Abstract
Studies on tick-pathogen-host interactions are helping to identify candidates for vaccines against ticks and tick-borne diseases and to discover potent bioactive tick molecules. The tick midgut is the main tissue involved in blood feeding and, moreover, the first organ to have contact with pathogens ingested through the blood meal. As little is known about the molecular biology of feeding and tick defence mechanisms against microorganisms, but important for understanding vector-pathogen interactions, we explored the early transcriptional changes in the midgut of Ornithodoros moubata after feeding and in response to challenge with the relapsing-fever spirochete Borrelia duttonii using the Ion S5XL platform. Besides transcripts with metabolic function and immune-related transcripts we discovered numerous putative and uncharacterized protein sequences. Overall, our analyses support previous studies and provides a valuable reference database for further functional proteomic analysis of midgut proteins of O. moubata.
1. Introduction
Numerous emerging infectious diseases are caused by zoonotic pathogens, including those transmitted by hematophagous arthropod vectors such as mosquitoes and ticks. Ticks and tick-borne diseases affect animal and human health worldwide and are often of great economic significance [1]. As tick control in livestock has to cope acaricide resistance and concerns are raised regarding the environmental impact of pesticides, new approaches for controlling ticks and preventing the transmission of pathogens are focusing on the molecular interactions between host, vector and pathogen. The aim is to identify promising candidates for vaccines against ticks and tick-borne diseases by reducing the tick infestation and preventing ticks from transmitting disease-causing pathogens [2].
In the late 1990s, a protective concealed antigen, the glycoprotein Bm86, was characterized from the midgut of the bovine tick Rhipicephalus microplus [3]. Based on the recombinant Bm86 antigen, the first and so far, only vaccine that have made it to the market was developed, which mainly reduce the average fertility of adult ticks due to a disturbed blood uptake [4]. Since then, intensive basic research focused on transcriptome and proteome profiling in tick salivary glands and midgut, as these structures represent key interfaces between host, vector and pathogen [5]. In a recent article, Tirloni et al. illustrate the dynamic changes in gene expression in embryos, ovaries, salivary glands, fat body and digestive cells during the parasitic phase and the embryonic development of R. microplus. The study reveals over 10,000 novel coding sequences that have highly tissue-specific expression profiles giving insights into the physiology of production and role of saliva, blood digestion, energy metabolism and development [6]. Beside organ-specific analysis of gene expression, special attention is paid to expression differences between female and male ticks as females are the key component of the tick life cycle and represent a suitable target for vaccine development. Oleaga et al. used two different mass spectrometry approaches to explore the sialoproteome of adult Ornithodoros moubata ticks. Quantification of salivary proteins by SWATH-MS indicated significant quantitative differences in the salivary proteome between male and female ticks, which should be considered when selecting vaccine targets, as suggested by the author [7]. Follow-up studies identified several major protein families that play important roles in tick development, feeding and digestion, host attachment and specificity, resistance to acaricides and response to abiotic factors [8,9]. Some of these proteins have recently been selected and their individual and joint vaccine efficacy were explored as protective candidate antigen in ixodid and argasid ticks [2,10,11]. However, no other anti-tick vaccine, except the Bm86-based one, have been commercialized so far due to different challenges discussed by de la Fuente and Estrada-Peña [12].
A positive side effect of using ‘omics technologies is the discovery of possibly bioactive molecules that are seen as promising source of novel therapeutics [13]. Tick saliva in particular, a complex cocktail of molecules that ticks inject into their hosts to inhibit pain, prevent blood clotting and suppress the host immune system, has potential for medical exploitation [14]. Several protein families with immunomodulatory activity, such as evasins, serpins, cystatins and lipocalins have been identified in ticks. Evasins, for example, are a class of cysteine-rich chemokine-binding proteins that modulate the host inflammatory response by binding to host chemokines. Francka et al. have shown, using the evasin ACA-01 from Amblyomma cajennense, that posttranslational modification trough sulfation can modulate binding affinity for proinflammatory chemokines and enhanced the ability to inhibit chemokine signaling through cognate receptors and may provide a strategy to optimize tick salivary proteins for anti-inflammatory applications [15]. However, the development of bioactive molecules into therapeutics is still in its infancy due to lack of follow-up studies and the high cost of drug development. Furthermore, the efficacy of proteins is based on a synergistic effect on compounds from a complex mixture, which is not captured by studies of individual genes or proteins [13].
From the medical and veterinary perspective hard ticks (Ixodidae) represent the most important tick family. They are responsible for transmitting a variety of viral, bacterial and protozoan agents of disease while taking a blood meal from their host. Some hard ticks can also elicit tick paralysis, caused by a salivary toxin released while feeding [1]. Hence it is not surprising that many studies are focusing on the family Ixodidae. Argasid ticks receive less attention despite their role as vectors and reservoirs of pathogens. Ornithodoros moubata is widespread throughout southern and eastern African countries [16]. This tick species is the main vector of the East African human relapsing fever causing agent Borrelia duttonii [17] and of African swine fever virus in pigs [18]. It has also been identified as competent vector for Coxiella burnetii [19] and West Nile virus [20].
