Abstract
Multidrug-resistant (MDR) Enterococcus faecium (Efm) infections continue to increase worldwide, although epidemiological studies remain scarce in lower middle-income countries. We aimed to explore which strains circulate in E. faecium causing human infections in Tunisian healthcare institutions in order to compare them with strains from non-human sources of the same country and finally to position them within the global E. faecium epidemiology by genomic analysis. Antibiotic susceptibility testing was performed and transfer of vancomycin-vanA and ampicillin-pbp5 resistance was performed by conjugation. WGS-Illumina was performed on Tunisian strains, and these genomes were compared with Efm genomes from other regions present in the GenBank/NCBI database (n = 10,701 Efm genomes available May 2021). A comparison of phenotypes with those predicted by the recent ResFinder 4.1-CGE webtool unveiled a concordance of 88%, with discordant cases being discussed. cgMLST revealed three clusters [ST18/CT222 (n = 13), ST17/CT948 strains (n = 6), and ST203/CT184 (n = 3)], including isolates from clinical, healthy-human, retail meat, and/or environmental sources in different countries over large time spans (10–12 years). Isolates within each cluster showed similar antibiotic resistance, bacteriocin, and virulence genetic patterns. pbp5-AmpR was transferred by VanA-AmpR-ST80 (clinical) and AmpR-ST17-Efm (bovine meat). Identical chromosomal pbp5-platforms carrying metabolic/virulence genes were identified between ST17/ST18 strains of clinical, farm animal, and retail meat sources. The overall results emphasize the role of high-resolution genotyping as provided by WGS in depicting the dispersal of MDR-Efm strains carrying relevant adaptive traits across different hosts/regions and the need of a One Health task force to curtail their spread.
1. Introduction
The number of Enterococcus faecium infections continues to increase worldwide, although the great asymmetry in their epidemiology among different regions [1,2] jeopardizes the effective control of the spread of multidrug-resistant (MDR) strains, such as vancomycin-resistant ones. Routes of transmission between different hosts are often unclear, and a better understanding of these dynamics is crucial to curb their dissemination by the early identification of strains or genetic entities with clinically relevant antibiotic resistance, virulence, and bacteriocins among other key adaptive features.
E. faecium historically emerged into distinct clones that are more adapted to the hospital setting. These clones are now epidemic in many hospitals worldwide, even though they are continuingly evolving into more highly adapted clones, challenging epidemiological and typing studies [2,3]. Whole-genome sequencing has provided the greatest resolution in establishing transmission pathways at a global scale, and the genetic intermixing between human infection E. faecium and wastewater, livestock, or pets has been previously documented [4,5,6]. Widespread genomic-surveillance studies suggest, however, that such identity between human clinical and non-clinical strains seems rare [7,8], despite the great bias of studies’ balance towards a greater focus on the hospital setting [9]. In fact, the main zoonotic risk was suggested to occur through the horizontal gene transfer of antimicrobial resistance or virulence genes [10].
Epidemiological data about E. faecium infections from lower middle-income countries and specific regions such as African countries remain generally scarce [11]. In Tunisia, in particular, epidemiological studies assessing antibiotic resistance, clonality, or other features of clinical E. faecium are limited [12,13,14], and those approaching their typing by WGS are absent. Among available studies, clinical vancomycin-resistant E. faecium (VREfm) have been reported in association with ST18, ST80, and other less disseminated clones and typical multi-resistance phenotypes. In this context, we aimed to characterize E. faecium strains that cause human infections in Tunisian healthcare institutions and to position them within the global E. faecium epidemiology. Whole-genome sequencing (WGS) was fundamental for tracing the dispersal of MDR E. faecium strains carrying clinically relevant features in an international context.
2. Materials and Methods
2.1. Bacterial Strain Collection
The clinical set of isolates we gathered from different healthcare institutions in Tunisia comprised 25 non-duplicate E. faecium isolates collected from February 2011 to February 2016 in two private clinics (North of Tunis) and one regional hospital (Gafsa City, Southwest). The epidemiological background of these isolates is shown in Table S1. Representative isolates of different phenotypes (including MDR and non-MDR: 5 AmpR, 3 VREfm, and 3 non-AmpR/VREfm) were selected for the establishment of clonal relationship by SmaI-PFGE. Briefly, genomic DNA was digested with SmaI (Takara Bio Inc., Shiga, Japan), and PFGE was performed by using the following conditions: 1 to 20 s for 26 h, 14 °C, and 6 V/cm2. Such PFGE profiles were compared to others previously obtained from isolates of food-producing animals and retail meat in different regions of Tunisia [15]. Eight Tunisian E. faecium isolates from different sources and in some cases presenting identical PFGE profiles between different hosts were selected for whole-genome sequencing: 3 from human clinical (342T, 349T, and 465T), 3 from retail bovine meat (361T, 365T, and 508T), and 2 from intensive farm cow milk (437T and 464T). This group of isolates was further compared to others that are publicly available in the GenBank database (see below), with identical strains comprising a second set of 24 E. faecium strains described below.
