Abstract
Bacteria thrive in ever-changing environments by quickly remodeling their transcriptome and proteome via complex regulatory circuits. Regulation occurs at multiple steps, from the transcription of genes to the post-translational modification of proteins, via both protein and RNA regulators. At the post-transcriptional level, the RNA fate is balanced through the binding of ribosomes, chaperones and ribonucleases. We aim to decipher the role of the double-stranded-RNA-specific endoribonuclease RNase III and to evaluate its biological importance in the adaptation to modifications of the environment. The inactivation of RNase III affects a large number of genes and leads to several phenotypical defects, such as reduced thermotolerance in Escherichia coli. In this study, we reveal that RNase III inactivation leads to an increased sensitivity to temperature shock and oxidative stress. We further show that RNase III is important for the induction of the heat shock sigma factor RpoH and for the expression of the superoxide dismutase SodA.
1. Introduction
In order to maintain fitness, bacteria sense environmental cues, and subsequently adapt their behavior by remodeling their transcriptome and proteome. Adaptation depends on regulatory mechanisms, which act at all levels of gene expression, from transcription to post-translation. At the post-transcriptional level, the fate of RNAs is decided by a plethora of effectors, e.g., small molecules, RNA-binding proteins and other RNAs. These regulators control the translation, protection and destabilization of messenger RNAs (mRNAs) via specific sequences and/or structural motifs (reviewed by [1,2,3]). Among these factors, ribonucleases (RNases) are major actors that ensure a tight control on gene expression (reviewed in [4]).
The endoribonuclease III (RNase III) domain, widely conserved in bacteria and eukaryotes, provides the specificity for double-stranded RNA (dsRNA) cleavage (reviewed in [5,6,7]). RNase III enzymes are involved in different processes, such as the repression and also activation of gene expression and maturation of stable and regulatory RNAs. However recent findings suggest that other functions could have emerged in some eukaryotic organisms, such as the direct involvement of DROSHA in the activation of transcription in humans [8]. In Escherichia coli, RNase III (encoded by the gene rnc) forms a dimer, and its canonical activity is to bind a dsRNA substrate that can either be intramolecular duplexes, formed within the same RNA molecule (e.g., hairpin loop) or intermolecular hybrids (e.g., antisense RNA bound to its complementary target) [7,9]. Upon binding to a specific target, the two RNase III monomers will adopt the active conformation and perform the cleavage of one or of both RNA strands, generating fragments whose ends are staggered by two bases on one strand compared to the other.
The role of RNase III in E. coli was first thought to be limited to the initial processing steps of rRNAs and to cleaving a few other specialized RNAs, such as SsrA and T7 mRNAs, to generate their mature and functional forms [10,11,12,13,14]. It later appeared that RNase III is more widely involved in the control of gene expression [15,16,17,18,19,20]. The dicistronic mRNA rpsO-pnp, encoding the ribosomal protein S15 and the exoribonuclease PNPase, was one of the first cases studied. RNase III cleavages in the 5′-untranslated region (UTR) of pnp initiate its degradation [16,18,21] and trigger further cleavages by RNase E, leading to the irreversible inactivation of the pnp mRNA [22]. As a consequence, inactivation of RNase III results in the overexpression of PNPase [23]. In a small number of cases, RNase III was also shown to be involved in the positive post-transcriptional regulation of a diverse set of genes, including gadE mRNA, encoding a regulator involved in acid resistance [24]; adhE mRNA encoding an alcohol dehydrogenase essential for fermentation [25]; eno mRNA encoding the metabolic enzyme enolase, which is also associated with the degradosome [26], cIII and N mRNAs from the phage λ [15,27] and the pre-mRNA of phage T7 [28]. Thus, in addition to its role in RNA decay, RNase III is also involved in the positive regulation of gene expression.
The application of RNA-seq methodologies has demonstrated an even wider role of RNase III in the control of gene expression [29,30,31]. One study, combining stability experiments with RNA-seq (i.e., sequencing after rifampicin treatment), reported 2054 RNase III-dependent cleavage sites (11 were verified on purified RNA fragments by in vitro RNase III cleavage assays) [32]. In another study, a tailored RNA-seq based approach coupled to an enrichment of small fragments in a mutant inactivated for RNase III (i.e., by the addition of a specific tag on 5′-monophosphate extremities after the degradation of 5′-triphosphate RNAs) detected 1003 cleavage sites in 615 targets [33]. Stead and colleagues observed, using a microarray-based experiment, that genes implicated in the response to heat constituted an important fraction of the total coding genes affected in the rnc mutant [30]. Another microarray study, performed in strains with reduced or increased RNase III levels, led to the identification of four genes negatively controlled by RNase III; proP, proU, bdm and betT, and revealed that RNase III activity is reduced during an osmotic shock and during a cold shock at the post-translational level (the latter caused by the binding of the YmdB protein to RNase III) [29,34,35,36,37]. A few other studies have addressed the roles of RNase III in E. coli physiology, revealing reduced thermotolerance and motility in an rnc mutant, while aminoglycoside-resistant mutant strains have been shown to rely on increased RNase III activity [38,39,40].
In this work, we show that the inactivation of RNase III is detrimental for survival at high and low temperatures and for resistance to oxidative stress. We observe that the induction of the heat shock sigma factor RpoH and the expression of the superoxide dismutase SodA are reduced upon inactivation of RNase III. We show that, during a heat shock, RNase III together with PNPase are required for the stabilization of the induced rpoH mRNA, while our results suggest that RNase III also plays a positive role in rpoH transcriptional induction. We further characterize the importance of RNase III and PNPase in the stabilization of sodA mRNA and highlight an additional role of RNase III in the transcriptional regulation of sodA.
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions
Strains, plasmids and primers used in this study are described in Table S1. RNase III, encoded by the rnc gene in E. coli, is co-expressed with the gene for the essential GTPase Era [41], and hence its deletion is not trivial, as demonstrated by the absence of an rnc mutant in the Keio collection [42]. We used the rnc105 mutation, a point mutation (G44D) leading to catalytic inactivation and reduced affinity for RNA [43] that has no polar effect on the transcription of the era gene, located downstream. Bacteria were grown in LB medium at 37 °C and sampled in mid-log phase (A650 ≈ 0.4) when not specified otherwise. The IBPC633 strain (carrying the rnc105 mutation linked to the nadB51::Tn10 (TetR) mutation in the N3433 background) transformed with the pRNC1 plasmid expressing RNase III from the Para promoter of the pKAN6 plasmid or the pKAN6 empty control vector [29] were grown in LB medium with 50 μg/mL kanamycin and induced by addition of arabinose at the indicated concentrations. N3433-Plac-sodA and IBPC633-Plac-sodA strains, transformed with the plasmid pBRlacIq, were grown in LB medium with 100 μg/mL ampicillin. Mutant alleles (pnp, crp and cytR) were moved to the N3433 and IBPC633 genetic background strains by P1 transduction.
2.2. Promoter Replacement and Strain Construction
The kmPcL genetic element, encoding the kanamycin resistance cassette followed by the Plac promoter, was amplified by PCR from the strain MG1655kmPcLyad with the primers mSodA3pClKan and Kan-pLac-SodA4 (Table S1) and inserted in front of the gene sodA at its native chromosomal location, as described in [44], with the following modifications. After purification, the cassette was electroporated into a strain containing an activated mini-λ expressing the λ-Red recombinase gene replacement system, allowing for the replacement by homologous recombination of the native promoter as described (Table S1) [45]. The modified Plac-sodA promoter was then moved by P1 transduction by selecting for resistance to kanamycin.