Compared to hard ticks, transcriptome [21,22,23,24,25,26,27,28] and proteome profiling [21,29,30,31,32] have been performed on a small number of soft tick species only, with only a few focusing on the midgut. The midgut embodies the primary organ for tick survival since it is responsible for blood digestion and absorption of nutrients [33]. Furthermore, the midgut is the first tissue where pathogens ingested with the blood meal are exposed to the tick’s internal tissues although there are not in direct contact with digestive enzymes as digestion in ticks takes place intracellular [34]. Oleaga et al. compared the midgut transcriptome of O. erraticus and O. moubata female ticks before and after a blood meal and identified genes whose expression are differentially regulated after feeding [23,25]. They provided novel information’s on argasid genes and proteins involved in the processes of blood digestion, nutrient transport and metabolism and response to the oxidative stress that is produced during blood digestion. Similar results were reported by Landulfo et al. where gut transcriptome analysis revealed several transcripts associated with hemoglobin digestion, such as serine, cysteine and aspartic proteases and metalloenzymes [24]. Phylogenetic analysis of digestive peptidases using transcriptomic data confirmed that most of them are clustered with cathepsin peptidases of other tick species. The midgut transcriptome profile of O. rostratus fourth instar nymphs described by Araujo et al. revealed that Defensin A and Ferritin were the most abundant transcripts represented ~17% of all RNA expressed in the midgut tissue [26]. However, transcriptomic studies exploring the tick immunity in response to pathogen infection are also limited to hard ticks (Amblyomma, Ixodes, Rhipicephalus) and focusing to a few but significant pathogens such as Lyme Borrelia burgdorferi [35] and Borrelia afzelii [36], Anaplasma spp. [37,38,39] and Rickettsia rickettsii [40].
In this study we want to characterize the early transcriptional changes in the midgut of O. moubata in the unfed, engorged and Borrelia duttonii-infected state. The aim is to identify transcripts that are differentially expressed in the respective physiological state and therefore play a key role. Our intention is to contribute to the understanding of the molecular mechanisms, in particular the molecular interaction between soft ticks and relapsing fever borrelia, as this knowledge forms the basis for the development of control strategies in general. To our knowledge, this is the first report of differential expression of midgut transcripts in response to infection with a relapsing fever borrelia in O. moubata.
2. Materials and Methods
2.1. Ticks Rearing and In Vitro Feeding
Colonies of O. moubata were maintained in the insectary within the biosafety level 2 facilities. The ticks were originally received in 2015 from the Department of Veterinary Medicine at the Freie Universität Berlin, where they were maintained over many years. All developmental stages were kept in darkness in an incubator (25 °C; 85% RH) within Drosophila cultivation tubes (Carl Roth, Karlsruhe, Germany) lined with filter paper to absorb coxal fluid and excretory waste. An artificial feeding system was applied to fed the ticks in vitro [41]. Therefore, a feeding chamber is placed on a heating plate adjusted to 36–38 °C, filled with fresh defibrinated pig blood obtained from a commercial source (LOT 35022500/01; Fiebig Nährstofftechnik, Düsseldorf, Germany) and stirred at 700 rpm to simulate a natural pulse. The ticks are placed in a second feeding chamber set on top, which is stretched with a butyric acid-treated Parafilm membrane. Ticks were allowed to feed until they were replete and had detached themselves from the membrane. Subsequently the fed ticks were incubated under the conditions outlined above to enable moulting and oviposition.
2.2. Experimental Setup and Tissue Collection
To address the question of differentially regulated genes through blood feeding and spirochetal infection, midgut samples of female O. moubata that were either unfed, engorged to repletion and infected with Borrelia duttonii were prepared. For each physiological condition, three biological replicates, represented each by single individuals, were processed. To obtain non-infected unfed females, unfed adult O. moubata which had not fed for six months were used. To obtain non-infected engorged females, unfed adult O. moubata were allowed to feed to repletion as described above. Swine blood of the same lot number was used to obtain B. duttonii-infected females, except that the blood was spiked with in vitro cultured B. duttonii at a final concentration of 2 × 106 spirochetes/mL, which corresponds to the spirochaetemia in the blood of an infected BALB/c laboratory mice during the first few days of infection [42]. Spirochetes were purchased from the German National Reference Center for Borrelia and cultured in Barbour-Stoenner-Kelly medium (BSK-H) containing 6% heat-inactivated rabbit serum under standard conditions. The species was confirmed by 16S rRNA gene sequencing. Tick midguts were dissected from unfed females and from fed females 24 h post feeding. Therefore, ticks were briefly washed with sterile phosphate buffer saline (PBS) before dissections. Tick dorsal integuments were cut with a surgical spring scissor and the body cavity was rinsed with PBS for better visualization. Midguts were then removed with forceps and washed briefly in PBS to remove contaminants.