2.2. Phenotypic and Molecular Techniques
Among the Tunisian clinical isolates, antimicrobial susceptibility testing (disk diffusion) and results’ interpretation against 13 antibiotics (Oxoid, Basingstoke, UK) were performed according to EUCAST (www.eucast.org; last accessed on 18 December 2020) or CLSI [16] guidelines when EUCAST clinical breakpoints were not available (chloramphenicol, erythromycin, and tetracycline). Minimum Inhibitory Concentrations (MICs) of ampicillin were determined by using E-test (Liofilchem, Italy) and interpreted according to EUCAST guidelines. High-level ampicillin resistance (AmpR) was considered for MIC values of ≥32 mg/L [17].
Genes coding for (i) resistance to vancomycin (vanA/vanB), (ii) replicases of plasmids commonly associated with vancomycin resistance in E. faecium (rep-pRUM, rep-pLG1, and rep-Inc18 (rep1/rep2)), and (iii) virulence or an increased risk of human infection by E. faecium (ptsD, orf1481, sgrA, IS16, hyl, esp, and complete acm) were screened by PCR, as previously described [18,19].
2.3. Whole-Genome Sequencing
Genomic DNA was extracted from 1 mL of overnight cultures in brain heart infusion broth by using a Wizard Genomic DNA Purification kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. WGS sequencing was performed by using an Illumina HiSeq platform (2 × 125 bp), according to standard Illumina protocols performed at GATC Biotech (Konstanz, Germany).
The assembled genomes (SPAdes (v.3.10.0)) were first screened for genes encoding antibiotic resistance (ABR), plasmid content, and MLST by using in silico genomic tools (ResFinder 4.0, PlasmidFinder 2.1, and MLST 2.0 tools, respectively) available at the Center for Genomic Epidemiology (CGE; http://www.genomicepidemiology.org; last accessed on 18 December 2020). Because the VirulenceFinder database (n = 26 genes typical of Enterococcus spp.) is not complete for E. faecium, we used a homemade database of 41 virulence factors important in this species [18] by using the MyDbFinder (BLAST) tool available at CGE. A second homemade database of 76 bacteriocin genes from Firmicutes [20] was also tested in these genomes.
High-resolution genotyping was performed by cgMLST by using E. faecium schemes from Ridom SeqSphere+ v. 7.2 software. Complex types (CT) were compared to those of 10.701 E. faecium genomes from the GenBank/NCBI database, and a minimum spanning tree based on cgMLST (1423 genes) analyses was performed with SeqSphere+ software.
PBP5 platforms of selected AmpR E. faecium were analyzed by genome mapping against TCGEHPH2 pbp5-containing contig (GenBank accession no. MBRI01000000) [21], and platforms were characterized by using Vector NTI advance v11 and EggNOG-mapper.
2.4. Transferability of Ampicillin Resistance
The transferability of ampicillin resistance was attempted in 7 AmpR E. faecium isolates of different clones and sources, as described [21]. Briefly, filter-mating assays were performed in brain heart infusion (BHI) agar not supplemented with antibiotics at 37 °C overnight by using a donor/recipient ratio of 1:1 and E. faecium GE1 as the recipient strain. Transconjugants were selected on BHI agar supplemented with antibiotics (ampicillin-10 mg/L, fusidic acid-25 mg/L, and rifampicin-30 mg/L) and incubated for 24 to 48 h (37 °C) to recover potential transconjugants.
3. Results and Discussion
3.1. Detection of a Small ST80 VanA-VREfm Outbreak and Other Ampicillin-Resistant Hospital Associated Clones Enriched in Virulence Markers
Clinical and epidemiological data about the 25 clinical E. faecium isolates collected in three different health institutions are described in Table S1. These 25 isolates were identified in samples from infection (n = 19) and gut colonization (n = 6) cases. Of the nineteen infection cases, fourteen were bloodstream infections (BSI), four were urinary tract infections (UTIs), and one was an infection of a surgical wound. Most E. faecium isolates came from intensive care unit patients (n = 13/19; 68%) and less in medical and surgical wards, whereas isolates from gut colonization screenings were obtained mostly from the gastroenterology ward (n = 5/6; 83%).
