamiana. Microorganisms 2022, 10, 707. https://doi.org/10.3390/microorganisms10040707
Zavala D, Fuenzalida I, Gangas MV, Peppino Margutti M, Bartoli C, Roux F, Meneses C, Herrera-Vásquez A, Blanco-Herrera F. Molecular and Genomic Characterization of the Pseudomonas syringae Phylogroup 4: An Emerging Pathogen of Arabidopsis thaliana and Nicotiana benthamiana. Microorganisms. 2022; 10(4):707. https://doi.org/10.3390/microorganisms10040707
Chicago/Turabian StyleZavala, Diego, Isabel Fuenzalida, María Victoria Gangas, Micaela Peppino Margutti, Claudia Bartoli, Fabrice Roux, Claudio Meneses, Ariel Herrera-Vásquez, and Francisca Blanco-Herrera. 2022. "Molecular and Genomic Characterization of the Pseudomonas syringae Phylogroup 4: An Emerging Pathogen of Arabidopsis thaliana and Nicotiana benthamiana" Microorganisms 10, no. 4: 707. https://doi.org/10.3390/microorganisms10040707
APA StyleZavala, D., Fuenzalida, I., Gangas, M. V., Peppino Margutti, M., Bartoli, C., Roux, F., Meneses, C., Herrera-Vásquez, A., & Blanco-Herrera, F. (2022). Molecular and Genomic Characterization of the Pseudomonas syringae Phylogroup 4: An Emerging Pathogen of Arabidopsis thaliana and Nicotiana benthamiana. Microorganisms, 10(4), 707. https://doi.org/10.3390/microorganisms10040707






