Predominance of Other Pathogenic Bacteria among Presumptive Tuberculosis Cases Attending Tuberculosis Clinics in Mwanza, Tanzania: A Cross-Sectional Laboratory-Based Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Population, Duration, and Setting
2.2. Enrollment of Participants, Data, and Sample Collections
2.3. Laboratory Procedures
2.3.1. Gram Staining of Sputum Samples for Assessing Sample Quality
2.3.2. Culture Procedures for Isolation of Other Pathogenic Bacteria from Presumptive TB Cases
2.3.3. Biochemical Identification Testing of Other Pathogenic Bacteria
2.3.4. Antibiotics Susceptibility Testing
2.3.5. GeneXpert for Detection of TB
2.3.6. Quality Control
2.3.7. Storage of Other Pathogenic Bacteria
2.4. Data Management and Analysis
2.5. Ethical Clearance Considerations
3. Results
3.1. Sociodemographic and Clinical Characteristics of Study Participants
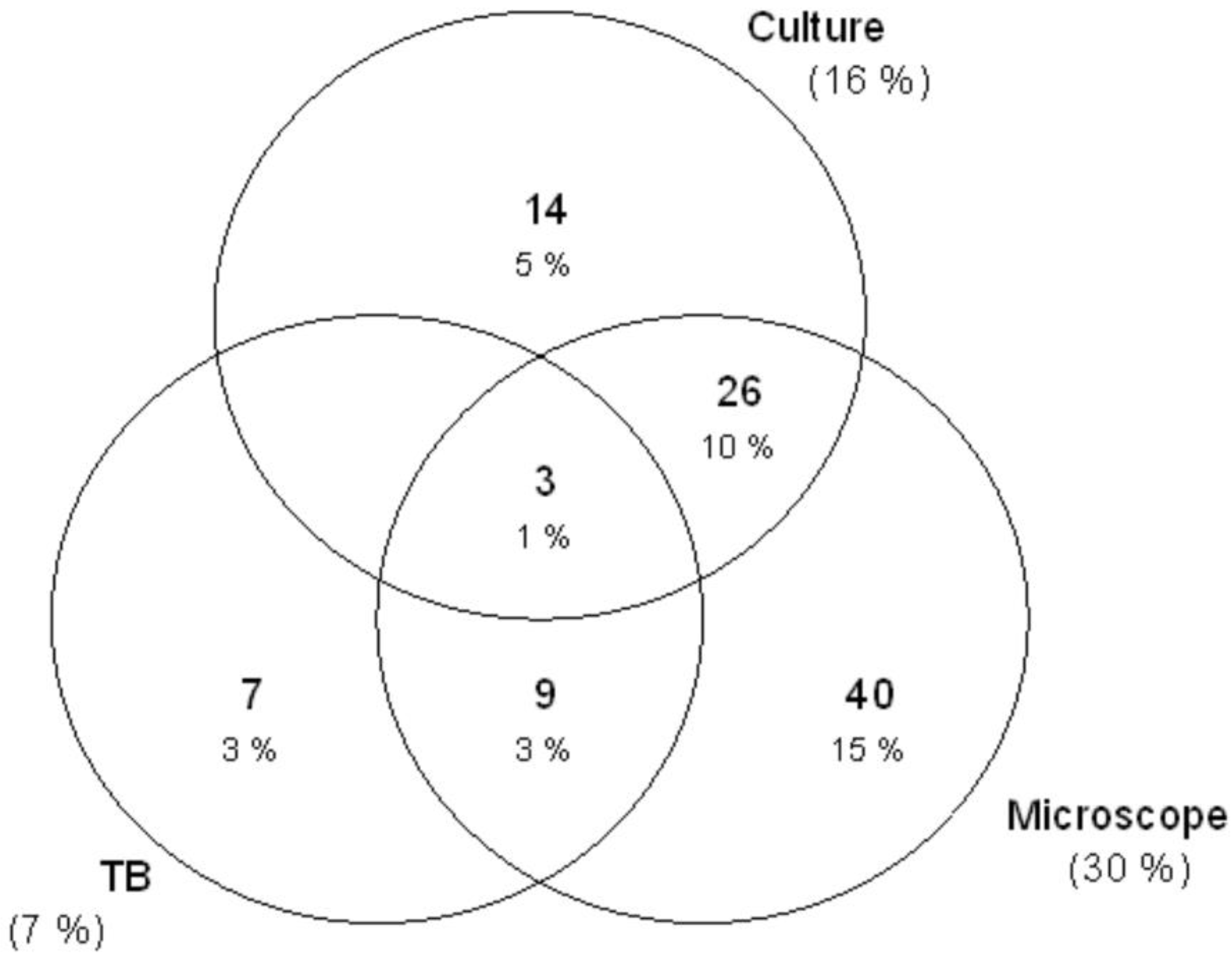
3.2. Microscope, Culture, and GeneXpert Results
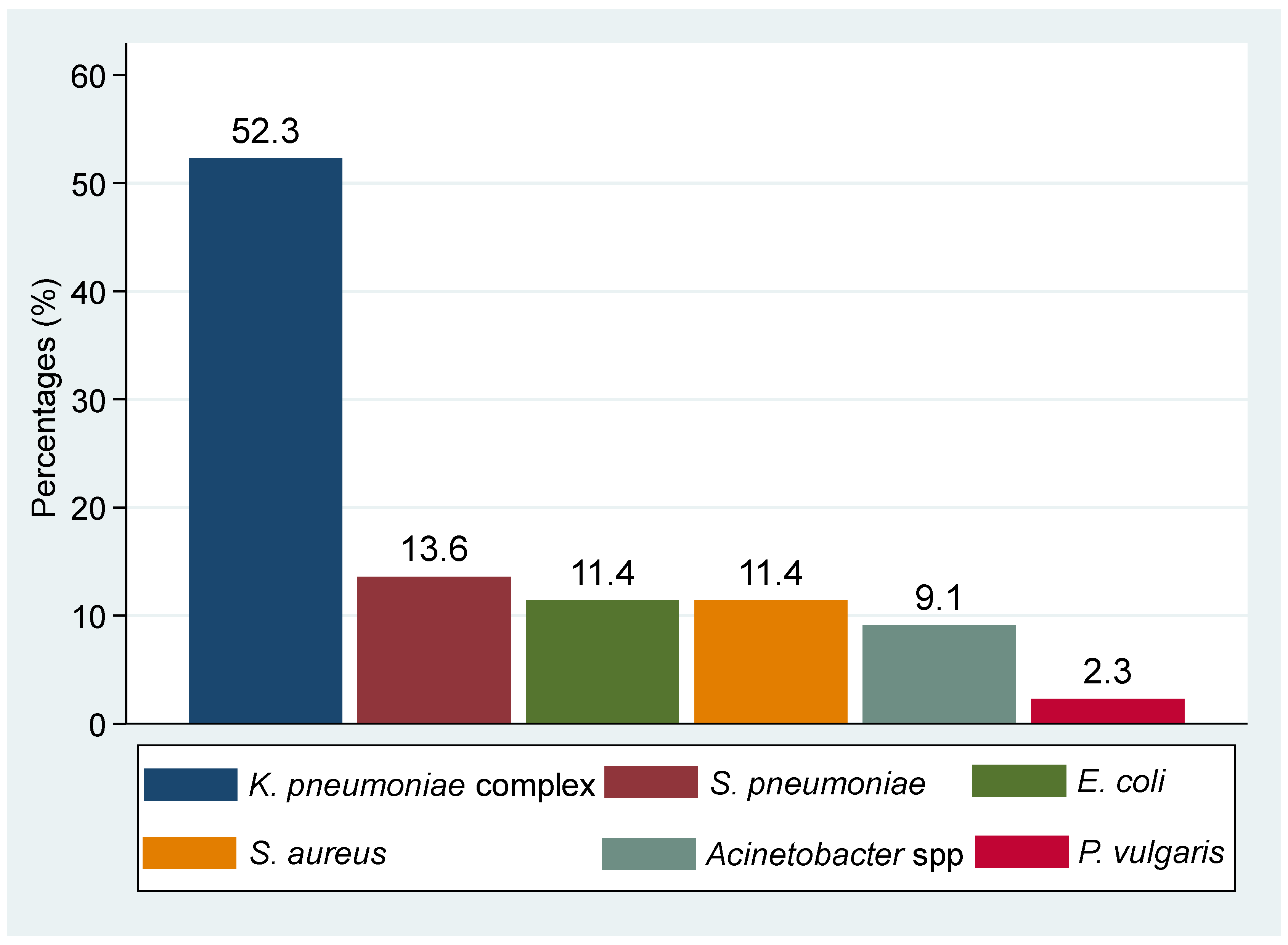
3.3. Percentages Resistance of Other Pathogenic Bacteria Isolated from Presumptive TB Cases
3.4. Factors Associated with a Culture Positive with Other Pathogenic Bacteria among Presumptive TB Cases
3.5. Factors Associated with Positive TB Results among Presumptive TB Cases
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Global Tuberculosis Report; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Arora, A.A.; Krishnaswamy, U.M.; Moideen, R.P.; Padmaja, M.S. Tubercular and bacterial coinfection: A case series. Lung India Off. Organ Indian Chest Soc. 2015, 32, 172. [Google Scholar] [CrossRef]
- Abdulkadir, B.; Abubakar, U.; Abdullahi, B.; Owuna, J.; Murtala, R.; Kabir, K.; Ibrahim, M. A survey of co-infection of some pathogenic bacteria with TB in patients attending Federal Medical Center Katsina, Nigeria. Bayero J. Pure Appl. Sci. 2019, 12, 209–214. [Google Scholar] [CrossRef]