2.3. RNA Extraction, Library Preparation and Sequencing
Washed midguts were snap frozen in liquid nitrogen and cryofractured individually using the cryoPREP impactor (Covaris, Brighton, UK). Total RNA was isolated from the pulverized midgut by adding 750 µL TRIzol LS Reagent (Life Technologies, Darmstadt, Germany) and 250 µL chloroform. After centrifugation at 13,000× g (4 °C) the upper aqueous phase was collected and used as input for the Agencourt RNAdvance Tissue Kit (Beckman Coulter, Krefeld, Germany) in combination with a KingFisher Flex System (Thermo Fisher Scientific, Darmstadt, Germany) for automated extraction. A DNAse I (Qiagen, Hilden, Germany) digestion was included prior to final elution. After that, an appropriate amount of ERCC spike-in mix (Life Technologies, Carlsbad, CA, USA) was combined with 2 µg total RNA for extraction of the poly(A) RNA fraction using the Dynabeads mRNA DIRECT Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Fragmentation and construction of the strand specific whole transcriptome library was done using the Ion Total RNA-Seq Kit v2.0 (Life Technologies, Carlsbad, CA, USA). The quality and integrity of the RNA, cDNA and final library were verified using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and appropriate chips and reagents. The final libraries were quantified using the KAPA Library Quantification Kit (Kapa Biosystems, Cape Town, South Africa) in combination with a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Feldkirchen, Germany). Subsequently, equimolar amounts of the library were then sequenced using an Ion S5XL system in combination with an Ion 540 chip (Life Technologies, Carlsbad, CA, USA) along with appropriate sequencing reagents according to the manufactures instructions.
2.4. De Novo Assembly of O. moubata Midgut Transcriptome
Raw Ion Torrent derived reads were initially quality trimmed and remaining specific adapter sequences as well as host specific rRNA sequences (reference: MF818022.1) were removed using the 454 Sequencing System Software (version 3.0; Roche). The trimmed reads from all biological replicates were combined and then used for a de novo assembly using SPAdes (version 3.15.3; [43]) running in Ion Torrent compatibility mode “--iontorrent” and SPAdes option “--rna” for RNA-Seq data.
Additionally, we received all publicly available Short Read Archive (SRA) datasets for the species O. moubata and individually trimmed them with trim-galore (version 0.6.7) and Cutadapt (version 3.5) using a quality phred score cutoff of 20 and adapter auto-detection. The combined and trimmed SRA datasets were then assembled using SPAdes (version 3.15.3) running in paired-end mode with option “--rna”. Subsequently, the de novo assembled transcript sequences from both, the Ion Torrent raw data obtained in this study and from the SRA, respectively, were combined. The quality of the individual de novo assemblies was benchmarked using BUSCO (version 5.2.2) along with the “arachnida_odb10” lineage dataset (creation date: 5 August 2020, number of genomes: 10, number of BUSCOs: 2934). In order to characterize the transcripts, a custom protein reference was created (n = 854,613 entries). In detail, manually reviewed (Swiss-Prot) and unreviewed entries (TrEMBL) of the UniProt Knowledgebase (https://www.uniprot.org/uniprot; accessed on 16 November 2021) were downloaded. Only TrEMBL entries belonging to the subphylum Chelicerata (NCBI taxonomy ID 6843) were accessed. The Swiss-Prot and TrEMBL protein sequences were clustered using cd-hit (version 4.7; [44]) with default options and a blast database was created. In order to characterize the transcripts, we then matched the de novo sequences to the clustered protein references using blastx (version 2.12.0+) with a cutoff E-value of 0.001.
2.5. Differential Expression Analysis of the Midgut Transcriptome
In order to quantify the expression of the RNA transcripts within each biological replicate of the different treatment groups the software Salmon (version 0.14.1; [45]) was used. Initially, a salmon index was created from the de novo assembled and filtered transcripts using a perfect hash and a k-mer size of 31. Subsequently, the trimmed reads were used for quantification using quasi-mapping mode and options for sequence-specific and fragment GC bias correction, alignment-based verification of mappings and 100 bootstrap samples per library. A tx2gene table was created using the annotation from the blastx analysis and the quantification results were further analyzed using DESeq2 (version 1.26.0; [46]) as available for R (version 3.6.2; https://www.r-project.org, accessed on 16 November 2021) in combination with RStudio (version 1.2.5033; https://www.rstudio.com, accessed on 16 November 2021). For initial data exploration, the read counts were normalized using the regularized log transformation (function rlog) and a principal components analysis was conducted (function plotPCA) using only the 2000 most variable transcripts. The dataset was tested for differential transcript expression (function dds) between the states “engorged vs. unfed—non-infected”, as well as “Borrelia duttonii-infected vs. non-infected—engorged”, using an adjusted p-value of 0.05 and a log2 foldchange of |1| as cutoffs. Log2 foldchanges were shrinked (function lfcShrink) prior to filtering and ranking using the “apeglm” algorithm [47]. Differentially expressed transcripts were subsequently categorized into GO terms using the UniProtKB Retrieve/ID mapping tool.
3. Results
3.1. De Novo Transcriptome Assembly and Functional Annotation of O. moubata Transcriptome
From a total of about 92 million Ion Torrent derived single-end reads from nine O. moubata midgut samples representing three biological treatments a de novo transcriptome was assembled. This transcriptome comprised 15,083 sequences with an average length of 1065.2 bp, a N50 of 1213 bp and a maximum contig length of 5729 bp (Table 1 and Table S1). Additionally, we used roughly 334.5 million Illumina derived paired-end reads from the SRA that represented samples from salivary glands of O. moubata under three different feeding conditions (n = 6) and samples from the O. moubata midgut after feeding (n = 2) (see Table S2). This de novo assembly resulted in 202,950 sequences with an average length of 1943.5 bp, a N50 of 5169 bp and a maximum contig length of 56,176 bp. A combined transcriptome using Ion Torrent derived reads from this study along with SRA derived data simultaneously was not possible, as this is explicitly not recommended by SPAdes developers. Therefore, we decided to combine both assemblies by simple merging them. We did not apply any clustering to reduce redundancy as read counts for duplicated transcripts will be collapsed based on the blastx results.