Multidrug-resistance phenotypes were only associated with isolates from infection cases (n = 12/19; 63%). MDR cases were detected in 80% of isolates from the hospital institution, were not detected in private clinic A and were detected in private clinic B only in three infection isolates. This observation is somewhat expected because private hospitals, on average, treat patients who have a lower risk of infection, whereas public hospitals provide services free of charge to all eligible patients (higher-stay hospitalization, more urgent cases, etc.). Three patients staying at the ICU of the private clinic B (2016) had BSI caused by identical vancomycin-resistant strains identified as ST80/CT1764 (same PFGE profile). The three isolates were MDR and expressed resistance to vancomycin (MIC > 32 mg/L; all vanA), teicoplanin, ampicillin, ciprofloxacin, erythromycin, and streptomycin, with two out of three additionally presenting resistance to gentamicin and quinupristin-dalfopristin (Table S1). They were also enriched in relevant virulence markers (esp, ptsD, IS16, orf1481, sgrA, and acm) that were previously linked to infection-derived and outbreak E. faecium strains globally [18]. VREfm ST80 strains have been previously identified from other hospitals in Tunis during 2012–2013 [14] and 2017 [13], and they are commonly found among clinical E. faecium from hospitalized human patients worldwide [2]. The unnoticed small outbreak detected in this work, together with the scarce number of published epidemiological studies detecting VREfm in healthcare institutions of Tunisia since 2007 [12,13,14,22], highlights the fact that VREfm numbers in this country may be higher than estimated. Indeed, a recent systematic review and meta-analysis developed by Alemayehu and Hailemariam [23] identified a high-pooled prevalence of VREfm enterococci in African countries, with Tunisia, despite everything, being one of the countries that most contributed to such analysis (just surpassed by South Africa and Ethiopia).
The remaining non-VREfm MDR isolates (n = 9) also exhibited resistance to antibiotics that are relevant in the treatment of enterococcal infections such as ampicillin (70%; MIC = 32 ≥ 256 mg/L) and gentamicin (20%) (Table S1). Resistance to erythromycin (100%), streptomycin (70%), ciprofloxacin (50%), tetracycline (30%), and quinupristin-dalfopristin (40%) were also detected at variable rates. Ten different PFGE profiles were established among this set of nine isolates, with two of them being identified as ST17 and ST18, which are both well-known major hospital-associated clones. As previously documented [18], ampicillin-resistant isolates were associated with a higher number of virulence genes that included ptsD, esp, IS16, orf1481, sgrA, and the complete acm and hyl genes (Table S1). Among the two ampicillin-susceptible isolates, one lacked virulence genes and the other only carried three virulence genes. In a previous study, including isolates colonizing patients at long-term care facilities [24], the predominant ampicillin-susceptible isolates only harbored sgrA gene coding for an adhesin involved in the formation of biofilms, thus reinforcing high-level resistance to ampicillin as a good marker of hospital-associated MDR E. faecium clones enriched in relevant putative virulence markers.
Plasmid types commonly linked to clinical E. faecium strains in previous studies [25,26] were identified among most isolates of this study (18/25; 72%). Exceptions greatly corresponded to non-MDR isolates from private clinic A or colonization isolates that lacked all rep types tested. The presence of pRUM-, Inc18-, and pLG1-like plasmids, all greatly associated with VREfm outbreaks in different countries [19], in the VanA-VREfm isolates, suggests that common plasmidomes circulate in clinical E. faecium, which are also from Tunisia. Until now, the rep from pRUM-like plasmids has been almost exclusively found in clinical isolates and mostly with vancomycin-resistant plasmids, and we here identified it only in the VREfm isolates from private clinic B and in a few MDR isolates from the hospital institution, thus confirming the role of this plasmid in the global spread of vancomycin resistance.
Although this set of isolates does not correspond to the full number of E. faecium isolates collected in the time period of the study in the three institutions included, which precludes to infer real antibiotic resistance rates and the full landscape of circulating strains and plasmids, all information from lower middle-income countries such as Tunisia is of value as the number of surveillance studies including clinical enterococci isolates is highly limited [12,13,14,27]. Enterococcal species not expressing resistance to ampicillin and/or vancomycin were not included in further analyses.
3.2. Reliability of In Silico Prediction of Phenotypes Based on Genomic Data by Using the ResFinder Webtool
We next compared the predicted phenotypes of sequenced Tunisian genomes, according to the ResFinder CGE webtool (version 4.1), with the antibiotic resistance patterns determined by disk diffusion in this study. Among all the 41 antibiotic genotype–phenotype cases (Table S2), most of them were concordant (n = 36/41; 88%). This value is, however, lower than that observed in the study analyzing 106 E. faecium isolates (92.8–96.2%) with ResFinder 4.0 [28]. Discordant cases were represented by tetracycline- and gentamicin-susceptible isolates harboring tet(M) genes (n = 2) and aac(6′)-Ii (n = 3), respectively. Gentamicin and tetracycline discordance cases were also reported by Bortolaia et al. [28], who described a low read depth for tet(M) genes.
A close inspection of discordant cases in our study showed that tet(M) genes were actually truncated, and aac(6′)-Ii is known to encode low-level chromosomal resistance to different aminoglycosides in E. faecium [29] and is often considered as an intrinsic gene. As we here tested high-level resistance to gentamicin (MIC > 128 mg/L), which makes sense in a clinical context, results are discordant because ResFinder seems to signal “resistant” intrinsic types of resistance. Therefore, the interpretation of phenotypic–genotypic results should be conducted carefully, with the knowledge on intrinsic resistance according to different bacterial species being essential [30].