2.3. RNA Extraction and Northern Blot Analysis
Total RNA was prepared using the hot-phenol procedure [46]. Total RNA (5 µg) was electrophoresed on 1% agarose, 1xTBE gels for analysis by northern blot [22,47]. Membranes were hybridized with RNA probes synthesized by T7 RNA polymerase with [α-32P] UTP yielding uniformly labeled RNAs [48]. The size of RNAs was estimated by comparison with migration of the RiboRuler High Range marker (Thermo Scientific, Thermo Fisher Scientific, Waltham, MA, USA). RNA stability was measured on cultures treated with rifampicin (500 μg/mL). Total RNA was extracted after 40 s (time 0) and at the indicated time points. Membranes were also probed for M1 RNA (or 5S rRNA for oldest experiments) used as charge control.
2.4. Western Blot
Total protein samples were collected by centrifugation and pellets were resuspended in SDS-loading buffer containing DTT (Biolabs), sonicated and heat denatured. Total protein was separated after denaturation on a 4–15% mini Protean TGX gel, and the proteins were transferred on a nitrocellulose membrane using Trans-Blot system (Biorad, Hercules, CA, USA). Membranes were blocked for 1 h prior to overnight incubation with the anti-SodA (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) or anti-RpoH (BioLegend, San Diego, CA, USA) antibodies that were diluted 1000-fold and 500-fold, respectively, in phosphate buffered saline (PBS) with 0.05% Tween 20. After washing and incubation with the secondary antibody, detection was performed using the Clarity Max reagent kit (Biorad, Hercules, CA, USA) and images acquired on a ChemiDoc (Biorad, Hercules, CA, USA). Protein sizes were estimated by comparison with migration of the PageRuler Plus 10–250 kDa ladder (Thermo Scientific, Thermo Fisher Scientific, Waltham, MA, USA). RpoH protein was identified as a single band on the membrane using monoclonal antibodies, whereas anti-SodA polyclonal antibodies revealed multiple bands. Therefore, the specific SodA protein was identified by comparison with a sample extracted from a sodA deleted strain (Figure S7C). Membranes were reprobed with anti-S1 antibodies (10,000-fold dilution) in order to use the ribosomal protein S1 as charge control.
2.5. In Vitro Processing by RNase III
A DNA template carrying a T7 promoter sequence was generated by PCR using primers mT7SodA and sodAterm (Table S1). The sodA template was transcribed using T7 RNA polymerase into the full-length sodA mRNA (772 nts), as described in [49]. RNA 5′-end labeling was performed with [γ-32P]-ATP using T4 polynucleotide kinase. Transcripts were incubated in 20 mM Tris acetate, pH 7.5, 10 mM magnesium acetate and 100 mM sodium acetate for 5 min at 37 °C and submitted to in vitro processing by RNase III of E. coli at 37 °C in the presence of 1 µg tRNA for 25 min with the indicated quantities of RNase III (Epicentre, Madison, WI, USA) [50]. After precipitation, addition of loading buffer and heat denaturation, samples were analyzed on 6% polyacrylamide (19/1)-urea 7 M-1xTBE gels along with 5′-radio-labeled MspI digested pBR322 (New England Biolabs, Ipswich, Massachusetts, United States).
2.6. Data Analysis
Northern blots were scanned using a Typhoon FLA 9500 scanner (GE Healthcare, Chicago, IL, USA). The resulting .gel images were quantified using the ImageQuantTL software version 8.1. The acquired images were uniformly adjusted for their contrast before being cropped and assembled. Bands of interest were quantified and the abundance of the studied transcripts was normalized by comparison to the abundance of the M1 or 5S RNA. Northern blots were performed in technical duplicates or more, as indicated in legends. For the stability assay presented in Table 1, normalized abundance of the studied mRNAs at the indicated times after rifampicin treatments were plotted using linear regression with a 90% confidence level, half-life and standard deviations calculated using the Microsoft Excel software. Western blots were quantified as .tif images by measuring raw integrated density and background normalization using the ImageJ software version 1.53c (NIH, https://imagej.nih.gov/ij/, last accessed on the 22 March 2022) [51]. The abundance of the protein of interest was then normalized compared to the S1 loading control. The acquired images were uniformly adjusted for their contrast before being cropped and assembled. Survival assays and growth curves were performed in biological duplicates and a representative experiment is shown. Images of survival plates were captured in .tif format on a GelDoc (Biorad), uniformly adjusted for their contrast before being cropped and assembled using the ImageJ software version 1.53c.

Table 1.
Effect of rnc and pnp mutations on the decay rates of rpoH and sodA mRNAs.
3. Results
3.1. Survival under Extreme Temperatures Depends on RNase III
We compared growth at different temperatures of a wild-type strain (N3433, referred to as wt) to its rnc105 derivative strain (IBPC633, referred to as rnc) by a droplet plating assay after a short heat shock (15 min at 45 °C). We observed comparable survival at 37 °C and at 30 °C, indicating that the number of viable cells is similar in the wt and rnc mutant (Figure 1A). In agreement with a previous study [38], the rnc mutant showed a decreased survival at 45 °C (Figure 1A). To demonstrate that RNase III inactivation is directly responsible for thermal sensitivity, we used the previously described pRNC1 plasmid, allowing for the ectopic expression of RNase III [29]. In the absence of induction, pRNC1 was shown to drive the expression of around 10% of the wt level and up to a 10-fold overexpression upon induction by arabinose [29]. In the rnc mutant, survival at 45 °C increases upon ectopic expression of RNase III from the pRNC1 plasmid, even in the absence of induction (Figure S1A), suggesting that a low level of wt RNase III is sufficient for phenotypic complementation.
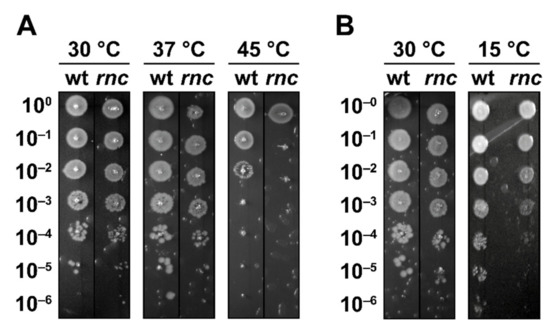
Figure 1.
RNase III is required for survival at high and low temperatures. (A) N3433 (wt) and its rnc105 (rnc) derivative were grown at 30 °C before being shifted to 45 °C for 15 min, and survival was assayed by droplet plating at 30, 37 and 45 °C. (B) Strains wt and rnc were grown at 30 °C and survival was assayed by droplet plating at 30 and 15 °C.
Previous studies showed that RNase III is post-translationally inhibited upon the induction of YmdB during a cold shock, resulting in the induction of PNPase [37], whose expression is essential for survival during a cold shock [52]. Remarkably, we observed that RNase III inactivation led to a reduced survival at 15 °C (Figure 1B), a phenotype that has not been reported before. YmdB perturbs RNase III catalytic activity and homodimer formation both in vitro and in vivo, although the RNase III-YmdB complex still binds dsRNAs [53]. Thus, one possible explanation is that, while it is necessary to reduce the RNase III catalytic activity during cold shock, the RNase III-YmdB complex could still exert a regulatory role and coordinate the induction of a stress response. In agreement with this hypothesis, the rnc70 mutation was shown to impair the endonucleolytic activity of RNase III without affecting its ability to bind dsRNAs whereas rnc105 (used in this study) was suggested to have a reduced affinity for dsRNA [43].