Table 1.
Transcriptome statistics. Raw reads from this study and from the Sequence Read Archive were assembled into contigs, representing the transcriptome. Transcriptomes were then combined and finally, reads from this study were mapped back to the combined contigs.
The BUSCO analysis showed that of the 2934 BUSCO groups searched, only 45.0% were detected and 44.4% were missing (Figure 1). Within the assembly from the SRA derived data BUSCO detected 97.8% of the BUSCO groups while 1.6% were missing. The combined transcriptome showed comparable results to SRA derived data with slightly higher duplication rate.
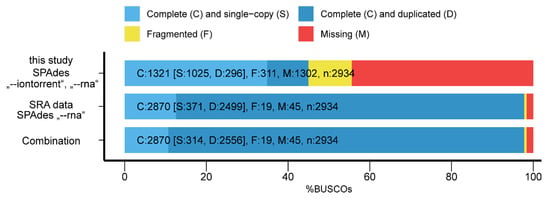
Figure 1.
BUSCO assessment results for the de novo transcriptomes.
3.2. Differential Expression Analysis of the Midgut Transcriptome
Initially, the overall variance within the dataset was inspected using a principal component analysis (Figure 2). Samples of the same group should maintain closest distance, while samples from different groups should be separated. Samples from the “non-infected—unfed” and “non-infected—engorged” groups clustered well together, while samples from the “Borrelia duttonii-infected—engorged” group were more spread. However, samples from both non-infected groups were clearly separated from each other, indicating differences based on starvation. Samples from the “Borrelia duttonii-infected” group were very diverse but different from the other samples.
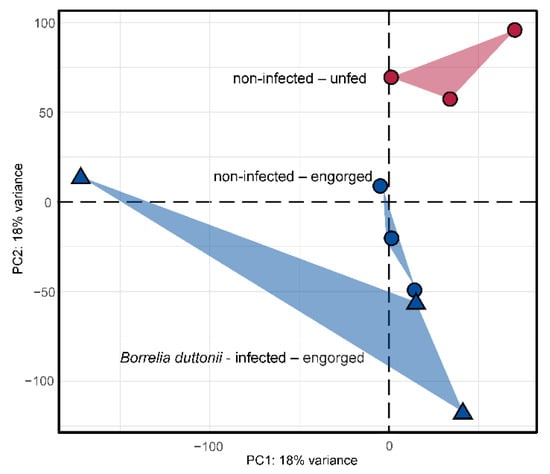
Figure 2.
Principle components analysis for a subset of 2000 transcripts showing the highest variance within the dataset. Read counts were normalized using the regularized log transformation prior to analysis. Samples from non-infected and Borrelia duttonii-infected samples are highlighted with circular or triangle shapes, respectively. Samples from O. moubata ticks that have either been allowed to fed to repletion or unfed are colored in blue and red, respectively.
The gene expression levels were then compared between two physiological states in more detail: “engorged (n = 3) vs. unfed (n = 3)—non-infected” and “Borrelia duttonii-infected (n = 3) vs. non-infected (n = 3)—engorged”. Cut off values were set to 0.05 for the adjusted p-value and >1 for the absolute shrinked log2 foldchange.
A total of 128 transcripts were significantly differentially expressed when comparing “engorged vs. unfed—non-infected” O. moubata ticks (71 and 57 are down- or up-regulated, respectively) (Table S3, Figure 3). Of the 128 transcripts, 108 transcripts were mapped to UniProtKB IDs with or without GO terms, of which 57 are down-regulated and 51 are up-regulated. When comparing “Borrelia duttonii-infected vs. non-infected—engorged” O. moubata ticks, 71 transcripts were significantly differentially expressed (34 and 37 are down- or up-regulated, respectively) (Table S3, Figure 3). Of the 71 transcripts, 59 were mapped to UniProtKB IDs with or without GO terms, of which 27 are downregulated and 32 are up-regulated. In total, 181 transcripts were differentially expressed, including 18 transcripts shared by both groups (Table S3).
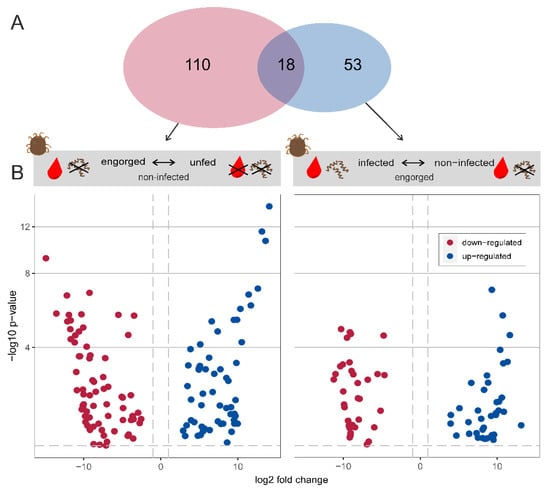
Figure 3.
Differential expression analysis. (A) Venn diagram showing the total number of differentially expressed transcripts in each experimental group. (B) Volcano plot showing differentially expressed transcripts between the states “engorged vs. unfed—non-infected” and “Borrelia duttonii-infected vs. non-infected—engorged”. Blue and red dots represent up- and down-regulated transcripts, respectively. The dashed lines indicate the used thresholds of 0.05 for adjusted p-value and |1| for log2 foldchange.
