The Escherichia coli Amino Acid Uptake Protein CycA: Regulation of Its Synthesis and Practical Application in l-Isoleucine Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Media
2.2. DNA Manipulation
2.3. Construction of Strains
2.4. Growth Curves
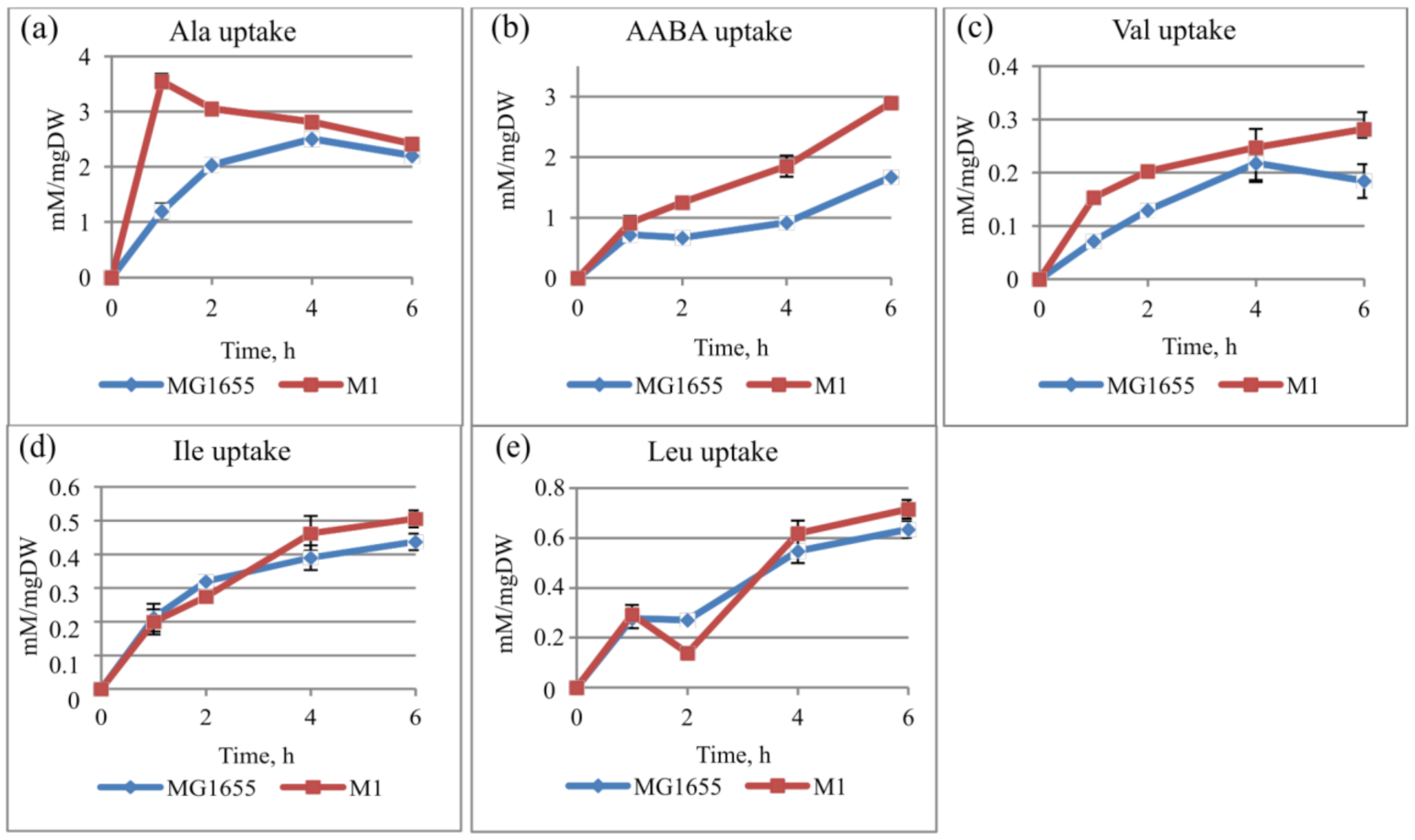
2.5. Amino Acid Uptake Assay
2.6. β-Galactosidase Activity Assay
2.7. Electromobility Shift Assay (EMSA)
2.8. CRP Extraction and Purification
2.9. Test-Tube Cultivation Conditions
3. Results
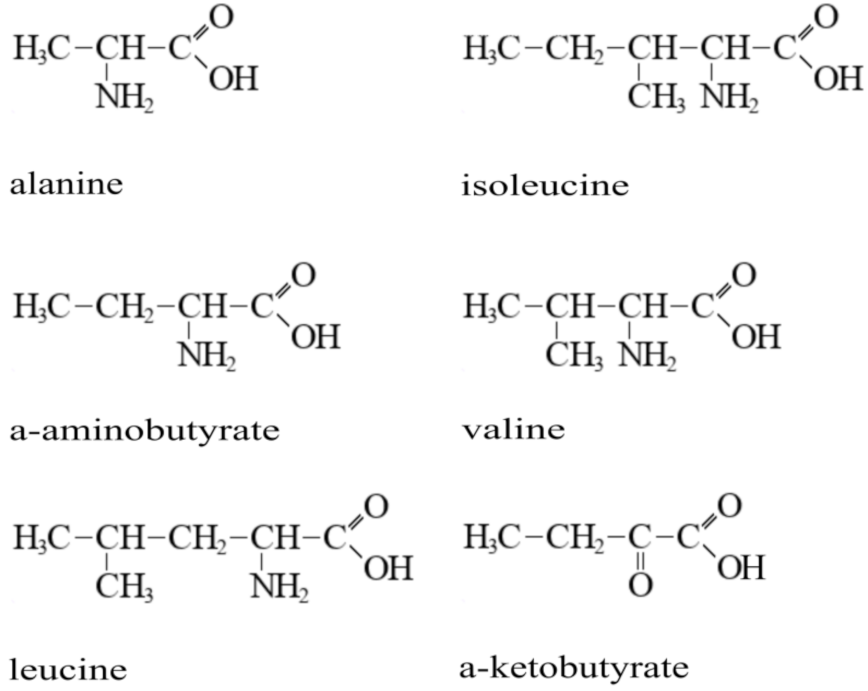
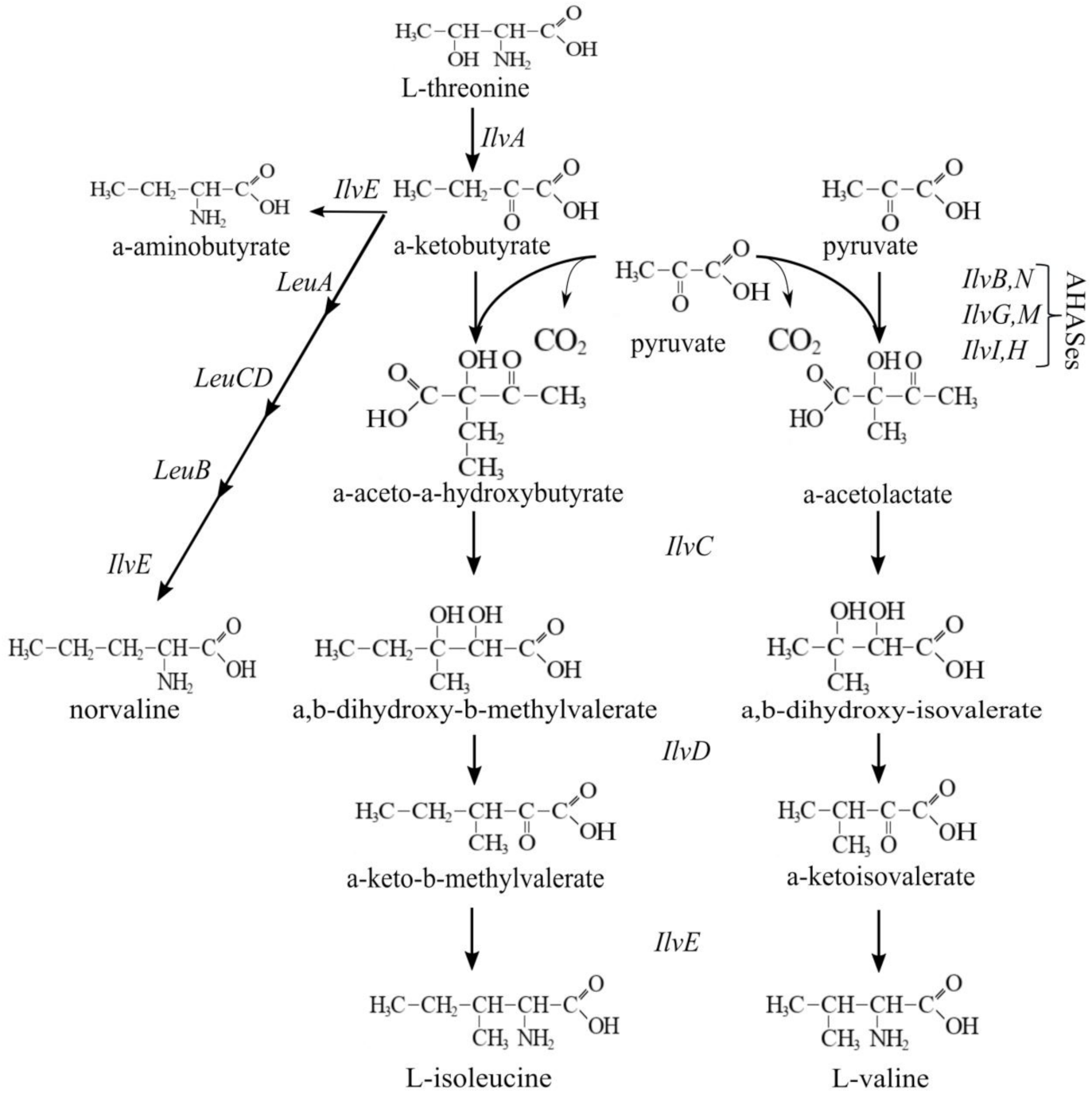
3.1. Novel Substrates of the CycA Transporter Include Branched-Chain Amino Acids
3.2. CycA Gene Expression Is Induced by Addition of Its Substrate(s) into Culture Medium
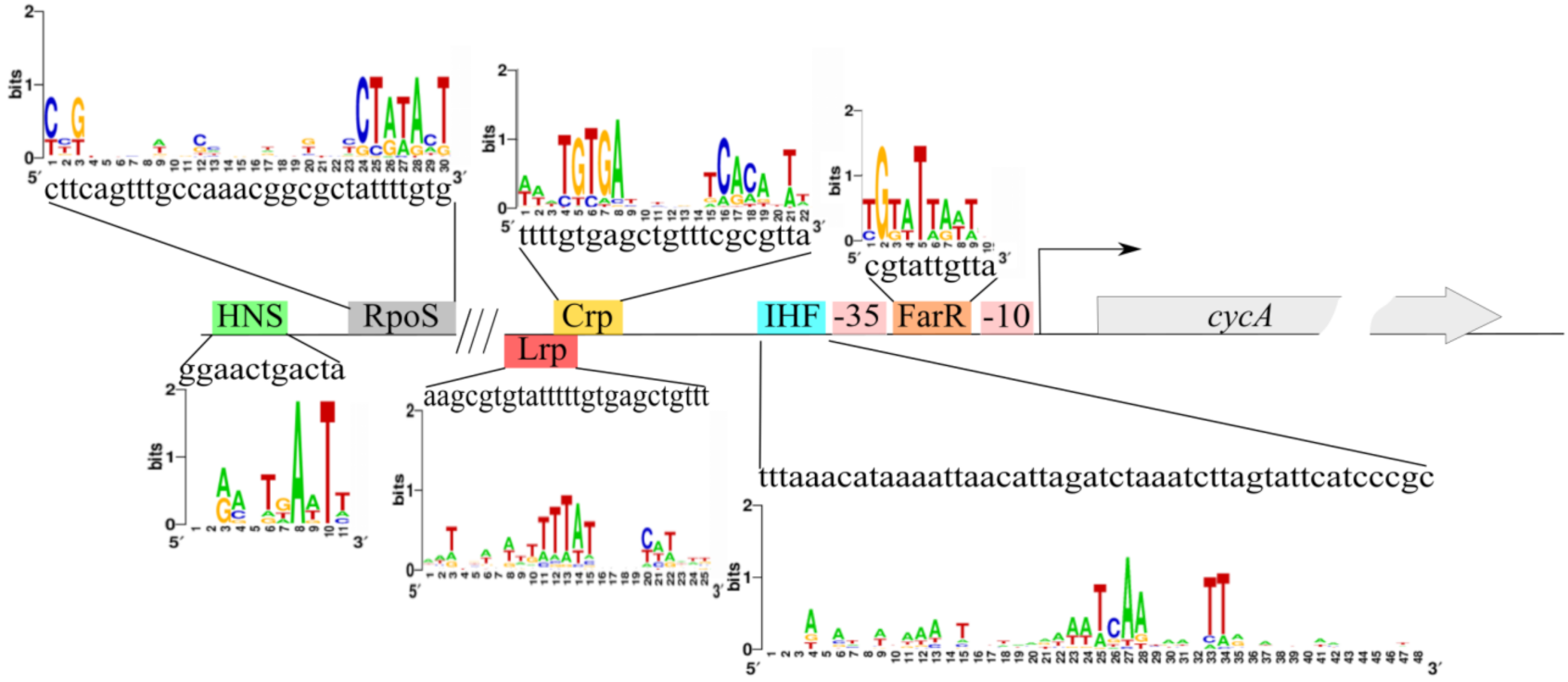
3.3. Prediction and Verification of Potential cycA Gene Expression Regulators
3.4. Crp Role in Regulation of cycA Gene Expression
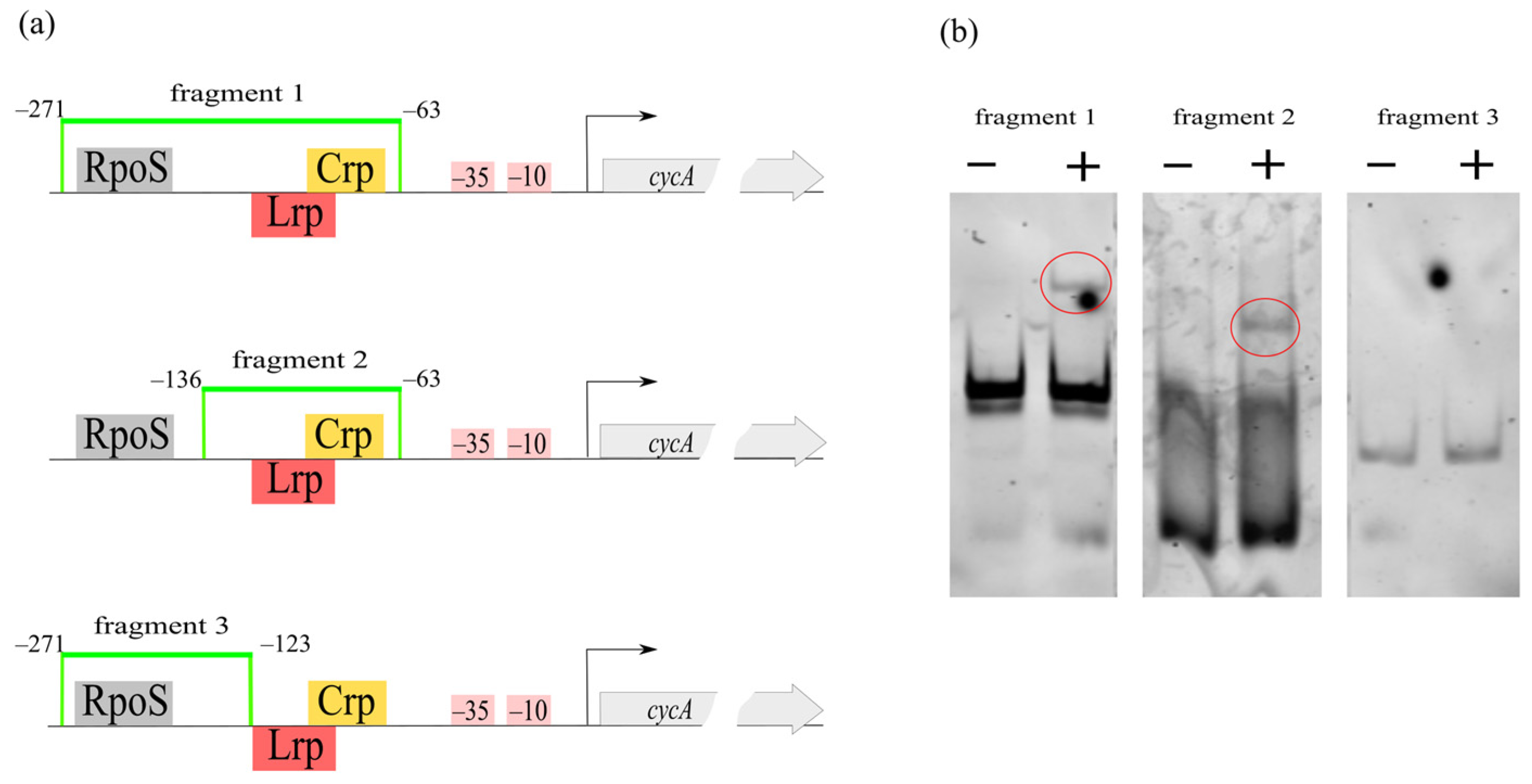
3.5. Binding of Protein Factors with the cycA Regulatory Region
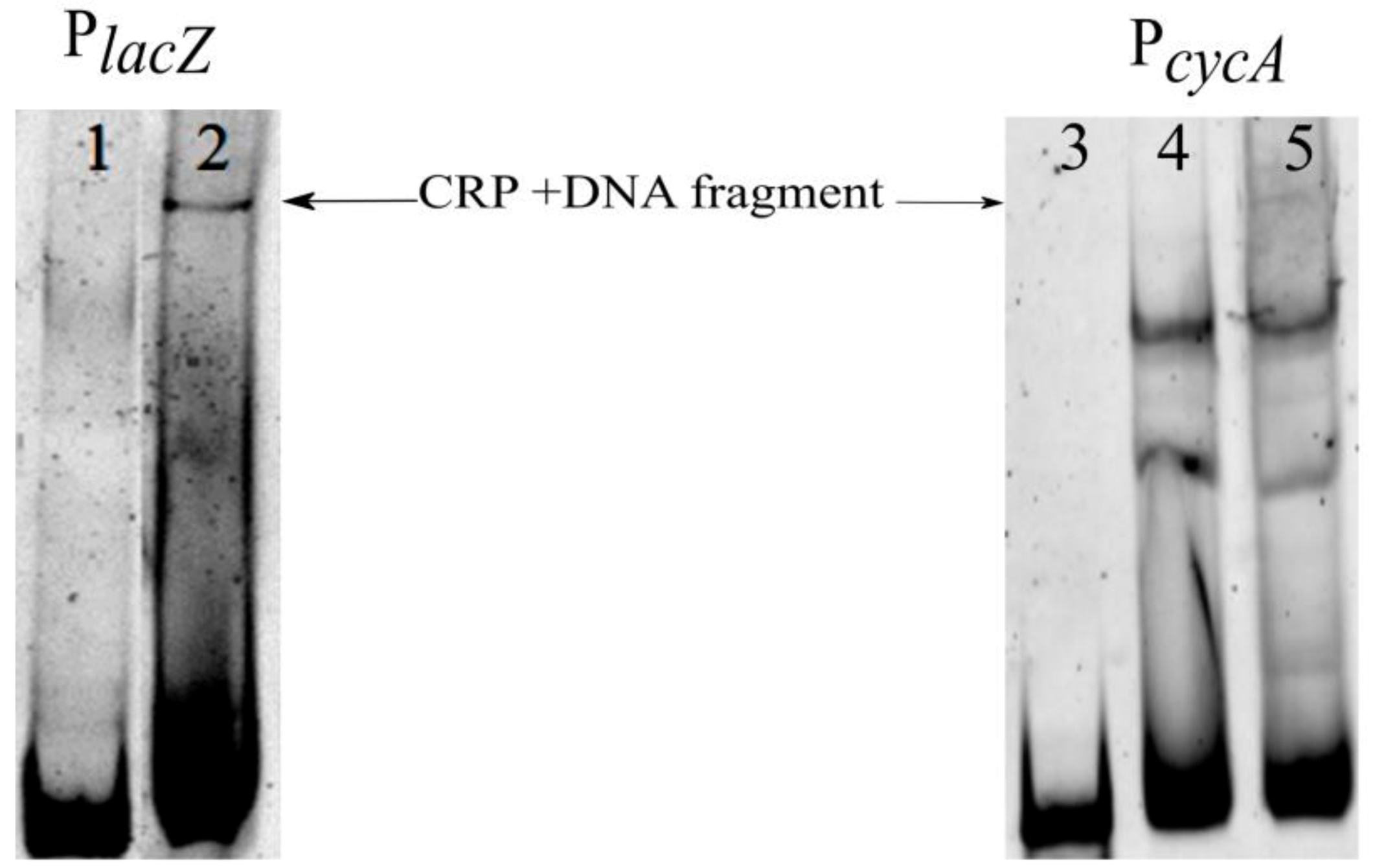
3.6. Crp Binds cycA Gene Regulatory Region
3.7. Overexpression of cycA Decreases Accumulation of Impurities by an l-Ile-Producing Strain
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, J.H.; Lee, S.Y. Fermentative Production of Branched Chain Amino Acids: A Focus on Metabolic Engineering. Appl. Microbiol. Biotechnol. 2010, 85, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; Shimizu, H. Recent Advances in Amino Acid Production by Microbial Cells. Curr. Opin. Biotechnol. 2016, 42, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Nielsen, J. Advancing Metabolic Engineering through Systems Biology of Industrial Microorganisms. Curr. Opin. Biotechnol. 2015, 36, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Contador, C.A.; Rizk, M.L.; Asenjo, J.A.; Liao, J.C. Ensemble Modeling for Strain Development of L-Lysine-Producing Escherichia coli. Metab. Eng. 2009, 11, 221–233. [Google Scholar] [CrossRef]
- Katashkina, J.I.; Hara, Y.; Golubeva, L.I.; Andreeva, I.G.; Kuvaeva, T.M.; Mashko, S.V. Use of the λ Red-recombineering method for genetic engineering of Pantoea ananatis. BMC Mol. Biol. 2009, 10, 34. [Google Scholar] [CrossRef]