In summary, these results demonstrate that RNase III is required by E. coli for sustained growth at high and at low temperatures.
3.2. RNase III Inactivation Increases Sensitivity to Hydrogen Peroxide
We challenged wt and rnc strains with hydrogen peroxide (H2O2), which is known to provoke a rapid, irreversible arrest of cell division due to the accumulation of DNA breaks [54]. A droplet plating assay after 10 min exposure to oxidative stress (from 5 mM H2O2) reveals a significant decrease in the survival of the rnc mutant (Figure 2A). Moreover, the ectopic expression of RNase III from pRNC1 increased the survival of the rnc mutant, even in the absence of induction, showing that even a low level of wt RNase III allows phenotypic complementation (Figure S1B), thus confirming that the expression of RNase III in the rnc105 (nadB51::Tn10) mutant can restore resistance to oxidative stress. We confirmed that the effect of H2O2 (0.8 mM in liquid culture) on the survival of the rnc mutant was due to an immediate growth arrest (Figure 2B). We also challenged the two strains with paraquat (0.1 mM), which is known to generate superoxide [54], and observed a reduction in the survival of the rnc mutant after 60 min of exposure by a droplet plating assay (Figure S1C). However, paraquat challenge in liquid culture led to only a slight reduction in the growth rate of the rnc mutant (Figure S1D). Thus, RNase III inactivation leads to an increased sensitivity to oxidative stress mediated by H2O2 and, to a lower extent, mediated by paraquat.
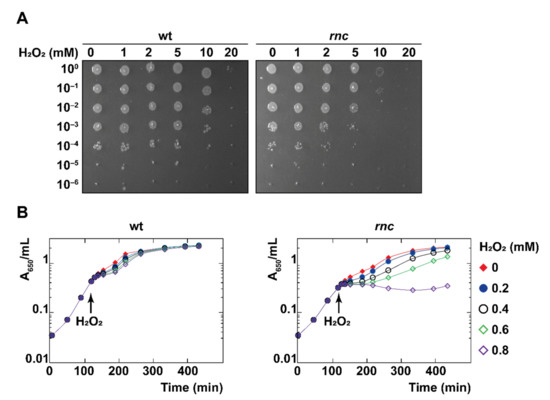
Figure 2.
RNase III is required for survival under oxidative stress. (A) Strains wt and rnc were grown until mid-log phase, exposed to H2O2 at the indicated concentrations for 10 min, and survival was assayed by droplet plating. (B) Strains wt and rnc were grown at 37 °C and H2O2 was added to the cultures in mid-log phase, as indicated by a black arrow.
3.3. The Induction of rpoH at High Temperature Depends on RNase III
A previous transcriptomic approach revealed that a large number of genes belonging to the gene ontology group “response to heat”, including members of the heat shock RpoH regulon, are differentially expressed upon RNase III inactivation [30]. The heat shock sigma factor σ 32 (RpoH) is a key regulator of the E. coli heat shock response and is essential for growth at temperatures higher than 20 °C [55,56]. The expression of rpoH is induced upon a temperature upshift (among other conditions, see Discussion) via both enhanced synthesis and stabilization [57,58]. In particular the rpoH mRNA harbors an extensive secondary structure, located downstream from the start codon, containing four stem loops that act as a thermosensor controlling the translational efficiency in order to maintain a low translation of rpoH mRNA at temperatures less than 45 °C [58,59]. To examine whether RNase III has a role in the heat adaptation of rpoH expression, we measured rpoH mRNA and RpoH protein levels in the early (i.e., less than 15 min) response to heat shock [60,61,62]. Whereas the abundance of the rpoH mRNA increases after the heat shock (3.5-fold after 15 min of heat shock, Figure 3A and Figure S2), the amplitude of the induction was reduced twofold upon RNase III inactivation (1.8-fold after 15 min of heat shock). At the protein level, RpoH induction peaked after 5 min in the wt strain (2.5-fold after 5 min of heat shock, Figure 3B and Figure S3) whereas, unexpectedly, the abundance of RpoH decreased in the rnc mutant (0.6-fold after 5 min of heat shock). In brief, RNase III activity is critical for the induction of rpoH expression at both mRNA and protein levels during a heat shock, i.e., it has a positive role in controlling rpoH expression.
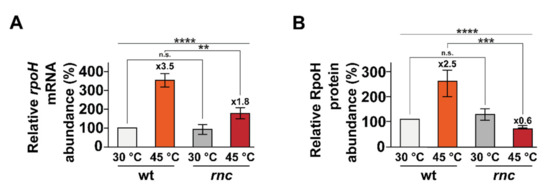
Figure 3.
rpoH induction upon temperature upshift is defective in the rnc mutant. Strains wt and rnc were grown at 30 °C and transferred to 45 °C. Total RNA and protein sampled before or after a heat shock were analyzed by northern blot (see Figure S2) and Western blot (see Figure S3). (A) Mean rpoH mRNA levels and (B) mean RpoH protein levels are indicated before (30 °C, in grey) and after (A) 15 min or (B) 5 min of heat shock (45 °C, in red). Values are means of 3 biological replicates and error bars are standard deviations. Statistical significance was determined by ANOVA (n.s. for p-values ≥ 0.05, ** for p-values ≤ 0.01, *** for p-values ≤ 0.001 and **** for p-values ≤ 0.0001).
3.4. RNase III and PNPase Stabilize rpoH mRNA during a Heat Shock
As we observed a significant reduction in rpoH expression in the rnc mutant during a heat shock, we investigated the importance of RNase III on rpoH mRNA stability after a shift at 45 °C. We observed that RNase III inactivation led to a slight reduction in rpoH stability after a 15 min heat shock from 30 to 45 °C (from 4.01 to 3.39 min, Table 1 and Figure S4A), suggesting that RNase III has only a modest effect on the stability of the rpoH mRNA during a heat shock. However, it should be remembered that the inactivation of RNase III leads to the accumulation of the exoribonuclease PNPase, as previously shown at the mRNA [22] and protein level [23,31]. We observed that the stability of rpoH mRNA after a heat shock in a PNPase inactivated mutant (pnp) is reduced (from 4.01 to 3.14 min, Table 1 and Figure S4A). Hence, as PNPase is also involved in the stabilization of the rpoH mRNA after a heat shock, we reasoned that the elevated abundance of PNPase in the rnc mutant could mask the effect of RNase III. Supporting this hypothesis, we observed that the inactivation of RNase III in a pnp mutant led to a further reduction in rpoH mRNA stability after the heat shock (from 4.01 min to 2.06 min, Table 1 and Figure S4A). Thus, the stability of the rpoH mRNA after a heat shock relies on both RNase III and PNPase, and both ribonucleases have positive roles in the expression of rpoH.
3.5. RNase III Acts Independently from Transcription Factors CRP and CytR
As the above results show that the reduced rpoH induction during a heat shock is only partly due to the role of RNase III in the stabilization of rpoH mRNA (which is, moreover, partially compensated by the increase in PNPase), it appears that RNase III plays an additional positive role in the transcriptional regulation of rpoH during a heat shock, which is likely to be indirect via other effectors. The transcription of rpoH was shown to be dependent on the sigma factors RpoD, RpoE, RpoN and RpoS and regulated by transcriptional factors DnaA, ZraR, CpxR, IHF, CRP and CytR (Ecocyc database [41]). Remarkably, the details of the transcriptional induction of rpoH during a heat shock remains, to our knowledge, non-elucidated [31]. We compared the induction level of the rpoH mRNA after 15 min heat shock from 30 to 45 °C in mutants inactivated for CRP and/or CytR and/or RNase III. We observed that, while both crp and cytR mutations slightly affected the expression of rpoH, the reduction in rpoH induction in the RNase III mutant could still be seen in the absence of either or both crp and cytR (Figure S5). Hence, the role of RNase III in the induction of rpoH expression during a heat shock is independent of the transcription factors CRP and CytR.