3.3. Identity of Ampicillin-Resistant Strains and Their Resistome, Virulome, and Bacteriocinome between Human Clinical, Animal, and Food Samples
Based on the cgMLST data retrieved from the analysis of the second set of 24 sequenced isolates (Figure 1), three main clusters were observed. Cluster 1 included ST18/CT222 strains (n = 13) from clinical, human, retail meat, and environmental sources in eight countries during 2000–2017, and they mostly carried the same antibiotic resistance genes encoding for aminoglycosides (aph(3′)-III and/or ant(6)-Ia), macrolide-lincosamide-streptogramins A/B (erm(B), msr(C), Inu(B), and/or Isa(E)), tetracyclines (tet(M), tet(L), and/or tet(O)), and trimethoprim (dfrG), with two French isolates harboring vancomycin-resistance genes, in addition to pbp5 mutations conferring ampicillin resistance (Table 1). Cluster 2 grouped ST17/CT948 strains (n = 6) obtained from hospitalized patients in the UK and Tunisia (2003–2014) together with isolates from farm animals and retail meat in Tunisia (2016). Interestingly, common pools of antibiotic resistance genes were observed for the oldest isolates (aac(6′)-aph(2″), aph(3′)-III, ant(6)-Ia, erm(A), erm(B), and/or msr(C)) (2003–2004) and the most recent ones (vanHAX, aph(3′)-III, ant(6)-Ia, erm(B), and/or msr(C)) (2014–2016), irrespective of their origin, and all showed common mutations compatible with ampicillin and quinolone resistance (Table 1). Cluster 3 grouped three ST203/CT184 strains obtained from hospitalized patients in UK/2007 and retail meat in Tunisia over 2010–2016 that showed similar antibiotic-resistance gene patterns (aac(6′)-aph(2″), aph(3′)-III, ant(6)-Ia, erm(B), msr(C), Inu(B), tet(M), and/or vanHAX) and mutations compatible with ampicillin and quinolone resistance (Table 1). Isolates from the three clusters, including those from animal/meat sources, were equally enriched in several different virulence markers (Table 2) involved in adhesion (e.g., acm and sgrA), the formation of biofilms and pili (empABC), intestinal colonization (e.g., ptsD), response to stress (gls genes), etc., that have been previously linked to an increased pathogenicity [18]. Only isolates from cluster 2 (ST17) and cluster 3 (ST203) carried hylEfm and ecbA, probably as a result of plasmid selection and clonal expansion, respectively.
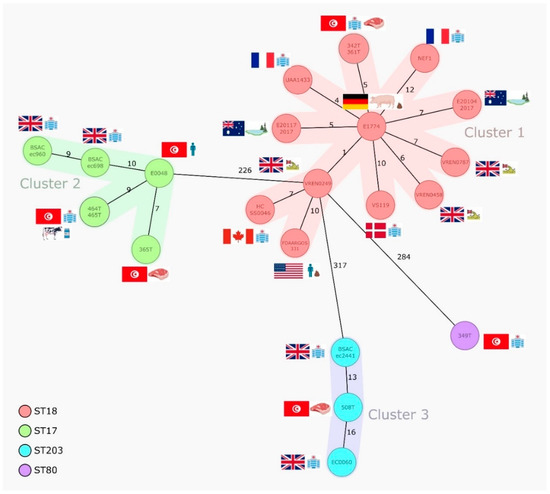
Figure 1.
Minimum spanning tree based on the cgMLST data from E. faecium isolates (n = 24) of different sources. The tree is based on cgMLST (1423 genes) analyses made with SeqSphere+ software. Each circle represents one allele profile. The numbers on the connecting lines represent the number of cgMLST allelic differences between two isolates. STs are shown in coloured circles (see legend). Colour shading around nodes indicates clusters of closely related isolates (≤20 SNPs).

Table 1.
Epidemiological data and antibiotic resistance genomic content (acquired genes and chromosomal mutations) in the 24 analysed strains.

Table 2.
Epidemiological data and virulence gene content in the 24 analysed strains.
Thirteen different bacteriocins out of the seventy-six tested were detected, with entA being common to all isolates, as has been reported in different studies and suggested to be part of the E. faecium core genome [31]. Despite the variability of bacteriocin genes found, each group of ST18, ST17, and ST203 strains shared identical bacteriocins with each other’s, thus revealing a positive association between clones and specific peptides potentially contributing for niche control [32].