- Attia, E.F.; Pho, Y.; Nhem, S.; Sok, C.; By, B.; Phann, D.; Nob, H.; Thann, S.; Yin, S.; Noce, R. Tuberculosis and other bacterial co-infection in Cambodia: A single center retrospective cross-sectional study. BMC Pulm. Med. 2019, 19, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Saldaña, N.G.; Bejarano, J.I.C.; Porras, M.H.; de la Garza, E.A.; Gutiérrez, S.F.; Gutiérrez, J.L.C.; Olguin, H.J. Co-infection with Streptococcus anginosus and Mycobacterium tuberculosis in an immunocompetent pediatric patient. A case report. BMC Pulm. Med. 2020, 20, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Meremo, A.; Kidenya, B.R.; Mshana, S.E.; Kabangila, R.; Kataraihya, J. High prevalence of tuberculosis among adults with fever admitted at a tertiary hospital in north-western Tanzania. Tanzan. J. Health Res. 2012, 14, 1–9. [Google Scholar]
- Mtwangambate, G.; Kalluvya, S.; Kidenya, B.; Kabangila, R.; Downs, J.; Smart, L.; Fitzgerald, D.; Peck, R. ‘Cough-triggered’ tuberculosis screening among adults with diabetes in Tanzania. Diabet. Med. 2014, 31, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Seni, J.; Kidenya, B.R.; Anga, M.; Kapesa, A.; Meda, J.R.; Mutakyawa, R.; Mkomwa, Z.H.; Marcel, F.; Changalucha, J.M.; Mshana, S.E. Incremental detection of pulmonary tuberculosis among presumptive patients by GeneXpert MTB/RIF® over fluorescent microscopy in Mwanza, Tanzania: An operational study. Healthc. Low-Resour. Settings 2015, 3, 10–13. [Google Scholar] [CrossRef]
- Sabi, I.; Kabyemera, R.; Mshana, S.; Kidenya, B.; Kasanga, G.; Gerwing-Adima, L.; Meremo, A.; Clowes, P.; Rachow, A.; Peck, R. Pulmonary TB bacteriologically confirmed by induced sputum among children at Bugando Medical Centre, Tanzania. Int. J. Tuberc. Lung Dis. 2016, 20, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Beyanga, M.; Kidenya, B.R.; Gerwing-Adima, L.; Ochodo, E.; Mshana, S.E.; Kasang, C. Investigation of household contacts of pulmonary tuberculosis patients increases case detection in Mwanza City, Tanzania. BMC Infect. Dis. 2018, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kishimbo, P.; Sogone, N.M.; Kalokola, F.; Mshana, S.E. Prevalence of gram-negative bacteria causing community acquired pneumonia among adults in Mwanza City, Tanzania. Pneumonia 2020, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Cochran, W.G. Sampling Techniques; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Bartlett, J.G.; Dowell, S.F.; Mandell, L.A.; File, T.M.; Jr Musher, D.M.; Fine, M.J. Practice Guidelines for the Management of Community-Acquired Pneumonia in Adults. Clin. Infect. Dis. 2000, 31, 347–382. [Google Scholar] [CrossRef] [PubMed]
- Koneman, E.W.; Allen, S.D.; Janda, W.; Schreckenberger, P.; Winn, W. Diagnostic Microbiology. The Nonfermentative Gram-Negative Bacilli Philedelphia; Lippincott-Raven Publishers: Philadelphia, PA, USA, 1997; pp. 253–320. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI: Pennsylvania, PA, USA, 2020. [Google Scholar]
- CDC. A New Tool to Diagnose Tuberculosis: The Xpert MTB/RIF Assay; CDC: Atlanta, GA, USA, 2016. [Google Scholar]
- MoHCDEC. Standard Treatment Guidelines and Essential Medicines List; MoHCDEC: Dar es Salaam, Tanzania, 2021; pp. 162–165. [Google Scholar]
- Kidenya, B.R.; Mshana, S.E.; Gerwing-Adima, L.; Kidola, J.; Kasang, C. Drug adherence and efficacy of smear microscopy in the diagnosis of pulmonary tuberculosis after 2 months of medication in North-western Tanzania. Int. J. Infect. Dis. 2017, 63, 43–47. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kang, H.K.; Jeong, B.-H.; Lee, H.; Park, H.Y.; Jeon, K.; Huh, H.J.; Ki, C.-S.; Lee, N.Y.; Koh, W.