3.6. Induction of Three Genes of the RpoH Regulon Is Defective in the rnc Mutant
To investigate whether the inactivation of RNase III has repercussions on the RpoH regulon, we analyzed the effect of heat shock on three genes belonging to the RpoH regulon: dnaK, encoding a heat shock protein (HSP) chaperone, ibpA, encoding a small HSP chaperone and lon, encoding a major protease. In the wt strain, the expression of dnaK, ibpA and lon increased after 15 min at 45 °C and subsequently decreased after 30 min of heat shock, demonstrating that their induction is transient at the RNA level (Figure 4). In the rnc mutant, the rpoH mRNA abundance was lower (twofold) than in the wt, but was still induced by the heat shock, whereas the dnaK, ibpA and lon mRNAs levels decreased, which is in agreement with the previous observation that the RpoH protein does not accumulate during a heat shock in the rnc mutant (Figure 3B). This establishes a temporal correlation between RpoH protein levels and the expression of its regulon. Thus, RNase III is required for a full induction of rpoH and for at least three genes of its regulon (dnaK, ibpA and lon) during a heat shock.
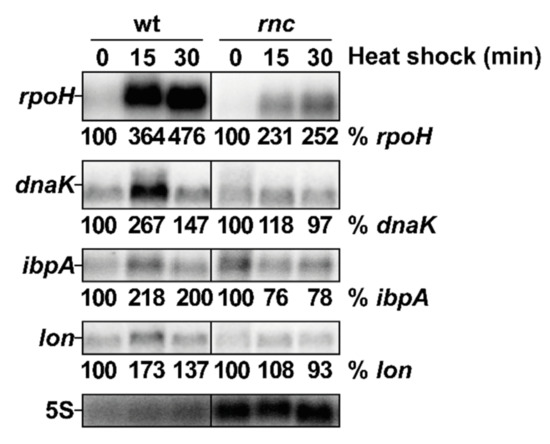
Figure 4.
RNase III positively controls the induction of rpoH and of three targets of the RpoH regulon after heat shock. Strains wt and rnc were grown at 30 °C and shifted to 45 °C. Total RNA samples were taken before (time 0) or at the indicated time after the heat shock and analyzed by northern blot. The membrane was probed successively (after removal of previous signals) for rpoH, dnaK, ibpA, lon and 5S. Quantification of the different transcripts is given as % of the indicated mRNA in the wt at 30 °C normalized to the 5S rRNA charge control.
3.7. RNase III Positively Regulates sodA Expression
RNase III is involved in the destabilization of the sodB mRNA, encoding the Fe2+-dependent superoxide dismutase B, upon binding by the sRNA RhyB under iron starvation [63]. More recently, the expression of sodA, encoding the major Mn2+-dependent cytoplasmic superoxide dismutase SodA, was observed to be strongly reduced in two independent rnc deletion mutants [31]. Thus, we investigated the role of RNase III in the regulation of sodA and in the resistance to oxidative stress. We first validated that sodA expression is reduced in the rnc mutant in the absence of stress at both mRNA (2.6-fold, Figure 5A and Figure S6) and protein levels (2.4-fold, Figure 5B and Figure S7). Furthermore, we observed that, after 10 min of oxidative stress in the presence of H2O2, sodA expression was slightly reduced at the mRNA level (Figure S6) but not at the protein level in the wt (Figure S7), while RNase III inactivation led to a strong reduction in sodA expression at both mRNA and protein level, similar to what could be observed in the absence of stress. Hence, RNase III is required for the expression of sodA in the absence of and during oxidative stress.
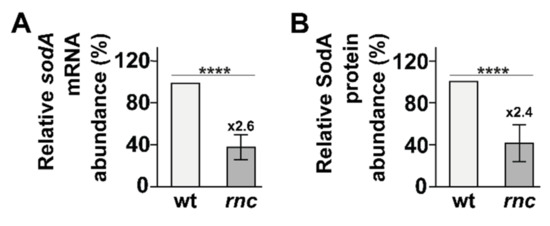
Figure 5.
RNase III positively controls sodA expression. Total RNA and protein were sampled from strains wt and rnc and analyzed by (A) northern blot (see Figure S6) and (B) Western blot (see Figure S7). (A) Mean sodA mRNA levels and (B) mean SodA protein levels are shown. Values are means of 3 biological replicates and error bars are standard deviations. Statistical significance was determined by ANOVA (**** for p-values ≤ 0.0001).
3.8. RNase III and PNPase Stabilize sodA mRNA
We then examined the stability of sodA mRNA and found that it was reduced in the rnc mutant (1.8-fold, Table 1 and Figure S4B). Since PNPase is upregulated in the rnc mutant strain, we examined whether the reduced stability of the sodA mRNA could be due to an increased expression of PNPase. The inactivation of PNPase destabilizes the sodA mRNA (2.3-fold, Table 1 and Figure S4B) and, together with the rnc mutation, leads to a further reduction (three-fold) in sodA mRNA stability. As RNase III has a role in the stabilization of sodA mRNA in the absence of stress, we investigated whether this effect could be due to an RNase III processing event that may stabilize the sodA mRNA. An in vitro RNase III cleavage assay showed that the full-length sodA mRNA is cleaved by RNase III at multiple positions, including one minor cleavage within the 5′-UTR (nts +28 relative to the transcription start site, Figure S8), which is similar to the unique in vivo cleavage site in the sodA mRNA previously identified [33]. In summary, RNase III is required for sodA mRNA stabilization and can cleave the sodA mRNA 5′-UTR in vitro, supporting previous in vivo observations. Hence, this suggests that, like in the case of the target mRNAs gadE [25] and adhE [25], RNase III can cleave the 5′-UTR of the sodA mRNA, which may lead to the stabilization of the transcript, either by protecting from other RNases or by improving the translation efficiency. The other cleavages observed in vitro, which were not detected in vivo, lie within the ORF and so might be protected from cleavage in vivo by translating ribosomes.
3.9. Transcriptional and Post-Transcriptional Regulation of the sodA Gene by RNase III
We next looked for an effect of RNase III on the transcription as well as on the post-transcriptional regulation of sodA. To separate these regulatory layers, the endogenous sodA promoter was replaced by the lacZ promoter, which allows for an appreciable expression of sodA mRNA from the Plac promoter in the absence of IPTG (Figure 6). Contrary to the case where sodA is expressed from its own promoter, the rnc mutation had little effect on the sodA mRNA level. However, when strongly overproduced after IPTG induction, there was a slight reduction in the sodA mRNA level in the rnc mutant. This limited effect of RNase III on sodA mRNA expressed from an exogenous promoter, compared to the strong positive effect on sodA mRNA levels, when it is expressed from its own promoter, implies that the major role of RNase III in the regulation of sodA expression occurs by a transcriptional activation of the sodA gene. This indirect effect could be due to the regulation of an upstream regulator of sodA expression.

Figure 6.
Transcriptional and post-transcriptional regulation of sodA by RNase III. N3433 Plac-sodA (wt) and IBPC633 Plac-sodA (rnc) strains with pBRlacIq expressing lacIq constitutively were induced (+) or not (−) with IPTG (0.1 mM) for 10 min. Total RNA was analyzed by northern blot; the membrane was probed for sodA and M1. Quantification of the sodA mRNA is given as % sodA mRNA in wt strain without IPTG.