- Hara, Y.; Kadotani, N.; Izui, H.; Katashkina, J.I.; Kuvaeva, T.M.; Andreeva, I.G.; Golubeva, L.I.; Malko, D.B.; Makeev, V.J.; Mashko, S.V.; et al. The Complete Genome Sequence of Pantoea ananatis AJ13355, an Organism with Great Biotechnological Potential. Appl. Microbiol. Biotechnol. 2012, 93, 331–341. [Google Scholar] [CrossRef]
- Rodriguez, A.; Martnez, J.A.; Flores, N.; Escalante, A.; Gosset, G.; Bolivar, F. Engineering Escherichia coli to Overproduce Aromatic Amino Acids and Derived Compounds. Microb. Cell Factories 2014, 13, 126. [Google Scholar] [CrossRef]
- Ginesy, M.; Belotserkovsky, J.; Enman, J.; Isaksson, L.; Rova, U. Metabolic Engineering of Escherichia coli for Enhanced Arginine Biosynthesis. Microb. Cell Factories 2015, 14, 29. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, S.Y. Towards Systems Metabolic Engineering of Microorganisms for Amino Acid Production. Curr. Opin. Biotechnol. 2008, 19, 454–460. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, Q.; Xu, Q.; Zhang, C.; Li, Y.; Fan, X.; Xie, X.; Chen, N. Systems Metabolic Engineering Strategies for the Production of Amino Acids. Synth. Syst. Biotechnol. 2017, 2, 87–96. [Google Scholar] [CrossRef]
- Shimizu, K. Metabolic Regulation of a Bacterial Cell System with Emphasis on Escherichia coli Metabolism. Int. Sch. Res. Not. 2013, 2013, 1–47. [Google Scholar]
- Burkovski, A.; Krämer, R. Bacterial Amino Acid Transport Proteins: Occurrence, Functions, and Significance for Biotechnological Applications. Appl. Microbiol. Biotechnol. 2002, 58, 265–274. [Google Scholar] [PubMed]
- Jack, D.L.; Paulsen, I.T.; Saier, M.H. The Amino Acid/polyamine/organocation (APC) Superfamily of Transporters Specific for Amino Acids, Polyamines and Organocations. Microbiology 2000, 146 Pt 8, 1797–1814. [Google Scholar] [CrossRef]
- Liu, Q.; Liang, Y.; Zhang, Y.; Shang, X.; Liu, S.; Wen, J.; Wen, T. YjeH Is a Novel Exporter of L-Methionine and Branched-Chain Amino Acids in Escherichia coli. Appl. Environ. Microbiol. 2015, 81, 7753–7766. [Google Scholar] [CrossRef] [PubMed]
- Steffes, C.; Ellis, J.; Wu, J.; Rosen, B.P. The lysP Gene Encodes the Lysine-Specific Permease. J. Bacteriol. 1992, 174, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Hersh, B.M.; Farooq, F.T.; Barstad, D.N.; Blankenhorn, D.L.; Slonczewski, J.L. A Glutamate-Dependent Acid Resistance Gene in Escherichia coli. J. Bacteriol. 1996, 178, 3978–3981. [Google Scholar] [CrossRef] [PubMed]