-J. Clinical significance of smear positivity for acid-fast bacilli after ≥5 months of treatment in patients with drug-susceptible pulmonary tuberculosis. Medicine 2016, 95, e4540. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Update: Nucleic acid amplification tests for tuberculosis. MMWR Morb. Mortal. Wkly. Rep. 2000, 49, 593–594. [Google Scholar]
- Dion, C.F.; Ashurst, J.V. Streptococcus Pneumoniae. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2017. [Google Scholar]
- Brooks, L.R.; Mias, G.I. Streptococcus pneumoniae’s virulence and host immunity: Aging, diagnostics, and prevention. Front. Immunol. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Żychska, M.; Witkowski, L.; Klementowska, A.; Rzewuska, M.; Kwiecień, E.; Stefańska, I.; Czopowicz, M.; Szaluś-Jordanow, O.; Mickiewicz, M.; Moroz, A.; et al. Rhodococcus equi-Occurrence in Goats and Clinical Case Report. Pathogens 2021, 10, 1141. [Google Scholar] [CrossRef] [PubMed]
- Van der Eerden, M.; Vlaspolder, F.; De Graaff, C.; Groot, T.; Jansen, H.; Boersma, W. Value of intensive diagnostic microbiological investigation in low-and high-risk patients with community-acquired pneumonia. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.M.; Bramley, A.M.; Jain, S.; Arnold, S.R.; Ampofo, K.; Self, W.H.; Williams, D.J.; Anderson, E.J.; Grijalva, C.G.; McCullers, J.A. Influence of antibiotics on the detection of bacteria by culture-based and culture-independent diagnostic tests in patients hospitalized with community-acquired pneumonia. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Loeb, M.; McGeer, A.; McArthur, M.; Walter, S.; Simor, A.E. Risk factors for pneumonia and other lower respiratory tract infections in elderly residents of long-term care facilities. Arch. Intern. Med. 1999, 159, 2058–2064. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-M.; Mou, C.-H.; Shen, T.-C.; Yang, C.-L.; Yang, M.-H.; Wu, F.-Y.; Sung, F.-C. Retrospective cohort evaluation on risk of pneumonia in patients with pulmonary tuberculosis. Medicine 2016, 95, e4000. [Google Scholar] [CrossRef] [PubMed]