3.10. Overexpression of SodA in the rnc Mutant Supports Growth during Oxidative Stress
As in the case of sodA expressed from its chromosomal location (Figure 2B), there was little effect of the addition of H2O2 (1 mM) to the wt strain in mid-exponential growth expressing the basal level of SodA from Plac-sodA (Figure 7, left). In the rnc strain, H2O2 immediately stopped growth, and adding IPTG to induce SodA at the same time as the H2O2 challenge was not sufficient for maintaining growth. However, the constant overexpression of SodA from a low cell density was sufficient for maintaining growth after the H2O2 challenge in the rnc mutant (Figure 7, right). In summary, constant SodA overexpression can restore resistance to H2O2 in the rnc strain, suggesting that the RNase III-mediated positive regulation of sodA expression is required to protect against oxidative stress.
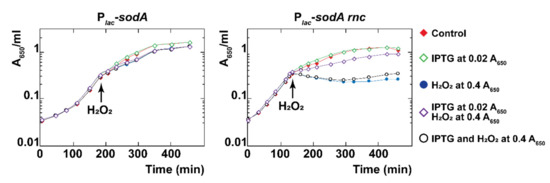
Figure 7.
Ectopic expression of sodA protects from oxidative stress in the rnc mutant. N3433 Plac-sodA and IBPC633 Plac-sodA strains containing the pBRlacIq expressing lacIq constitutively were grown to mid-log phase, and IPTG (0.1 mM) and H2O2 (1 mM) were added as indicated. Red filled diamonds show the control with no additions; empty green diamonds show IPTG added at the beginning of the growth; blue filled circles show H2O2 added at mid-log phase; empty purple diamonds show IPTG added at the beginning of the growth and H2O2 at mid-log phase; empty black circles show IPTG added together with H2O2 at mid-log phase.
4. Discussion
Recent global approaches, aimed at quantifying the importance of RNase III, have suggested that the range of RNase III targets had been considerably underestimated [29,30,31,32,33]. In this study, we demonstrate that the inactivation of RNase III increases sensitivity to heat and cold shock and to oxidative stress. We then show that the induction of rpoH during a heat shock is greatly impaired upon RNase III inactivation due, in part, to the requirement of RNase III, together with PNPase, in the stabilization of the rpoH mRNA. In addition, our results suggest that RNase III is required for the transcriptional activation of rpoH independently from the known transcriptional regulators of rpoH expression, CRP and CytR. Furthermore, we show that RNase III also positively regulates the expression of sodA at both the transcriptional and post-transcriptional levels. Significantly, we demonstrate that SodA overexpression restores growth to the rnc mutant under H2O2 stress. RpoH and SodA are, thus, new examples showing that RNase III is a positive regulator of gene expression. These new roles of RNase III are summarized in Figure 8.
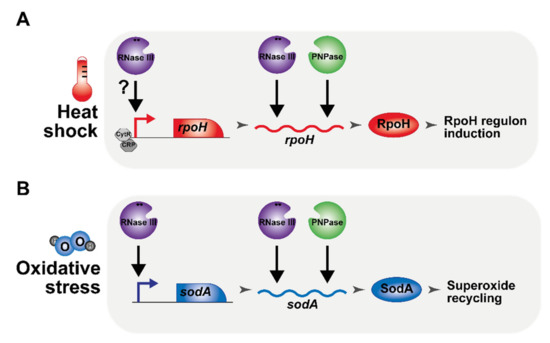
Figure 8.
Schematic regulatory roles of RNase III in the adaptation to heat and to oxidative stress. (A) RNase III (purple) and PNPase (green) are required for the induction of rpoH (red) independently from the transcription factors CRP (dark grey) and CytR (grey). (B) RNase III and PNPase are important for the expression of the superoxide dismutase SodA (blue). Positive regulatory events (direct or indirect) are represented by a black arrow.
4.1. Role of RNase III in Thermotolerance
E. coli adapts to heat shock through the transient induction of the RpoH regulon, leading to a short-term increase in HSPs in order to reduce heat shock damage (e.g., by expressing chaperones to counteract protein misfolding) [56]. Remarkably, it has been shown that RpoH and its regulon are induced under several other stress conditions, including carbohydrate starvation, hyperosmotic shock and growth in conditions of extracellular alkaline pH and in the presence of ethanol [56,64,65,66]. Furthermore, RpoH is required for long-term growth at 45 °C, and this thermotolerance remains poorly characterized. A proteomic analysis comparing E. coli strains grown in a bioreactor at 37 °C versus 45 °C revealed that among the most differentially expressed genes are the RpoH-dependent HSP DnaJ and the oxidative stress response factors SodA, AhpC and Dps [67]. Another study suggested that the early and transient peak of HSP expression plays an important role in the long-term remodeling of gene expression [68]. Other members of the RpoH regulon include ribosomal effectors, such as L31, and YbeY, an RNase involved in 16S rRNA maturation and quality control [69] that may suggest that thermotolerance also requires remodeling of the translation machinery. This hypothesis is corroborated by the fact that DnaJ, DnaK and other HSPs are instrumental in the maturation of ribosomes at 44 °C [70]. Thus, the reduced thermotolerance of the rnc mutant could result from the combined effects of the reduced rpoH induction and also from an alteration of rRNA maturation.
4.2. Importance of RNase III in the Positive Regulation of Gene Expression
RNase III is involved in mRNA destabilization, as in the case of the rpsO-pnp operon, where a dsRNA cleavage in the intergenic region enables the degradation of the pnp mRNA via RNase E [22]. Examples of destabilization by RNase III include, but are not limited to, the rnc-era operon [16], proU [35], betT [34], bdm [29], proP [36] and rng [39] mRNAs. However, RNase III was also shown to play a role in mRNA stabilization, as in the case of the mRNA adhE [25], where a dsRNA cleavage permits efficient translation and increases the mRNA stability. During acid stress, the asRNA ArrS is induced and binds to the gadE mRNA [24], triggering a maturation event by RNase III, leading to an increased translation and stability. In the case of the precursor mRNA of the T7 phage [28,71], RNase III was shown to proceed to single strand cleavages, releasing individual mRNAs with a stable hairpin in their 3′-ends. Other cases of positive regulation by RNase III have been identified, but the mechanism has not been elucidated, such as eno [26], ahpC, pflB, yajQ [32] and sucA [72] mRNAs. The importance of RNase III in the positive regulation of gene expression is further suggested by two independent transcriptomic analyses showing that 23% (120 out of 511 [30]) and 47% (87 out 187 [29]) of the genes whose expression was altered in an rnc mutant were downregulated. Furthermore, the identification of RNase III cleavage sites in vivo revealed that the RNase III targetome is even larger than previously suspected (615 targeted RNAs, including the 5′-UTR of sodA mRNA) [33]. In this work, we show that rpoH (during a heat shock) and sodA are new examples demonstrating a role of RNase III in the positive regulation of gene expression. The identification of new targets positively regulated by RNase III is limited by two aspects: first, RNase III inactivation leads to a large increase in PNPase expression, which could mask the direct role of RNase III in gene expression. Second, since our results and previous studies show that RNase III is implicated in various stress responses (heat and cold shock, oxidative stress, osmotic choc [29] and antibiotic resistance [39]), further studies aimed at identifying targets of RNase III would benefit from transcriptomic and proteomic analyses performed under stress conditions comparing strains inactivated for both RNase III and PNPase.