- Kutukova, E.A.; Livshits, V.A.; Altman, I.P.; Ptitsyn, L.R.; Zyiatdinov, M.H.; Tokmakova, I.L.; Zakataeva, N.P. The yeaS (leuE) Gene of Escherichia coli Encodes an Exporter of Leucine, and the Lrp Protein Regulates Its Expression. FEBS Lett. 2005, 579, 4629–4634. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Shi, T.; Sun, G.; Gao, N.; Zhao, X.; Guo, X.; Ni, X.; Yuan, Q.; Feng, J.; et al. CRISPR-Assisted Rational Flux-Tuning and Arrayed CRISPRi Screening of an L-Proline Exporter for L-Proline Hyperproduction. Nat. Commun. 2022, 13, 891. [Google Scholar] [CrossRef]
- Doroshenko, V.; Airich, L.; Vitushkina, M.; Kolokolova, A.; Livshits, V.; Mashko, S. YddG from Escherichia coli Promotes Export of Aromatic Amino Acids. FEMS Microbiol. Lett. 2007, 275, 312–318. [Google Scholar] [CrossRef]
- Yamada, S.; Awano, N.; Inubushi, K.; Maeda, E.; Nakamori, S.; Nishino, K.; Yamaguchi, A.; Takagi, H. Effect of Drug Transporter Genes on Cysteine Export and Overproduction in Escherichia coli. Appl. Environ. Microbiol. 2006, 72, 4735–4742. [Google Scholar] [CrossRef]
- Brown, K. Formation of Aromatic Amino Acid Pools in Escherichia coli K-12. J. Bacteriol. 1970, 104, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Hasegawa, A.; Matsubara, K.; Date, T.; Okada, T.; Kiritani, K. Cloning and Nucleotide Sequence of the brnQ Gene, the Structural Gene for a Membrane-Associated Component of the LIV-II Transport System for Branched-Chain Amino Acids in Salmonella Typhimurium. Jpn. J. Genet. 1988, 63, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.; Yang, Y.; Hong, J.; Lum, D. Cloning and Sequencing of a Gene Encoding a Glutamate and Aspartate Carrier of Escherichia coli K-12. J. Bacteriol. 1990, 172, 3214–3220. [Google Scholar] [CrossRef]
- Pi, J.; Wookey, P.J.; Pittard, A. Cloning and Sequencing of the pheP Gene, Which Encodes the Phenylalanine-Specific Transport System of Escherichia coli. J. Bacteriol. 1991, 173, 3622–3629. [Google Scholar] [CrossRef][Green Version]
- Piperno, J.R.; Oxender, D.L. Amino Acid Transport Systems in Escherichia coli K12. J. Biol. Chem. 1968, 243, 5914–5920. [Google Scholar] [CrossRef]