The comparison with the user-defined protein reference, including 854,613 protein sequences, made it possible to annotate 150 of the 181 differentially expressed transcripts. The remaining 31 transcripts showed no significant matches with other sequences within the databases searched, suggesting that many of the differentially expressed transcripts are previously unknown. Possible reasons for the lack of sequence homology can be artefacts of the assembly process, poor assembly quality, taxonomic isolation of the species and the resulting lack of data. The formation of new genes through duplication and/or transposition mechanisms has already been described [48,49]. The blastx annotated sequences were next functionally characterized using the information available in the Gene Ontology (GO) database and classified according to their molecular function and biological process. Figure 4 shows the classification of differentially expressed transcripts in the midgut of O. moubata in the analysis of “engorged vs. unfed—non-infected” and “Borrelia duttonii-infected vs. non-infected—engorged”. In the molecular Function category of “engorged vs. unfed—non-infected”, the most commonly annotated functions were catalytic activity (n = 46) and binding activity (n = 40), which is consistent with results observed in the midgut transcriptome of O. mimon [24], O. moubata [23] and O. erraticus [25]. Similar results were also documented by other authors in hard ticks [50,51,52,53]. The remaining annotated functions were divided as follows: transporter activity (n = 6), molecular function regulator (n = 3) and ATPase (n = 2). Similarly, the classification of the transcripts according to biological processes resulted in five categories. The following, more frequently occurring categories corresponded to transcripts that were involved in cellular processes (n = 31) and metabolic processes (n = 25). The remaining functional annotations were less numerous, the most common being biological regulation (n = 6), localization (n = 6) and response to stimulus (n = 4).
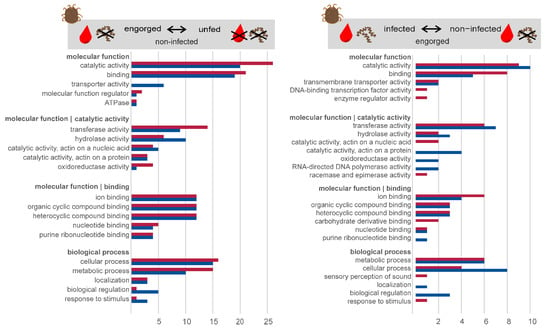
Figure 4.
Classification of differentially expressed transcripts. Gene ontology annotation of up-regulated (red) and down-regulated (blue) transcripts in the midgut of O. moubata in the comparison “engorged vs. unfed—non-infected”, and “Borrelia duttonii-infected vs. non-infected—engorged”. The categories molecular function and biological process are shown.
The classification of differentially expressed transcripts in the midgut of O. moubata in the analysis “Borrelia duttonii-infected vs. non-infected—engorged” shows that only about half as many transcripts are differentially regulated (n = 71). In the molecular function category, the most frequently annotated functions were catalytic activity (n = 19) and binding activity (n = 13). The remaining annotated functions were divided as follows: transmembrane transporter activity (n = 4), DNA-binding transcription factor activity (n = 1) and enzyme regulator activity (n = 1). Similarly, classification of transcripts by biological processes resulted in seven categories. The following more frequent categories corresponded to transcripts involved in cellular processes (n = 12), metabolic processes (n = 12) and biological regulation (n = 3). The remaining functional annotations were less numerous and were divided into four categories, the most frequent being involved in localization (n = 1), sensory perception of sound (n = 1), homeostatic process (n = 1) and response to stimulus (n = 1).
Some selected transcripts, possibly related to feeding and digestion of the blood meal, were examined in more detail after annotation and functional characterization with respect to assignment to specific enzyme classes. Table 2 shows transcripts that are related to the digestion of the blood meal and nutrient uptake. These are mainly oxidoreductases, transferases and hydrolases. Furthermore, among the 59 differentially expressed and annotated transcripts from the analysis of “Borrelia duttonii-infected vs. non-infected—engorged”, we sought to identify those transcripts that might be differentially regulated in response to infection with B. duttonii. As no distinctly immune-related transcripts were significantly differentially expressed, we then searched our assembly for non-differentially expressed immune-related transcripts. This was done by comparison with a user-defined protein reference (n = 1535) extracted from UniProtKB using the organism group “Acari” and the following gene ontology terms: defence, dorin, lysozyme, oxidative stress response, antimicrobial peptide, innate immune response. Out of 1535, 72 proteins are included in our assembly and their expression levels in the midgut of unfed, engorged and B. duttonii-infected O. moubata groups are shown in Figure S1. Among these protein matches, four transcripts, namely defensin, putative lysozyme, Dorin M and OMFREP, showed notably higher but non-significant expression levels in the B. duttonii-infected group (Figure 5).

Table 2.
Selected genes predicted to be involved in protein metabolism, carbohydrate metabolism and transport, lipid metabolism and transport and response to oxidative stress. It is shown the Fold Change Value. Experimental groups 1 and 2 refer to “engorged vs. unfed—non-infected” and “Borrelia duttonii-infected vs. non-infected—engorged”, respectively.
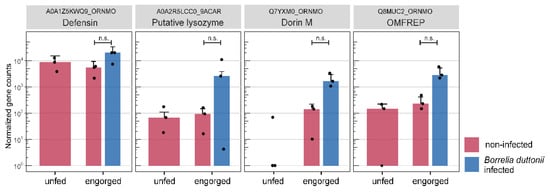
Figure 5.