4.3. RNase III Is a General Stress Response Regulator and a Potential Target to Control Bacterial Virulence
In E. coli, the post-translational inhibition of RNase III by the protein YmdB is required for the resistance to osmotic choc and leads to an increase in biofilm formation through the stabilization of bdm, proV, proW, proX, proP and betT mRNAs [29,34,35,36]. On the contrary, RNase III activity is required for resistance to low concentrations of aminoglycosides via the repression of RNase G expression [39]. In addition, we show that RNase III is required for survival under heat and cold shock and for oxidative resistance. Remarkably, in Listeria monocytogenes, RNase III was similarly shown to be critical for resistance to heat and cold shock, to high and low pH, to oxidative stress and for survival in macrophages [73]. Hence, RNase III is an important factor in the cell’s ability to coordinate the cellular response toward different stress conditions.
The general stress response has been defined as the activation of a resistance response to multiple stresses after the sensing of a single stress, for which, the sigma factor RpoS is the primary regulator [73]. It is noteworthy that previous studies have reported a role for RNase III in the regulation of RpoS. Nevertheless, there were discrepancies in the outcome of this regulation in the different studies, which may result from differences in the experimental conditions and/or multiple regulatory mechanisms involving RNase III [31,74,75,76]. However, as in the case of RpoS [74], the absence of RNase III under standard conditions already affects the expression of a wide range of catabolic genes, as well as stress functions. RNase III can thus be considered as another general stress response regulator [75] and an important component of the complex mechanisms determining the ability of E. coli to adapt to ever-changing environmental conditions. Furthermore, the function of RNase III as a general stress response factor is likely to be conserved in other organisms, as demonstrated in the case of L. monocytogenes [73].
RNase III is a well-conserved protein among bacteria and was shown to be important for virulence in a wide range of pathogenic organisms, such as Staphylococcus aureus in the infection of mice [77], Salmonella enterica serovar Typhimurium [78] and Enterococcus fecalis [79] in the infection of Galleria mellonella. Hence, RNase III could represent an interesting target for therapeutic purposes. However, we want to stress that two contra-indicatory points should be considered: first RNase III inhibition in E. coli increases the sensitivity to temperature and oxidative stress but also increases resistance to osmotic choc and favors biofilm formation, which may lead to adverse outcomes that could preclude clinical uses. Second, targeting RNase III will be difficult to limit to a single species and will affect a whole range of species due to the strong conservation of RNase III in bacteria (reviewed in [5]). Thus, the use of RNase III modulators would have to be carefully assessed in a clinical setting.
5. Conclusions
We have extended our understanding of E. coli stress responses by showing that RNase III is required for the expression of RpoH after a heat shock and for the expression of SodA, thus allowing bacteria to cope with oxidative stress. Hence, RNase III is an important factor in the cell’s ability to coordinate the cellular response toward different stress conditions.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10040699/s1, Table S1: Strains, plasmids and primers, Figure S1: RNase III is required for heat shock and oxidative stress resistance, Figure S2: rpoH induction upon temperature upshift is defective in the rnc mutant at the mRNA level, Figure S3: rpoH induction upon temperature upshift is defective in the rnc mutant at the protein level, Figure S4: Effect of rnc and pnp mutations on the decay rates of rpoH and sodA mRNAs, Figure S5: RNase III inactivation reduces rpoH induction after heat shock independently from CRP and CytR, Figure S6: RNase III positively controls sodA expression at the mRNA level, Figure S7: RNase III positively controls sodA expression at the protein level, Figure S8: In vitro cleavage of sodA mRNA by RNase III and uncropped gels from Figure 4, Figure 6, Figures S3, S5 and S7. References References [80,81,82,83,84] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, methodology, validation, writing—original draft preparation, writing—review and editing, visualization, M.L. and E.H.; supervision, project administration, funding acquisition, E.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Centre National de la Recherche Scientifique (UMR8261), Université de Paris, Agence Nationale de la Recherche (asSUPYCO, ANR-12-BSV6-0007-03 to E.H.), (RIBECO ANR-18-CE43-0010 to E.H.) and the “Initiative d’Excellence” program from the French State (Grant “DYNAMO,” ANR-11-LABX-0011). ML is supported by JSPS Postdoctoral Fellowship for Research in Japan (P22709).
Acknowledgments
We are indebted to J. Plumbridge for discussions and critical reading of the manuscript. We thank C. Beloin, M. Guillier and K. Lee for providing strains and plasmids and L. Kuhn, P. Hammann and A. Maes for their help during preliminary experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hör, J.; Matera, G.; Vogel, J.; Gottesman, S.; Storz, G. Trans-Acting Small RNAs and Their Effects on Gene Expression in Escherichia coli and Salmonella enterica. EcoSal Plus 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Kavita, K.; de Mets, F.; Gottesman, S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018, 42, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Vogel, J. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018, 16, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Trinquier, A.; Durand, S.; Braun, F.; Condon, C. Regulation of RNA processing and degradation in bacteria. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194505. [Google Scholar] [CrossRef]
- Lejars, M.; Kobayashi, A.; Hajnsdorf, E. RNase III, Ribosome Biogenesis and Beyond. Microorganisms 2021, 9, 2608. [Google Scholar] [CrossRef]
- Nicholson, A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. WIREs RNA 2014, 5, 31–48. [Google Scholar] [CrossRef]
- Court, D.L.; Gan, J.; Liang, Y.H.; Shaw, G.X.; Tropea, J.E.; Costantino, N.; Waugh, D.S.; Ji, X. RNase III: Genetics and function; structure and mechanism. Annu. Rev. Genet. 2013, 47, 405–431. [Google Scholar] [CrossRef]
- Lee, D.; Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 2018, 15, 186–193. [Google Scholar] [CrossRef]
- Li, Z.; Deutscher, M.P. Exoribonucleases and Endoribonucleases. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef]
- Gegenheimer, P.; Apirion, D. Precursors to 16S and 23S ribosomal RNA from a ribonuclear III-strain of Escherichia coli contain intact RNase III processing sites. Nucleic Acids Res. 1980, 8, 1873–1891. [Google Scholar] [CrossRef]