- Robbins, J.C.; Oxender, D.L. Transport Systems for Alanine, Serine, and Glycine in Escherichia coli K-12. J. Bacteriol. 1973, 116, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Reitz, R.H.; Slade, H.D.; Neuhaus, F.C. The Biochemical Mechanisms of Resistance by Streptococci to the Antibiotics D-Cycloserine and O-Carbamyl-D-Serine. Biochemistry 1967, 6, 2561–2570. [Google Scholar] [CrossRef]
- Cosloy, S.D. D-Serine Transport System in Escherichia coli K-12. J. Bacteriol. 1973, 114, 679–684. [Google Scholar] [CrossRef]
- Wargel, R.J.; Shadur, C.A.; Neuhaus, F.C. Mechanism of D-Cycloserine Action: Transport Systems for D-Alanine, D-Cycloserine, L-Alanine, and Glycine. J. Bacteriol. 1970, 103, 778–788. [Google Scholar] [CrossRef]
- Schneider, F.; Krämer, R.; Burkovski, A. Identification and Characterization of the Main Beta-Alanine Uptake System in Escherichia coli. Appl. Microbiol. Biotechnol. 2004, 65, 576–582. [Google Scholar] [CrossRef]
- Saier, M.H. Families of Transmembrane Transporters Selective for Amino Acids and Their Derivatives. Microbiology 2000, 146 Pt 8, 1775–1795. [Google Scholar] [PubMed]
- Pulvermacher, S.C.; Stauffer, L.T.; Stauffer, G.V. Role of the sRNA GcvB in Regulation of cycA in Escherichia coli. Microbiology 2009, 155, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.-K.; Barrett, C.L.; Knight, E.M.; Park, Y.S.; Palsson, B.Ø. Genome-Scale Reconstruction of the Lrp Regulatory Network in Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 19462–19467. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Constantinidou, C.; Hobman, J.L.; Minchin, S.D. Identification of the CRP Regulon Using in Vitro and in Vivo Transcriptional Profiling. Nucleic Acids Res. 2004, 32, 5874–5893. [Google Scholar] [CrossRef]
- Franchini, A.G.; Ihssen, J.; Egli, T. Effect of Global Regulators RpoS and Cyclic-AMP/CRP on the Catabolome and Transcriptome of Escherichia coli K12 during Carbon-and Energy-Limited Growth. PLoS ONE 2015, 10, e0133793. [Google Scholar] [CrossRef][Green Version]
- Aquino, P.; Honda, B.; Jaini, S.; Lyubetskaya, A.; Hosur, K.; Chiu, J.G.; Ekladious, I.; Hu, D.; Jin, L.; Sayeg, M.K.; et al. Coordinated Regulation of Acid Resistance in Escherichia coli. BMC Syst. Biol. 2017, 11, 1. [Google Scholar] [CrossRef]