Immune related transcripts. Boxplots show the normalized gene counts of transcripts that have been related to tick immune response. The different sample groups are highlighted by color: unfed (red), engorged (red) and Borrelia duttonii-infected (blue). Error bars represent the standard deviation and dots represent individual samples; ns: not significant.
4. Discussion and Conclusions
Differential expression analysis is used to detect quantitative changes between experimental groups or conditions and to identify biologically significant differentially expressed genes. Here, we analyzed differentially expressed midgut-specific genes from unfed, engorged and Borrelia dutonii-infected O. moubata and further characterized genes involved in the metabolism of nutrients and in defense response and response to oxidative stress soon after feeding and infection with the relapsing fever spirochete B. duttonii.
4.1. Tick Metabolism
The differences in the feeding, mating and oviposition strategies of the two families Argasidae and Ixodidae are reflected in the process of digestion. While digestion in argasid ticks begins after the tick dropped off the host, the processes of feeding and digestion in ixodid ticks are not separated in time. Digestion in female argasid ticks is also dependent on mating status, which may occur before or after feeding, with no differences in the amount of blood ingested. While mated females digest the blood meal, develop eggs, oviposit and then are ready to repeat the gonotrophic cycle, virgin females may store undigested haemoglobin as a nutrient reserve until mating occurs [54]. By contrast, hard ticks feed only once per life-stage and females die several days after oviposition. What both families have in common, however, is that digestion takes place in the acidic intracellular compartments of the intestinal epithelium [55], unlike in blood-feeding insects, which feed in the neutral pH of the intestinal lumen and digest blood rapidly [56].
By analysing the mialome of O. moubata we have identified 128 transcripts that are differentially expressed between “unfed” and “engorged” O. moubata, suggesting that they are probably involved in protein digestion, carbohydrate metabolism and transport, lipid metabolism and transport and in response to oxidative stress associated with blood feeding (Table 2). Of these, 108 (84.4%) transcripts were annotated and the remaining 20 (15.6%) transcripts were classified as unknown as they did not yield any hits within the searched database. This leads to the assumption that these (unknown) transcripts reflect (i) genes that have not been transcribed in the few soft tick studies conducted so far and are therefore still undiscovered. For example, Oleaga et al. [23] analysed the mialome of unfed O. moubata females and that of fed females 48 h post-feeding, versus 24 h post-feeding in our study, resulting in a different gene expression profile; (ii) genes that correspond to new genes that are unique to the O. moubata strain used due to many years of inbreeding. Charrier et al. [57] studied polymorphism in a highly inbred laboratory strain of Ixodes ricinus and showed that the laboratory strains have less heterozygosity and differ strongly from wild ticks. In this context it is interesting to mention that Pereira De Oliveira et al. observed higher susceptibility for ASFV infection in a long-time lab-reared colony of O. moubata compared to field collected Ornithodoros sp. [58]. They hypothesized that lab-reared ticks are highly inbred with fixed alleles and have a poor microbiome, resulting in a poorly stimulated or even immunologically naïve immune system compared to ticks collected from the field; (iii) represent non-coding RNA (ncRNA) sequences. Non-coding RNAs are known to participate in vector-parasite-host-interaction, either by modulate vertebrate host responses and favour vector survival or to be regulated in the host by parasites to favour parasite survival [59]. Many other studies on both, hard and soft ticks, also described ncRNAs in transcriptomic studies, but their regulatory functions in the processes of blood digestion is unknown [23,50].
In contrast to previously published studies, the number of differentially expressed genes in our study is significantly lower. This can be explained by biological reasons due to no altered expression level in “unfed” and “engorged” condition, or by technical reason due to conservative filtering approaches on RNA-seq data. As shown in the immune-related transcripts (Figure 5), interindividual differences in gene expression can also prevent a statistically significant result. As expected, the most important categories comprised transcripts for the common ‘housekeeping’ genes, i.e., oxidoreductases, hydrolases and transferases. At least one transcript showed identity with a metalloaminopeptidase from UniProtKB entries of O. moubata (A0A1Z5KYM0). In general, metalloproteases have been found in various tick species, including hard ticks and soft ticks and are among the most abundant type of proteases in ticks, which have a broad spectrum of activity that includes fibrinogenolysis [60] and blood protein digestion [61]. Most of them have been described in the salivary glands/saliva, where they are thought to play an important role in inflammation, immunomodulation, fibrinolysis, blood protein digestion, nociception, vitellogenesis, remodelling of extracellular matrix and pathogen transmission [62]. Oleaga et al. identified several transcripts with metallopeptidase activity in the midgut of O. erraticus and O. moubata by proteomic and transcriptomic approach which likely involved in haemoglobin and albumin digestion [23,31]. A recent study, Perner et al. identified zinc-dependent metalloproteases as the predominant salivary immunogens and showed that neutralisation of tick Zn-dependent metalloproteases by rabbit immunoglobulins or inhibition by phosphoramidon prevented ticks from initiating and continuing natural feeding, but not in vitro on an artificial membrane system [63]. They conclude that metalloproteases present in tick saliva target the components of the host hemostatic and defence systems that are absent during artificial membrane feeding.