- Baum, S.G. Laboratory evaluation of infectious disease emergencies. In Clinical Methods: The History, Physical, and Laboratory Examinations; Butterworths: London, UK, 2011. [Google Scholar]
- Lee, Y.J.; Shin, S.; Roh, E.Y.; Yoon, J.H.; Kim, D.K.; Chung, H.S.; Lee, C.-H. Acceptability of sputum specimens for diagnosing pulmonary tuberculosis. J. Korean Med. Sci. 2015, 30, 733–736. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Frequency (n)/mean (±SD) | Percentages (%) | |
|---|---|---|---|
| Mean (±SD) age in years | 36.6 (±19.7) | - | |
| Gender | Female | 106 | 40.2 |
| Male | 158 | 59.8 | |
| Healthcare of enrolment | BMC | 109 | 41.3 |
| SRRH | 93 | 35.2 | |
| SDDH | 62 | 23.5 | |
| Patient category | Inpatient | 9 | 3.4 |
| Outpatient | 255 | 96.6 | |
| History of fever over past 3 months | Yes | 53 | 20.1 |
| No | 211 | 79.9 | |
| History of antibiotic use | Yes | 7 | 2.7 |
| No | 257 | 97.3 | |
| Type of antibiotic used (N = 7) | Amoxicillin | 5 | 71.4 |
| Other | 2 | 28.6 | |
| Participant with chronic disease | Yes | 33 | 12.5 |
| No | 231 | 87.5 | |
| Type of chronic disease (N = 36) * | HIV | 30 | 83.3 |
| Other ** | 6 | 16.7 | |
| Current symptom of upper respiratory tract | Yes | 264 | 100.0 |
| No | 0 | 0.0 | |
| Previous history of TB | Yes | 10 | 3.8 |
| No | 254 | 96.2 | |
| Antibiotic Agents Tested | Gram-Negative Bacteria n (%) | Gram-Positive Bacteria n (%) |
|---|---|---|
| AMP | 32 (96.9) | NA |
| SXT | 12 (36.4) | 10 (90.9) |
| TE | 13 (39.4) | 6 (54.5) |
| CN | 8 (24.2) | 3 (27.3) |
| CIP | 15 (45.4) | 4 (36.4) |
| TZP | 13 (39.4) | NA |
| CRO | 9 (27.3) | NA |
| MEM | 3 (9.1) | NA |
| AK | 2 (6.1) | NA |