- Federowicz, S.; Kim, D.; Ebrahim, A.; Lerman, J.; Nagarajan, H.; Cho, B.; Zengler, K.; Palsson, B. Determining the Control Circuitry of Redox Metabolism at the Genome-Scale. PLoS Genet. 2014, 10, e1004264. [Google Scholar] [CrossRef]
- Sambrook, J.R.D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Livshits, V.A.; Vitushkina, M.V.; Mashko, S.V.; Doroshenko, V.G.; Biryukova, I.V.; Katashkina, Z.I.; Skorokhodova, A.Y.; Belareva, A.V. Process for Producing L-Amino Acid Using Escherichia. EP1449918 B1, 21 November 2002. [Google Scholar]
- Geraskina, N.V.; Sycheva, E.V.; Samsonov, V.V.; Eremina, N.S.; Hook, C.D.; Serebrianyi, V.A.; Stoynova, N.V. Engineering Escherichia coli for Autoinducible Production of L-Valine: An Example of an Artificial Positive Feedback Loop in Amino Acid Biosynthesis. PLoS ONE 2019, 14, e0215777. [Google Scholar] [CrossRef]
- Brikun, I.; Suziedelis, K.; Stemmann, O.; Zhong, R.; Alikhanian, L.; Linkova, E.; Mironov, A.; Berg, D.E. Analysis of CRP-CytR Interactions at the Escherichia coli Udp Promoter. J. Bacteriol. 1996, 178, 1614–1622. [Google Scholar] [CrossRef]
- Livshits, V.A.; Doroshenko, V.G.; Mashko, S.V.; Akhverdian, V.Z.; Kozlov, Y.I. Amino Acid Producing Strains Belonging to the Genus Escherichia and Method for Producing Amino Acid. EP1318196 B1, 20 April 2001. [Google Scholar]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Katashkina, J.; Skorokhodova, A.Y.; Zimenkov, D.; Gulevich, A.Y.; Minaeva, N.; Doroshenko, V.; Biryukova, I.; Mashko, S. Tuning the Expression Level of a Gene Located on a Bacterial Chromosome. Mol. Biol. 2005, 39, 719–726. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.C.; Costantino, N.; Court, D.L. E. Coli Genome Manipulation by P1 Transduction. Curr. Protoc. Mol. Biol. 2007, 79, 1–17. [Google Scholar] [CrossRef] [PubMed]
- LaRossa, R.A.; Van Dyk, T.K.; Smulski, D.R. Toxic Accumulation of Alpha-Ketobutyrate Caused by Inhibition of the Branched-Chain Amino Acid Biosynthetic Enzyme Acetolactate Synthase in Salmonella Typhimurium. J. Bacteriol. 1987, 169, 1372–1378. [Google Scholar] [CrossRef]
- Miller, J. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Hellman, L.M.; Fried, M.G. Electrophoretic Mobility Shift Assay (EMSA) for Detecting Protein-Nucleic Acid Interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- Aiba, H.; Fujimoto, S.; Ozaki, N. Molecular Cloning and Nucleotide Sequencing of the Gene for E. Coli cAMP Receptor Protein. Nucleic Acids Res. 1982, 10, 1345–1361. [Google Scholar] [CrossRef]