- King, T.C.; Schlessinger, D. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J. Biol. Chem. 1983, 258, 12034–12042. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Miczak, A.; Apirion, D. Maturation of precursor 10Sa RNA in Escherichia coli is a two-step process: The first reaction is catalyzed by RNase III in presence of Mn2+. Biochimie 1990, 72, 791–802. [Google Scholar] [CrossRef]
- Srivastava, R.A.; Srivastava, N.; Apirion, D. Characterization of the RNA processing enzyme RNase III from wild type and overexpressing Escherichia coli cells in processing natural RNA substrates. Int. J. Biochem. 1992, 24, 737–749. [Google Scholar] [CrossRef]
- Robertson, H.D.; Dunn, J.J. Ribonucleic acid processing activity of Escherichia coli Ribonuclease III. J. Biol. Chem. 1975, 250, 3050–3056. [Google Scholar] [CrossRef]
- Kameyama, L.; Fernandez, L.; Court, D.L.; Guarneros, G. RNase lll activation of bacteriophage λ N synthesis. Mol. Microbiol. 1991, 5, 2953–2963. [Google Scholar] [CrossRef]
- Bardwell, J.C.; Régnier, P.; Chen, S.M.; Nakamura, Y.; Grunberg-Manago, M.; Court, D.L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989, 8, 3401–3407. [Google Scholar] [CrossRef]
- Takata, R.; Izuhara, M.; Planta, R.J. Differential degradation of the Escherichia coli polynucleotide phosphorylase mRNA. Nucleic Acids Res. 1989, 17, 7441–7451. [Google Scholar] [CrossRef]
- Portier, C.; Dondon, L.; Grunberg-Manago, M.; Régnier, P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′ end. EMBO J. 1987, 6, 2165–2170. [Google Scholar] [CrossRef]
- Barry, G.; Squires, C.; Squires, C.L. Attenuation and processing of RNA from the rplJL-rpoBC transcription unit of Escherichia coli. Proc. Natl. Acad. Sci. USA 1980, 77, 3331–3335. [Google Scholar] [CrossRef]
- Régnier, P.; Portier, C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J. Mol. Biol. 1986, 187, 23–32. [Google Scholar] [CrossRef]
- Régnier, P.; Grunberg-Manago, M. RNase III cleavages in non-coding leaders of Escherichia coli transcripts control mRNA stability and genetic expression. Biochimie 1990, 72, 825–834. [Google Scholar] [CrossRef]
- Hajnsdorf, E.; Carpousis, A.J.; Regnier, P. Nucleolytic inactivation and degradation of the RNase III processed pnp message encoding polynucleotide phosphorylase of Escherichia coli. J. Mol. Biol. 1994, 239, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Robert-Le Meur, M.; Portier, C.E. coli polynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 1992, 11, 2633–2641. [Google Scholar] [CrossRef] [PubMed]
- Aiso, T.; Kamiya, S.; Yonezawa, H.; Gamou, S. Overexpression of an antisense RNA, ArrS, increases the acid resistance of Escherichia coli. Microbiology 2014, 160, 954–961. [Google Scholar] [CrossRef]
- Aristarkhov, A.; Mikulskis, A.; Belasco, J.G.; Lin, E.C. Translation of the adhE transcript to produce ethanol dehydrogenase requires RNase III cleavage in Escherichia coli. J. Bacteriol. 1996, 178, 4327–4332. [Google Scholar] [CrossRef][Green Version]
- Lee, M.; Joo, M.; Sim, M.; Sim, S.H.; Kim, H.L.; Lee, J.; Ryu, M.; Yeom, J.H.; Hahn, Y.; Ha, N.C.; et al. The coordinated action of RNase III and RNase G controls enolase expression in response to oxygen availability in Escherichia coli. Sci. Rep. 2019, 9, 17257. [Google Scholar] [CrossRef]
- Altuvia, S.; Kornitzer, D.; Kobi, S.; Oppenheim, A.B. Functional and structural elements of the mRNA of the cIII gene of bacteriophage lambda. J. Mol. Biol. 1991, 218, 723–733. [Google Scholar] [CrossRef]
- Dunn, J.J.; Studier, F.W. T7 Early RNAs and Escherichia coli Ribosomal RNAs are Cut from Large Precursor RNAs In Vivo by Ribonuclease III. Proc. Natl. Acad. Sci. USA 1973, 70, 3296–3300. [Google Scholar] [CrossRef]
- Sim, S.-H.; Yeom, J.-H.; Shin, C.; Song, W.-S.; Shin, E.; Kim, H.-M.; Cha, C.-J.; Han, S.H.; Ha, N.-C.; Kim, S.W.; et al. Escherichia coli ribonuclease III activity is downregulated by osmotic stress: Consequences for the degradation of bdm mRNA in biofilm formation. Mol. Microbiol. 2010, 75, 413–425. [Google Scholar] [CrossRef]
- Stead, M.B.; Marshburn, S.; Mohanty, B.K.; Mitra, J.; Castillo, L.P.; Ray, D.; van Bakel, H.; Hughes, T.R.; Kushner, S.R. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2011, 39, 3188–3203. [Google Scholar] [CrossRef]
- Huang, L.; Deighan, P.; Jin, J.; Li, Y.; Cheung, H.-C.; Lee, E.; Mo, S.S.; Hoover, H.; Abubucker, S.; Finkel, N.; et al. Tombusvirus p19 Captures RNase III-Cleaved Double-Stranded RNAs Formed by Overlapping Sense and Antisense Transcripts in Escherichia coli. mBio 2020, 11, e00485-20. [Google Scholar] [CrossRef] [PubMed]
- Gordon, G.C.; Cameron, J.C.; Pfleger, B.F. RNA Sequencing Identifies New RNase III Cleavage Sites in Escherichia coli and Reveals Increased Regulation of mRNA. mBio 2017, 8, e00128-17. [Google Scholar] [CrossRef] [PubMed]
- Altuvia, Y.; Bar, A.; Reiss, N.; Karavani, E.; Argaman, L.; Margalit, H. In vivo cleavage rules and target repertoire of RNase III in Escherichia coli. Nucleic Acids Res. 2018, 46, 10380–10394. [Google Scholar] [CrossRef] [PubMed]
- Sim, M.; Lim, B.; Sim, S.H.; Kim, D.; Jung, E.; Lee, Y.; Lee, K. Two tandem RNase III cleavage sites determine betT mRNA stability in response to osmotic stress in Escherichia coli. PLoS ONE 2014, 9, e100520. [Google Scholar] [CrossRef]
- Kavalchuk, K.; Madhusudan, S.; Schnetz, K. RNase III initiates rapid degradation of proU mRNA upon hypo-osmotic stress in Escherichia coli. RNA Biol. 2012, 9, 98–109. [Google Scholar] [CrossRef][Green Version]
- Lim, B.; Lee, K. Stability of the Osmoregulated Promoter-Derived proP mRNA Is Posttranscriptionally Regulated by RNase III in Escherichia coli. J. Bacteriol. 2015, 197, 1297–1305. [Google Scholar] [CrossRef]
- Kim, K.S.; Manasherob, R.; Cohen, S.N. YmdB: A stress-responsive ribonuclease-binding regulator of E. coli RNase III activity. Genes Dev. 2008, 22, 3497–3508. [Google Scholar] [CrossRef]
- Apirion, D.; Neil, J.; Watson, N. Consequences of losing ribonuclease III on the Escherichia coli cell. Mol. Gen. Genet. 1976, 144, 185–190. [Google Scholar] [CrossRef]
- Song, W.; Kim, Y.-H.; Sim, S.-H.; Hwang, S.; Lee, J.-H.; Lee, Y.; Bae, J.; Hwang, J.; Lee, K. Antibiotic stress-induced modulation of the endoribonucleolytic activity of RNase III and RNase G confers resistance to aminoglycoside antibiotics in Escherichia coli. Nucleic Acids Res. 2014, 42, 4669–4681. [Google Scholar] [CrossRef]
- Apirion, D.; Watson, N. Ribonuclease III is involved in motility of Escherichia coli. J. Bacteriol. 1978, 133, 1543–1545. [Google Scholar] [CrossRef]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martínez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2016, 45, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Fernandez, L.; Kameyama, L.; Inada, T.; Nakamura, Y.; Pappas, A.; Court, D.L. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III-the effect of dsRNA binding on gene expression. Mol. Microbiol. 1998, 28, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Korea, C.-G.; Badouraly, R.; Prevost, M.-C.; Ghigo, J.-M.; Beloin, C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone–usher fimbriae with distinct surface specificities. Environ. Microbiol. 2010, 12, 1957–1977. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006, 59, 231–247. [Google Scholar] [CrossRef]