| Polymyxin-B | 6 (18.2) | NA |
| E | NA | 7 (63.6) |
| CD | NA | 6 (54.5) |
| FOX (S. aureus, n = 5) | NA | 1 (20.0) |
| P (S. pneumoniae, n = 6) | NA | 0 (0.0) |
| VA | NA | 0 (0.0) |
| LZD | NA | 0 (0.0) |
| Characteristics | Culture Results | Pearson Chi2 Test | |||
|---|---|---|---|---|---|
| Negative n (%) | Positive n (%) | X2 | p-Value | ||
| Gender | Female | 87 (82.1) | 19 (17.9) | ||
| Male | 134 (84.8) | 24 (15.2) | 0.3479 | 0.555 | |
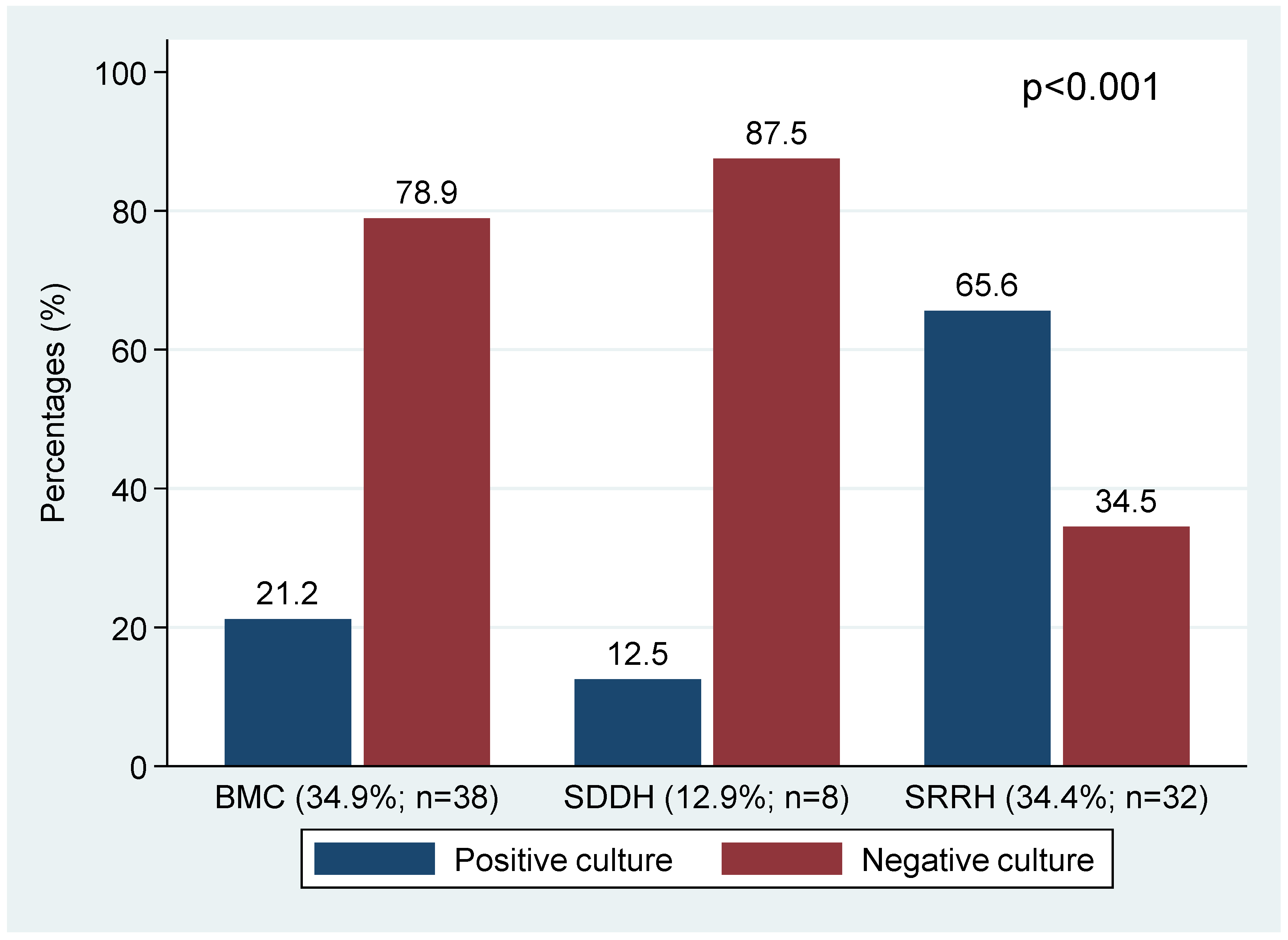
| Healthcare facility of enrolment | BMC | 95 (87.2) | 14 (12.8) | ||
| SRRH | 66 (70.9) | 27 (29.1) | |||
| SDDH | 60 (96.8) | 2 (3.2) | 19.7845 | <0.001 | |
| History of fever | No | 173 (81.9) | 34 (18.1) | ||
| Yes | 48 (90.6) | 5 (9.4) | 2.2847 | 0.131 | |
| History of antibiotic use | No | 214 (83.3) | 43 (16.7) | ||
| Yes | 7 (100.0) | 0 (0.0) | 1.3991 | 0.237 | |
| Chronic disease condition | No | 196 (84.6) | 35 (15.2) | ||
| Yes | 25 (75.8) | 8 (24.2) | 1.7502 | 0.186 | |
| Microscope inflammation | Contamination | 172 (92.5) | 14 (7.5) | ||
| Inflammation | 49 (62.8) | 29 (37.2) | 35.4386 | <0.001 | |
| TB status | Negative | 205 (83.7) | 40 (16.3) | ||
| Negative | 16 (84.2) | 3 (15.8) | 0.0037 | 0.951 | |
| Characteristics | TB/GeneXpert Results | Pearson Chi2 Test | |||
|---|---|---|---|---|---|
| Negative n (%) | Positive n (%) | X2 | p-Value | ||
| Gender | Female | 101 (95.3) | 5 (4.7) | ||
| Male | 144 (91.1) | 14 (8.9) | 1.6309 | 0.202 | |
| Healthcare facility of enrolment | BMC | 104 (95.4) | 5 (4.6) | ||
| SRRH | 79 (84.9) | 14 (15.1) | |||
| SDDH | 62 (100.0) | 0 (0.0) | 14.5150 | 0.001 | |
| History of fever | No | 196 (92.9) | 15 (7.1) | ||
| Yes | 49 (92.5) | 4 (7.6) | - | 1.000 * | |
| History of antibiotic use | No | 239 (93.0) | 18 (7.0) | ||
| Yes | 6 (84.7) | 1 (14.3) | - | 0.411 * | |
| Chronic disease condition | No | 212 (91.8) | 19 (8.2) | ||