- Wickstrum, J.R.; Egan, S.M. Ni+-Affinity Purification of Untagged cAMP Receptor Protein. Biotechniques 2002, 33, 728–730. [Google Scholar] [CrossRef]
- Rabinovitz, M.; Finkleman, A.; Reagan, R.L.; Breitman, T.R. Amino Acid Antagonist Death in Escherichia coli. J. Bacteriol. 1969, 99, 336–338. [Google Scholar] [CrossRef]
- Hart, B.R.; Blumenthal, R.M. Unexpected Coregulator Range for the Global Regulator Lrp of Escherichia coli and Proteus Mirabilis. J. Bacteriol. 2011, 193, 1054–1064. [Google Scholar] [CrossRef]
- Robison, K.; Church, G. Dpinteract: A Database on Dna-Protein Interactions. 1994. Available online: http://arep.med.harvard.edu/dpinteract/ (accessed on 18 October 2021).
- Mironov, A.A.; Alexandrov, N.; Bogodarova, N.Y.; Grigorjev, A.; Lebedev, V.; Lunovskaya, L.; Truchan, M.; Pevzner, P.A. DNASUN: A Package of Computer Programs for the Biotechnology Laboratory. Bioinformatics 1995, 11, 331–335. [Google Scholar] [CrossRef]
- Updegrove, T.B.; Zhang, A.; Storz, G. Hfq: The Flexible RNA Matchmaker. Curr. Opin. Microbiol. 2016, 30, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-H.; Hsu, C.-C.; Yang, C.-D.; Ju, Y.-W.; Chen, Y.-P.; Tseng, C.-P. Negative Effect of Glucose on ompA mRNA Stability: A Potential Role of Cyclic AMP in the Repression of Hfq in Escherichia coli. J. Bacteriol. 2011, 193, 5833–5840. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reznikoff, W. Catabolite Gene Activator Protein Activation of Lac Transcription. J. Bacteriol. 1992, 174, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, T.; Katayama, T.; Suzuki, H.; Kumagai, H. Identification of the LIV-I/LS System as the Third Phenylalanine Transporter in Escherichia coli K-12. J. Bacteriol. 2004, 186, 343–350. [Google Scholar] [CrossRef]
- Anderson, J.J.; Oxender, D.L. Genetic Separation of High- and Low-Affinity Transport Systems for Branched-Chain Amino Acids in Escherichia coli K-12. J. Bacteriol. 1978, 136, 168–174. [Google Scholar] [CrossRef]
- Zimmer, D.P.; Soupene, E.; Lee, H.L.; Wendisch, V.F.; Khodursky, A.B.; Peter, B.J.; Bender, R.A.; Kustu, S. Nitrogen Regulatory Protein C-Controlled Genes of Escherichia coli: Scavenging as a Defense against Nitrogen Limitation. Proc. Natl. Acad. Sci. USA 2000, 97, 14674–14679. [Google Scholar] [CrossRef]