A transcript (A0A293LQ15) up-regulated in the process of blood digestion was functionally annotated as aspartic-type endopeptidase. These enzymes are involved in the proteolytic pathway of hemoglobin degradation in the digestive vesicles in the midgut cells of hard ticks [64]. Since proteins are the largest and most important content in vertebrate blood, transcripts with lipid and carbohydrate transport and metabolism are differentially expressed in lower numbers. Furthermore, Grandjean et al. describes that lipids and carbohydrates are detectable in the cells two days after the blood meal and can also be stored within the cell [65,66]. Accordingly, the expression of genes involved in the metabolism of carbohydrates and lipids after blood feeding could start at a later stage or maintain a constant level of expression regardless of nutritional status. A transcript which was up-regulated in the course of Borrelia duttonii-infection was functionally annotated as putative chitinase (A0A2R5LGB3). In many blood-feeding arthropods, such as mosquitoes and ticks, the midgut is covered by a peritrophic membrane (PM), primarily composed of chitin, along with glycoproteins and other polycarbohydrates. Beside its function as a structural barrier, the PM can play a critical role in the arthropod innate immune defense against invading pathogens [67]. In Ixodes scapularis, it was shown that the disruption of the PM affect the persistence of Borrelia burgdorferi spirochetes in the gut [68]. On the other hand, parasites such as Plasmodium and Leishmania are known to secrete chitinases that enable their movement and also aid their dissemination through the gut [69,70]. In O. moubata and O. erraticus, enzymes have recently been identified that are thought to be involved in chitin metabolism and in the formation and maintenance of the peritrophic membrane during the digestive process [23,25].
The haemoglobin digestion releases a significant amount of haem and iron, which imbalance the production of reactive oxygen species and antioxidant defences, known as oxidative stress [71]. In ticks, excessive haem is detoxified by aggregation in specialised organelles in the midgut epithelium termed haemosomes [72]. In the midgut transcriptome, we identified up to 10 antioxidants and proteins involved in detoxification processes. Half of which were either up- or down-regulated after feeding which support to maintain a redox homeostasis in the cells. Interestingly, the transcript Cytochrome P450 (A0A1Z5LEL5) was differently expressed in both experimental groups. In the “engorged vs. unfed—non-infected” analysis, Cytochrome P450 was up-regulated after feeding, whereas in the “Borrelia duttonii-infected vs. non-infected—engorged” analysis it was down-regulated in the B. duttonii-infected group. Cytochrome P450s are members of a family of genes whose expression levels can rapidly change in response to external environmental stresses [51]. Cytochrome P450 enzymes have already been identified in saliva of O. moubata, Amblyomma maculatum, I. ricinus [30,73,74] and in ixodid ticks after bacterial and fungal challenge [51,75]. Thus, the P450s are also assumed to participate in detoxification during the infection response process in the tick gut [51]. Beside the heme binding proteins, we identified enzymes involved in the glutathione catabolic process and proteins with sulfotransferase activity. Putative gamma-glutamyltransferase are present in cell membranes and involved in cross cell membrane trafficking of amino acids and peptides as well as glutathione metabolism [76]. Glutathione peroxidases are antioxidant enzymes that catalyse the glutathione-dependent reduction of organic peroxides and hydrogen peroxide and are part of the cellular defence against the effects of oxidative stress. In our study, the expression of glutathione peroxidase is about 12 times higher in fed ticks than in the unfed state and down-regulated in the Borrelia duttonii-infected group compared to the fed group. Bourret et al. documented higher expression values in the salivary glands of Ornithodoros turicata infected with Borrelia turicatae [28]. The upregulation of such oxidases upon infection may represent an attempt by ticks to control pathogens by producing hydrogen peroxide. Narasimhan et al. showed that salp25D, a glutathione peroxidase homologue in the salivary glands of Ixodes scapularis, detoxified reactive oxygen species at the vector-pathogen-host interface, thereby providing a survival advantage to B. burgdorferi at the tick feeding site in mice [77]. However, what direct or indirect influence the redox balance has on B. duttonii infection in O. moubata midgut is unknown. It is also possible that the microbiome balance in the midgut is affected, which again may have an influence on pathogen transmission.