| Yes | 33 (100.0) | 0 (0.0) | - | 0.143 * | |
| Microscope inflammation | Contamination | 179 (96.2) | 7 (3.8) | ||
| Inflammation | 66 (84.6) | 12 (15.4) | 11.1120 | 0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buchera, F.S.; Silago, V.; Japhet, G.; Mtemisika, C.I.; Damiano, P.; Nyawale, H.A.; Mushi, M.F.; Mirambo, M.M.; Seni, J.; Mshana, S.E. Predominance of Other Pathogenic Bacteria among Presumptive Tuberculosis Cases Attending Tuberculosis Clinics in Mwanza, Tanzania: A Cross-Sectional Laboratory-Based Study. Microorganisms 2022, 10, 703. https://doi.org/10.3390/microorganisms10040703
Buchera FS, Silago V, Japhet G, Mtemisika CI, Damiano P, Nyawale HA, Mushi MF, Mirambo MM, Seni J, Mshana SE. Predominance of Other Pathogenic Bacteria among Presumptive Tuberculosis Cases Attending Tuberculosis Clinics in Mwanza, Tanzania: A Cross-Sectional Laboratory-Based Study. Microorganisms. 2022; 10(4):703. https://doi.org/10.3390/microorganisms10040703
Chicago/Turabian StyleBuchera, Florencia S., Vitus Silago, Geofrey Japhet, Conjester I. Mtemisika, Prisca Damiano, Helmut A. Nyawale, Martha F. Mushi, Mariam M. Mirambo, Jeremiah Seni, and Stephen E. Mshana. 2022. "Predominance of Other Pathogenic Bacteria among Presumptive Tuberculosis Cases Attending Tuberculosis Clinics in Mwanza, Tanzania: A Cross-Sectional Laboratory-Based Study" Microorganisms 10, no. 4: 703. https://doi.org/10.3390/microorganisms10040703
APA StyleBuchera, F. S., Silago, V., Japhet, G., Mtemisika, C. I., Damiano, P., Nyawale, H. A., Mushi, M. F., Mirambo, M. M., Seni, J., & Mshana, S. E. (2022). Predominance of Other Pathogenic Bacteria among Presumptive Tuberculosis Cases Attending Tuberculosis Clinics in Mwanza, Tanzania: A Cross-Sectional Laboratory-Based Study. Microorganisms, 10(4), 703. https://doi.org/10.3390/microorganisms10040703