The ST80/CT1764 VanA-VREfm strain (349T) did not show CT homologs at the GenBank according to our analysis, but we could identify this CT in two E. faecium from Germany/2018 of the SeqSphere database. As expected, genomic analysis revealed that this strain was also enriched in antibiotic resistance and virulence genes (Table 1 and Table 2). vanA-Tn1546 was located in the same contig as the replicase identical to that of pRUM, confirming the circulation of vanA-carrying pRUM-like plasmids in hospitalized patients from Tunisia [19]. A ST18/CT2661 MDR-AmpR strain (437T) recovered from the milk of a farm cow was also not linked to any other publicly available strain or to clinical Tunisian strains, but as it was enriched in clinically relevant antibiotic resistance and virulence genes, we provide its data in Table 1 and Table 2 as well.
3.4. Transferability of Ampicillin Resistance and Identity of Pbp5-Carrying Genetic Platforms
Two out of the seven Tunisian AmpR isolates tested (28%) were able to transfer ampicillin resistance to the E. faecium strain GE1 under our experimental conditions. The positive cases corresponded to the VanA-VRE ST80 clinical strain and a ST17 MDR strain from bovine meat (both Tunisian) transferring at 2.8 × 10−7 and 1.4 × 10−8 rates, respectively. In addition to ampicillin, rifampicin, and fusidic acid, the transconjugant of the VanA-VREfm strain acquired resistance to vancomycin, teicoplanin, erythromycin, and streptomycin, whereas different transconjugants obtained from the ST17 bovine strain were resistant to erythromycin, tetracycline, and gentamicin or streptomycin.
All 24 genomes analyzed had several mutations in the pbp5 gene (Table S3) that have been associated with pbp5 clade A1R, which mostly comprises ampicillin-resistant isolates [21]. Even though most data seem to indicate that there are no consistent amino acid changes correlating with specific increases in the MICs of ampicillin [33], a recent study by Darehkordi et al. established a link between high ampicillin MICs, high PBP5 expression levels, and specific pbp5 mutations [34]. A comparison between pbp5-containing platforms (associated with AmpR) of our sequenced genomes with a large transferable pbp5-containing platform that we have previously described in a clinical isolate [21] allowed the partial identification of highly similar genetic platforms carrying different metabolic and adaptive features, including virulence genes (e.g., sgrA or fms2 important in biofilm formation) in AmpR-Efm of different origins (Figure 2). The aforementioned ST17 bovine Tunisian strain able to transfer AmpR shared a ~6 kb pbp5 Type I platform, which we described previously as the predominant prototype Type I platform not containing indels [21], with the clinical ST125 Portuguese strain (urine). A larger pbp5 platform of about 10 kb containing variable insertion sequences (ISEf1, ISEm1, or IS1542) at different positions was shared by four Tunisian strains: two clinical ST17 (465T) and ST18 (342T) strains, one ST18 (361T) from bovine meat, and one ST17 (464T) from cow milk (Figure 2).
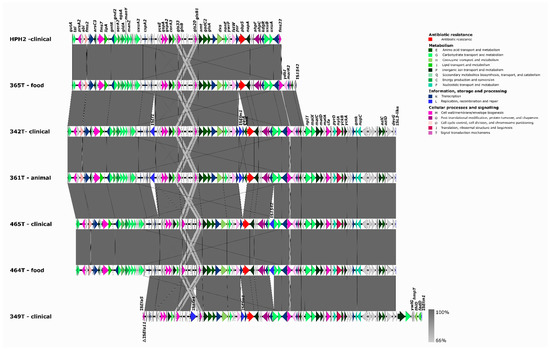
Figure 2.
Representation of partial transferable chromosomal genetic platforms containing pbp5. Mapping and annotation (Geneious Prime version 2020.2.2) of partial transferable pbp5 chromosomal genetic platforms of Tunisian AmpR-E. faecium of this study [365T/ST17 (68,260 bp), 342T/ST18 (99,914 bp), 361T/ST18 (99,914 bp), 465T/ST17 (97,354 bp), 464T/ST17 (97,354 bp), and 349T/ST80 (84,987 bp)] by using the pbp5-containing contig of the ampicillin-resistant transconjugant TCGEHPH2 (63,575 bp; GenBank accession no. MBRI01000000) [21]. The contigs identified were assembled by using Vector NTI advance v11, and the platform was annotated by using EggNOG-mapper.
The mean rates of ampicillin resistance among E. faecium of animal origin seem low in Europe [10] and abroad [35], although such data are generally based on centralized surveillance studies (e.g., DANMAP in Denmark) or studies that do not include ampicillin selection during sample processing. Regardless of these numbers and the fact that the public health risk from AmpR E. faecium due to the veterinary use of penicillins in food-producing animals is suggested as lower than that from their use in human medicine [10,36], our results reinforce the ability of E. faecium to transfer large chromosomal pbp5 platforms along with other resistance and virulence determinants independently of strain origin. A link to hospital-associated clones was, however, noted as in previous studies [21]. As such, the driving force that beta-lactams can exert toward AMR in different environments, such as the animal production setting where aminopenicillins can be heavily used and where antimicrobial use has been suggested as the major risk factor for selection of AMR [37], should not be discarded. Moreover, the hypothesis that the transferability of pbp5-containing platforms under laboratory conditions may be underestimated in comparison to natural conditions, together with evidence that genomic rearrangements of large DNA fragments can occur in the region upstream of pbp5 [21,38], suggest the possibility that E. faecium adapts to changing environments and that AmpR E. faecium rates among farm animals may be also undervalued.