- Alvarez, A.F.; Georgellis, D. In Vitro and in Vivo Analysis of the ArcB/A Redox Signaling Pathway. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 471, pp. 205–228. [Google Scholar]
- Kroner, G.; Wolfe, M.; Freddolino, P. Escherichia coli Lrp Regulates One-Third of the Genome via Direct, Cooperative, and Indirect Routes. J. Bacteriol. 2019, 201, e00411-18. [Google Scholar] [CrossRef]
- Calvo, J.M.; Matthews, R.G. The Leucine-Responsive Regulatory Protein, a Global Regulator of Metabolism in Escherichia coli. Microbiol. Rev. 1994, 58, 466–490. [Google Scholar] [CrossRef]
- Ghrist, A.C.; Stauffer, G.V. The Escherichia coli Glycine Transport System and Its Role in the Regulation of the Glycine Cleavage Enzyme System. Microbiology 1995, 141 Pt 1, 133–140. [Google Scholar] [CrossRef]
- Busby, S.; Ebright, R.H. Transcription Activation by Catabolite Activator Protein (CAP). J. Mol. Biol. 1999, 293, 199–213. [Google Scholar] [CrossRef]
- Zampieri, M.; Hörl, M.; Hotz, F.; Müller, N.F.; Sauer, U. Regulatory Mechanisms Underlying Coordination of Amino Acid and Glucose Catabolism in Escherichia coli. Nat. Commun. 2019, 10, 3354. [Google Scholar] [CrossRef] [PubMed]





| Strain or Plasmid | Description | Reference or Source |
|---|---|---|
| MG1655 | Escherichia coli K12, wild-type | VKPM a B6195 |
| BW25113 cat-PL-yddG | E. coli K12, cat-PL-yddG | [39] |
| M1 | MG1655 cat-PL-cycA | This work |
| M2 | MG1655 cat-PcycA-lacZ | This work |
| K12 2Δ PL-ilvBNfbr | E. coli K12 ∆ilvGM ∆ilvIH PL-ilvBNfbr | [40] |
| M3 | K12 2Δ PL-ilvBNfbr cat-PcycA-lacZ | This work |
| BW25113 ΔgcvB | E. coli K12, ΔgcvB | KEIO collection b |
| M4 | BW25113 ΔgcvB cat-PcycA-lacZ | This work |
| BW25113 Δhns | E. coli K12, Δhns | KEIO collection b |
| M5 | BW25113 Δhns cat-PcycA-lacZ | This work |
| BW25113 ΔihfA | E. coli K12, ΔihfA | KEIO collection b |
| M6 | BW25113 ΔihfA cat-PcycA-lacZ | This work |
| BW25113 Δcrp | E. coli K12, Δcrp | KEIO collection b |
| M7 | BW25113 Δcrp cat-PcycA-lacZ | This work |
| BW25113 ΔfarR | E. coli K12, ΔfarR | KEIO collection b |
| M8 | BW25113 ΔfarR cat-PcycA-lacZ | This work |
| BW25113 ΔrpoS | E. coli K12, ΔrpoS | KEIO collection b |
| M9 | BW25113 ΔrpoS cat-PcycA-lacZ | This work |