- Braun, F.; Hajnsdorf, E.; Regnier, P. Polynucleotide phosphorylase is required for the rapid degradation of the RNase E-processed rpsO mRNA of Escherichia coli devoid of its 3′ hairpin. Mol. Microbiol. 1996, 19, 997–1005. [Google Scholar] [CrossRef]
- Hajnsdorf, E.; Regnier, P.E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol. 1999, 286, 1033–1043. [Google Scholar] [CrossRef]
- Hajnsdorf, E.; Regnier, P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. USA 2000, 97, 1501–1505. [Google Scholar] [CrossRef]
- Folichon, M.; Allemand, F.; Régnier, P.; Hajnsdorf, E. Stimulation of poly(A) synthesis by E. coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005, 272, 454–463. [Google Scholar] [CrossRef]
- Fontaine, F.; Gasiorowski, E.; Gracia, C.; Ballouche, M.; Caillet, J.; Marchais, A.; Hajnsdorf, E. The small RNA SraG participates in PNPase homeostasis. RNA 2016, 22, 1560–1573. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Awano, N.; Inouye, M.; Phadtare, S. RNase Activity of Polynucleotide Phosphorylase Is Critical at Low Temperature in Escherichia coli and Is Complemented by RNase II. J. Bacteriol. 2008, 190, 5924–5933. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, S.; Alfonso-Prieto, M.; Carnevale, V.; Redhu, S.K.; Klein, M.L.; Nicholson, A.W. Combined computational and experimental analysis of a complex of ribonuclease III and the regulatory macrodomain protein, YmdB. Proteins 2015, 83, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Oxidative Stress. EcoSal Plus 2009, 3. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, G.; Blankschien, M.; Herman, C.; Gross, C.A.; Rhodius, V.A. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006, 20, 1776–1789. [Google Scholar] [CrossRef]
- Jenkins, D.E.; Auger, E.A.; Matin, A. Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J. Bacteriol. 1991, 173, 1992–1996. [Google Scholar] [CrossRef]
- Guo, M.S.; Updegrove, T.B.; Gogol, E.B.; Shabalina, S.A.; Gross, C.A.; Storz, G. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014, 28, 1620–1634. [Google Scholar] [CrossRef]
- Morita, M.T.; Tanaka, Y.; Kodama, T.S.; Kyogoku, Y.; Yanagi, H.; Yura, T. Translational induction of heat shock transcription factor sigma32: Evidence for a built-in RNA thermosensor. Genes Dev. 1999, 13, 655–665. [Google Scholar] [CrossRef]
- Morita, M.; Kanemori, M.; Yanagi, H.; Yura, T. Heat-Induced Synthesis of ς32 in Escherichia coli: Structural and Functional Dissection of rpoH mRNA Secondary Structure. J. Bacteriol. 1999, 181, 401–410. [Google Scholar] [CrossRef]
- Arsène, F.; Tomoyasu, T.; Bukau, B. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 2000, 55, 3–9. [Google Scholar] [CrossRef]
- Erickson, J.W.; Vaughn, V.; Walter, W.A.; Neidhardt, F.C.; Gross, C.A. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1987, 1, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Straus, D.B.; Walter, W.A.; Gross, C.A. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature 1987, 329, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Afonyushkin, T.; Večerek, B.; Moll, I.; Bläsi, U.; Kaberdin, V.R. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005, 33, 1678–1689. [Google Scholar] [CrossRef]
- Bianchi, A.A.; Baneyx, F. Hyperosmotic shock induces the σ32 and σE stress regulons of Escherichia coli. Mol. Microbiol. 1999, 34, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Taglicht, D.; Padan, E.; Oppenheim, A.B.; Schuldiner, S. An alkaline shift induces the heat shock response in Escherichia coli. J. Bacteriol. 1987, 169, 885–887. [Google Scholar] [CrossRef] [PubMed]
- VanBogelen, R.A.; Kelley, P.M.; Neidhardt, F.C. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J. Bacteriol. 1987, 169, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Lüders, S.; Fallet, C.; Franco-Lara, E. Proteome analysis of the Escherichia coli heat shock response under steady-state conditions. Proteome Sci. 2009, 7, 36. [Google Scholar] [CrossRef]
- Riehle, M.M.; Bennett, A.F.; Long, A.D. Changes in Gene Expression Following High-Temperature Adaptation in Experimentally Evolved Populations of E. coli. Physiol. Biochem. Zool. 2005, 78, 299–315. [Google Scholar] [CrossRef][Green Version]
- Rasouly, A.; Ron, E.Z. Interplay between the heat shock response and translation in Escherichia coli. Res. Microbiol. 2009, 160, 288–296. [Google Scholar] [CrossRef]
- Al Refaii, A.; Alix, J.-H. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol. Microbiol. 2009, 71, 748–762. [Google Scholar] [CrossRef]
- Mayer, J.E.; Schweiger, M. RNase III is positively regulated by T7 protein kinase. J. Biol. Chem. 1983, 258, 5340–5343. [Google Scholar] [CrossRef]
- Kim, M.; Kim, M.; Kim, K.S. YmdB-mediated down-regulation of sucA inhibits biofilm formation and induces apramycin susceptibility in Escherichia coli. Biochem. Biophys. Res. Commun. 2017, 483, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiao, M.; Meng, Q.; Qiao, J.; Wu, Y.; Guo, J.; Wang, X.; Li, J.; Zhang, X.; Cai, X. Listeria monocytogenes in RNase III rncS. Kafkas Univ. Vet. Fak. Derg. 2019, 26, 269–277. [Google Scholar] [CrossRef]
- Freire, P.; Amaral, J.D.; Santos, J.M.; Arraiano, C.M. Adaptation to carbon starvation: RNase III ensures normal expression levels of bolA1p mRNA and σS. Biochimie 2006, 88, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Basineni, S.R.; Madhugiri, R.; Kolmsee, T.; Hengge, R.; Klug, G. The influence of Hfq and ribonucleases on the stability of the small non-coding RNA OxyS and its target rpoS in E. coli is growth phase dependent. RNA Biol. 2009, 6, 584–594. [Google Scholar] [CrossRef]
- Resch, A.; Afonyushkin, T.; Lombo, T.B.; McDowall, K.J.; Bläsi, U.; Kaberdin, V.R. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 2008, 14, 454–459. [Google Scholar] [CrossRef][Green Version]
- Lioliou, E.; Sharma, C.M.; Caldelari, I.; Helfer, A.C.; Fechter, P.; Vandenesch, F.; Vogel, J.; Romby, P. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012, 8, e1002782. [Google Scholar] [CrossRef]
- Viegas, S.C.; Mil-Homens, D.; Fialho, A.M.; Arraiano, C.M. The Virulence of Salmonella enterica Serovar Typhimurium in the Insect Model Galleria mellonella Is Impaired by Mutations in RNase E and RNase III. Appl. Environ. Microbiol. 2013, 79, 6124–6133. [Google Scholar] [CrossRef]
- Salze, M.; Muller, C.; Bernay, B.; Hartke, A.; Clamens, T.; Lesouhaitier, O.; Rincé, A. Study of key RNA metabolism proteins in Enterococcus faecalis. RNA Biol. 2020, 17, 794–804. [Google Scholar] [CrossRef]
- Goldblum, K.; Apirion, D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J. Bacteriol. 1981, 146, 128–132. [Google Scholar]
- Regnier, P.; Hajnsdorf, E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3' stabilizing stem and loop structure. J. Mol. Biol. 1991, 217, 283–292. [Google Scholar] [PubMed]
- Reuven, N.B.; Deutscher, M.P. Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. Faseb J. 1993, 7, 143–148. [Google Scholar] [PubMed]
- Korshunov, S.; Imlay, J.A. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol. 2010, 75, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).