4. Conclusions
Clinical multidrug-resistant E. faecium clones circulating in Tunisian patients are closely related to Tunisian strains across the food chain as well as to foreign clinical E. faecium originating from other countries and continents. Identical multidrug- and ampicillin-resistant E. faecium strains carrying markers associated with an increased risk of human infection were found in clinical and animal sources from Tunisia or other countries, emphasizing the global and continuous transmission of relevant strains across different hosts and settings. Previous observations suggest that reducing the acquisition of hospital-associated strains by patients entering the hospital is needed [3], but the possibility of acquiring those strains from community contexts including the food chain cannot be discarded. Our study also adds evidence to the exchange of similar ampicillin resistance genetic platforms between different strains of different hosts, a genetic event that may be more common than expected in response to environmental stimuli such as the use of aminopenicillins in food-producing animals.
Future effective genomic surveillance of MDR enterococci or other bacterial pathogens must also consider plasmids that are pivotal in the acquisition and transfer of resistance genes between bacterial strains or even species. Our data extend the distribution of key hospital-associated E. faecium clones to variable sources of the African continent and highlight the role of high-resolution genotyping, as provided by whole-genome sequencing in depicting the dispersal of MDR E. faecium strains and the need of a transdisciplinary One Health approach to curtail their dissemination.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10030632/s1, Table S1: Epidemiological data and characterization of clinical Enterococcus faecium isolates from Tunisia, Table S2: Reliability of in silico prediction of phenotypes based on genomic data by using the ResFinder webtool. Table S3: Description of PBP5 alleles amino acid changes and correlation with epidemiological data.
Author Contributions
A.R.F. designed the study; acquired and interpreted phenotypic, molecular, and whole-genome sequencing data; and wrote the manuscript. H.E. and B.D. performed the characterization of isolates. M.S.A. and A.H. collected epidemiological information. A.P.T., C.N. and A.C.A.-S. performed the genomic analysis. A.P.T., C.N. and L.P. revised the manuscript for scientific content. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by national funds from FCT—Fundação para a Ciência e a Tecnologia, I.P.—in the scope of project UIDP/04378/2020 and UIDB/04378/2020 of the Research Unit on Applied Molecular Biosciences—UCIBIO; project LA/P/0140/2020 of the Associate Laboratory Institute for Health and Bioeconomy—i4HB; and the exploratory project EXPL/SAU-INF/0261/2021. Additionally, we would like to acknowledge the support of AgriFood XXI I&D&I project (NORTE-01-0145-FEDER-000041) co-financed by the European Regional Development Fund (ERDF) through NORTE 2020 (Programa Operacional Regional do Norte 2014/2020). A.R.F. and A.C.A.-S. gratefully acknowledge the Junior Research Position (CEECIND/02268/2017—Individual Call to Scientific Employment Stimulus 2017) and the UI/BD/151317/2021 fellowship, respectively, both granted by FCT/MCTES through national funds. A.P.T. was supported by the Sara Borrell Research Grant (CD18/00123) from ISCIII and co-funded by ERDF/ESF, “A way to make Europe”/“Investing in your future”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under BioProject accession number PRJNA800622 (Biosample accession numbers: SAMN25270785-SAMN25270792).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updates 2018, 40, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Pereira, A.P.; Novais, C.; Peixe, L. Multidrug-resistant high-risk Enterococcus faecium clones: Can we really define them? Int. J. Antimicrob. Agents 2021, 57, 106227. [Google Scholar] [CrossRef]
- Van Hal, S.J.; Willems, R.J.L.; Gouliouris, T.; Ballard, S.A.; Coque, T.M.; Hammerum, A.M.; Hegstad, K.; Pinholt, M.; Howden, B.P.; Malhotra-Kumar, S.; et al. The interplay between community and hospital Enterococcus faecium clones within health-care settings: A genomic analysis. Lancet Microbe 2022, 3, E133–E141. [Google Scholar] [CrossRef]