| BW25113 Δlrp | E. coli K12, Δlrp | KEIO collection b |
| M10 | BW25113 Δlrp cat-PcycA-lacZ | This work |
| M11 | MG1655 cat-PcycA-5′-UTRlacZ-lacZ | This work |
| M12 | MG1655 Δcrp cat-PcycA-5′-UTRlacZ-lacZ | This work |
| SA500/pHA5 | E. coli K12, rpsL his su− [pHA5, contains crp gene] | [41] |
| 44-3-15 Scr | Ile producing strain | [42] |
| M13 | 44-3-15 Scr cat-PL-cycA | This work |
| pKD46 | oriR101, repA101ts, araC, ParaB-[γ, β, exo of phage λ], ApR; used as a donor of λRed-genes to provide λRed-dependent recombination | [43] |
| pMW118-CmR | oriR101, repA, MCS, ApR, CmR λattR-cat-λattL; used as a donor of λXis/Int-excisable CmR marker | [44] |
| Ile-Producing Strain | Ile, mM | Impurities, % of Ile | ||
|---|---|---|---|---|
| Nva | Ala | AABA | ||
| 44-3-15 Scr | 2.0 ± 0.2 | 390 ± 2 | 260 ± 3 | 362 ± 15 |
| 44-3-15 Scr kan-PL-cycA (M13) | 0.8 ± 0.1 | 110 ± 3 0 | <39 | <12 |
| Ile-Producing Strain | Ile, mM | Val, % of Ile |
|---|---|---|
| 44-3-15 Scr | 105 ± 3 | 26.2 ± 0.7 |
| 44-3-15 Scr kan-PL-cycA (M13) | 107 ± 8 | 19 ± 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hook, C.; Eremina, N.; Zaytsev, P.; Varlamova, D.; Stoynova, N. The Escherichia coli Amino Acid Uptake Protein CycA: Regulation of Its Synthesis and Practical Application in l-Isoleucine Production. Microorganisms 2022, 10, 647. https://doi.org/10.3390/microorganisms10030647
Hook C, Eremina N, Zaytsev P, Varlamova D, Stoynova N. The Escherichia coli Amino Acid Uptake Protein CycA: Regulation of Its Synthesis and Practical Application in l-Isoleucine Production. Microorganisms. 2022; 10(3):647. https://doi.org/10.3390/microorganisms10030647
Chicago/Turabian StyleHook, Christine, Natalya Eremina, Petr Zaytsev, Daria Varlamova, and Nataliya Stoynova. 2022. "The Escherichia coli Amino Acid Uptake Protein CycA: Regulation of Its Synthesis and Practical Application in l-Isoleucine Production" Microorganisms 10, no. 3: 647. https://doi.org/10.3390/microorganisms10030647
APA StyleHook, C., Eremina, N., Zaytsev, P., Varlamova, D., & Stoynova, N. (2022). The Escherichia coli Amino Acid Uptake Protein CycA: Regulation of Its Synthesis and Practical Application in l-Isoleucine Production. Microorganisms, 10(3), 647. https://doi.org/10.3390/microorganisms10030647