- Freitas, A.R.; Finisterra, L.; Tedim, A.P.; Duarte, B.; Novais, C.; Peixe, L. Linezolid- and Multidrug-Resistant Enterococci in Raw Commercial Dog Food, Europe, 2019–2020. Emerg. Infect. Dis. 2021, 27, 2221–2224. [Google Scholar] [CrossRef] [PubMed]
- Arredondo-Alonso, S.; Top, J.; McNally, A.; Puranen, S.; Pesonen, M.; Pensar, J.; Marttinen, P.; Braat, J.C.; Rogers, M.R.C.; van Schaik, W.; et al. Plasmids Shaped the Recent Emergence of the Major Nosocomial Pathogen Enterococcus faecium. mBio 2020, 11, e03284-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouliouris, T.; Raven, K.E.; Moradigaravand, D.; Ludden, C.; Coll, F.; Blane, B.; Naydenova, P.; Horner, C.; Brown, N.M.; Corander, J.; et al. Detection of vancomycin-resistant Enterococcus faecium hospital-adapted lineages in municipal wastewater treatment plants indicates widespread distribution and release into the environment. Genome Res. 2019, 29, 626–634. [Google Scholar] [CrossRef] [Green Version]
- Gouliouris, T.; Raven, K.E.; Ludden, C.; Blane, B.; Corander, J.; Horner, C.S.; Hernandez-Garcia, J.; Wood, P.; Hadjirin, N.F.; Radakovic, M.; et al. Genomic Surveillance of Enterococcus faecium Reveals Limited Sharing of Strains and Resistance Genes between Livestock and Humans in the United Kingdom. mBio 2018, 9, e01780-18. [Google Scholar] [CrossRef] [Green Version]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.A.; Strong, K.; Cork, S.C.; McAllister, T.A.; Liljebjelke, K.; Zaheer, R.; Checkley, S.L. The Role of Whole Genome Sequencing in the Surveillance of Antimicrobial Resistant Enterococcus spp.: A Scoping Review. Front. Public Health 2021, 9, 599285. [Google Scholar] [CrossRef] [PubMed]
- Committee for Medicinal Products for Veterinary Use (CVMP). Reflection Paper on the Use of Aminopenicillins and Their Beta-Lactamase Inhibitor Combinations in Animals in the European Union: Development of Resistance and Impact on Human and Animal Health; EMA/CVMP/AWP/842786/2015; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziri, R.; Lozano, C.; Said, L.B.; Bellaaj, R.; Boudabous, A.; Slama, K.B.; Torres, C.; Klibi, N. Multidrug-resistant enterococci in the hospital environment: Detection of novel vancomycin-resistant E. faecium clone ST910. J. Infect. Dev. Ctries. 2016, 10, 799–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziri, R.; El Kara, F.; Barguellil, F.; Ouzari, H.I.; El Asli, M.S.; Klibi, N. Vancomycin-Resistant Enterococcus faecium in Tunisia: Emergence of Novel Clones. Microb. Drug Resist. 2019, 25, 469–474. [Google Scholar] [CrossRef]
- Elhani, D.; Klibi, N.; Dziri, R.; Hassan, M.B.; Asli Mohamed, S.; Said, L.B.; Mahjoub, A.; Slama, K.B.; Jemli, B.; Bellaj, R.; et al. vanA-containing E. faecium isolates of clonal complex CC17 in clinical and environmental samples in a Tunisian hospital. Diagn. Microbiol. Infect. Dis. 2014, 79, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Elghaieb, H.; Freitas, A.R.; Abbassi, M.S.; Novais, C.; Zouari, M.; Hassen, A.; Peixe, L. Dispersal of linezolid-resistant enterococci carrying poxtA or optrA in retail meat and food-producing animals from Tunisia. J. Antimicrob. Chemother. 2019, 74, 2865–2869. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Joste, V.; Gydé, E.; Toullec, L.; Courboulès, C.; Talb, Y.; Riverain-Gillet, E.; Pangon, B.; Amara, M.; Patel, R. Enterococcus faecium and Ampicillin Susceptibility Determination: Overestimation of Resistance with Disk Diffusion Method Using 2 Micrograms of Ampicillin? J. Clin. Microbiol. 2019, 57, e01467-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Coque, T.M.; Peixe, L. Distribution of putative virulence markers in Enterococcus faecium: Towards a safety profile review. J. Antimicrob. Chemother. 2018, 73, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Tedim, A.P.; Francia, M.V.; Jensen, L.B.; Novais, C.; Peixe, L.; Sánchez-Valenzuela, A.; Sundsfjord, A.; Hegstad, K.; Werner, G.; et al. Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986–2012). J. Antimicrob. Chemother. 2016, 71, 3351–3366. [Google Scholar] [CrossRef] [Green Version]
- Tedim, A.P.; Freitas, A.R.; Lanza, V.F.; Novais, C.; Coque, T.M.; Peixe, L. Towards the elucidation of bacteriocins role in shaping the ecology and evolution of Enterococcus faecium. In Proceedings of the 29th European Congress of Clinical Microbiology & Infectious Diseases. (ECCMID 2019), Amsterdam, The Netherlands, 13–16 April 2019; p. 2755. [Google Scholar]
- Novais, C.; Tedim, A.P.; Lanza, V.F.; Freitas, A.R.; Silveira, E.; Escada, R.; Roberts, A.P.; Al-Haroni, M.; Baquero, F.; Peixe, L.; et al. Co-diversification of Enterococcus faecium Core Genomes and PBP5: Evidences of pbp5 Horizontal Transfer. Front. Microbiol. 2016, 7, 1581. [Google Scholar] [CrossRef]
- Abbassi, M.S.; Znazen, A.; Mahjoubi, F.; Hammami, A.; Hassen, A.B. Emergence of vancomycin-resistant Enterococcus faecium in Sfax: Clinical features and molecular typing. Médecine Et Mal. Infect. 2007, 37, 240–241. [Google Scholar] [CrossRef]
- Alemayehu, T.; Hailemariam, M. Prevalence of vancomycin-resistant enterococcus in Africa in one health approach: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 20542. [Google Scholar] [CrossRef]
- Freitas, A.R.; Novais, C.; Duarte, B.; Pereira, A.P.; Coque, T.M.; Peixe, L. High rates of colonisation by ampicillin-resistant enterococci in residents of long-term care facilities in Porto, Portugal. Int. J. Antimicrob. Agents 2018, 51, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Novais, C.; Tedim, A.P.; Francia, M.V.; Baquero, F.; Peixe, L.; Coque, T.M. Microevolutionary events involving narrow host plasmids influences local fixation of vancomycin-resistance in Enterococcus populations. PLoS ONE 2013, 8, e60589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wardal, E.; Kuch, A.; Gawryszewska, I.; Żabicka, D.; Hryniewicz, W.; Sadowy, E. Diversity of plasmids and Tn1546-type transposons among VanA Enterococcus faecium in Poland. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 313–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belhaj, M.; Boubaker, I.B.-B.; Redjeb, S.B.; Bouchami, O. Molecular characterisation of high-level ampicillin-resistant Enterococcus faecium isolates from hospitalised patients in Tunis. Int. J. Antimicrob. Agents 2008, 32, 284–286. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Costa, Y.; Galimand, M.; Leclercq, R.; Duval, J.; Courvalin, P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 1993, 37, 1896–1903. [Google Scholar] [CrossRef] [Green Version]
- EUCAST. Intrinsic Resistance and Exceptional Phenotypes, Expert Rules. Version 3.2. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Expert_Rules/2020/Intrinsic_Resistance_and_Unusual_Phenotypes_Tables_v3.2_20200225.pdf (accessed on 18 December 2020).
- De Maat, V.; Arredondo-Alonso, S.; Willems, R.J.L.; van Schaik, W. Conditionally essential genes for survival during starvation in Enterococcus faecium E745. BMC Genom. 2020, 21, 568. [Google Scholar] [CrossRef]
- Almeida-Santos, A.C.; Novais, C.; Peixe, L.; Freitas, A.R. Enterococcus spp. as a Producer and Target of Bacteriocins: A Double-Edged Sword in the Antimicrobial Resistance Crisis Context. Antibiotics 2021, 10, 1215. [Google Scholar] [CrossRef]
- Galloway-Peña, J.R.; Rice, L.B.; Murray, B.E. Analysis of PBP5 of early U.S. isolates of Enterococcus faecium: Sequence variation alone does not explain increasing ampicillin resistance over time. Antimicrob. Agents Chemother. 2011, 55, 3272–3277. [Google Scholar] [CrossRef] [Green Version]
- Darehkordi, H.; Saffari, F.; Mollaei, H.R.; Ahmadrajabi, R. Amino acid substitution mutations and mRNA expression levels of the pbp5 gene in clinical Enterococcus faecium isolates conferring high level ampicillin resistance. Apmis 2019, 127, 115–122. [Google Scholar] [CrossRef]
- Kim, M.H.; Moon, D.C.; Kim, S.-J.; Mechesso, A.F.; Song, H.-J.; Kang, H.Y.; Choi, J.-H.; Yoon, S.-S.; Lim, S.-K. Nationwide Surveillance on Antimicrobial Resistance Profiles of Enterococcus faecium and Enterococcus faecalis Isolated from Healthy Food Animals in South Korea, 2010 to 2019. Microorganisms 2021, 9, 925. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A. Quantitative human health risk assessments of antimicrobial use in animals and selection of resistance: A review of publicly available reports. Rev. Sci. Tech. 2012, 31, 261–276. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar] [CrossRef] [PubMed]
- Montealegre, M.C.; Roh, J.H.; Rae, M.; Davlieva, M.G.; Singh, K.V.; Shamoo, Y.; Murray, B.E. Differential Penicillin-Binding Protein 5 (PBP5) Levels in the Enterococcus faecium Clades with Different Levels of Ampicillin Resistance. Antimicrob. Agents Chemother. 2017, 61, e02034-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