4.2. Tick Immune System
In contrast to the expression patterns in the analysis of “engorged vs. unfed—non-infected”, only half as many transcripts (n = 71) were significantly differentially expressed in the analysis “Borrelia duttonii-infected vs. non-infected—engorged”, of which 34 transcripts are down-regulated and 37 transcripts are up-regulated. Of these, 59 (83.1%) transcripts were annotated and the remaining 12 (16.9%) transcripts were classified as unknown as they did not yield any hits within the searched database. According to the GO annotation, many transcripts are assigned to be integral components of membranes. Integral membrane proteins have a range of important functions, including channelling or transporting molecules across the membrane or acting as cell receptors. As no distinctly immune-related transcripts were significantly differentially expressed, we then searched our assembly for non-differentially expressed immune-related transcripts and identified four transcripts which are slightly up-regulated after infection with Borrelia duttonii. Among these transcripts, two transcripts were annotated as lectins (Q8MUC2, Q7YXM0). It is known that lectins in ticks are involved in the humoral immune response by non-specific pathogen-recognizing defence systems [78]. Dorin M was the first lectin purified from the haemolymph of the soft tick O. moubata and later classified as a member of the fibrinogen-related protein family [79]. This lectin shows preferential binding specificity for sialic acid, N-acetyl-d-hexosamines and sialoglycoproteins [80]. Since sialic acid has been identified as a constituent of bacterial cell walls including the spirochete Borrelia burgdorferi, the authors assume that the plasma lectin dorin M may also take part in mechanisms of transmission of other spirochetes including B. duttonii. Beside dorin M, two more lectins (OmGalec, OMFREP) have been described in the haemocytes, midgut, ovaries and salivary glands of soft ticks of the genus Ornithodoros [81]. Studies characterising lectins in ticks of the genus Ixodes have shown that so-called ixoderins are important for the phagocytosis of Gram-negative bacteria and yeasts by haemocytes in the haemolymph and that their expression is up-regulated after injection of microorganisms, wounding or after blood ingestion [82]. This suggests that lectins in ticks are crucial immune proteins that cause phagocytosis or lysis of microorganisms and that these proteins generally up-regulated in the course of feeding on a potentially infected host. The third transcript which was up-regulated in “B. duttonii-infected” O. moubata was annotated as putative lysozyme (A0A2R5LCC0). Lysozymes are antimicrobial peptides which are stimulated by the ingestion of blood. They have already been isolated in the midgut of soft ticks of the genus Ornithodoros and their role in bacterial defence has been described [25,26,83,84]. In two other studies by Tanaka et al. and Johns et al. [85,86], increased expression of lysozymes were observed when hard ticks of the species Haemaphysalis longicornis and Dermacentor variabilis were exposed to bacteria, also suggesting possible roles of lysozyme as innate immunity of ticks against microorganisms. Apart from the endogenous enzymes like lysozymes, it is proposed that Ixodidae [87] and Argasidae [88] apparently can use the hemoglobin of the host as an antimicrobial agent (hemocidins) in the midgut [34]. The fourth transcript was annotated as defensin (A0A1Z5KWQ9). These small proteins are acting as antimicrobial peptides in vertebrates, invertebrates and plants and serve as a defence against microbial pathogens, primarily bacteria, but also fungi and toxins. In a study by Araujo et al. a defensin that showed ~80% amino acid identity with O. moubata defensin A was by far the most abundant immunity related transcript in the midgut of Ornithodoros rostratus fourth instar nymphs [26]. Interestingly, Nakajima et al. found that synthetic defensin A from O. moubata showed antibacterial activity against many Gram-positive bacteria but not Gram-negative bacteria. Since they did not include Gram-negative Borrelia species in the antibacterial activity assay, we cannot determine whether defensin expression is induced by infection with Borrelia duttonii in our experiments.
Using next-generation sequencing, we obtained a de novo assembly of the midgut transcriptome that significantly increased the genomic resources available for O. moubata. Further, our study is the first RNA-seq study analysing infection-induced changes in gene expression in the midgut of a tick species in the family Argasidae. Our results support previous findings and contribute to a better knowledge of the complexities of the tick metabolism and immune system and pave the way for further study and characterization of proteins that modulate blood digestion and microbial infections in the agasid tick O. moubata.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10030525/s1, Table S1. Basic summary of analysed samples, treatments and sequencing results. Table S2. Additional sequencing data from Ornithodoros moubata, that was included into the de novo assembly. Table S3. List of significantly differentially expressed transcripts. Figure S1. Expression data for selected immune-related transcripts in Ornithodoros moubata.
Author Contributions
Conceptualization, M.S. and C.S.; methodology, M.S., F.P. and D.H.; software, F.P.; validation, M.S. and F.P.; formal analysis, M.S. and F.P.; investigation, M.S.; resources, M.S., F.P. and D.H.; data curation, F.P.; writing—original draft preparation, M.S. and F.P.; writing—review and editing, M.S., F.P., C.S. and D.H.; visualization, M.S. and F.P.; supervision, C.S.; project administration, M.S.; funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Federal Ministry of Education and Research (BMBF, Germany), grant number 01KI1804.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw sequencing data along with deduced Salmon read count tables, de novo assembly and substantial metadata are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress, accessed on 21 February 2022) under the accession number E-MTAB-11507.
Acknowledgments
We would like to thank René Klein (deceased) and Patrick Zitzow for excellent technical assistance in tick colony maintenance and for sequencing, respectively. We thank Volker Fingerle and Cecilia Hitzo-Teufel for providing the Borrelia duttonii culture and Oliver Tauchmann for maintenance of Borrelia duttonii in vitro. The author thanks the reviewers for valuable feedback on an early draft of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, J.; Kopacek, P.; Lew-Tabor, A.; Maritz-Olivier, C. Strategies for new and improved vaccines against ticks and tick-borne diseases. Parasite Immunol. 2016, 38, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Willadsen, P.; Riding, G.A.; McKenna, R.V.; Kemp, D.H.; Tellam, R.L.; Nielsen, J.N.; Lahnstein, J.; Cobon, G.S.; Gough, J.M. Immunologic control of a parasitic arthropod. Identification of a protective antigen from Boophilus microplus. J. Immunol. 1989, 143, 1346–1351. [Google Scholar] [PubMed]
- De la Fuente, J.; Almazan, C.; Canales, M.; de la Lastra, J.M.P.; Kocan, K.M.; Willadsen, P. A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Anim. Health Res. Rev. 2007, 8, 23–28. [Google Scholar] [CrossRef]
- Lew-Tabor, A.E.; Rodriguez Valle, M. A review of reverse vaccinology approaches for the development of vaccines against ticks and tick borne diseases. Ticks Tick Borne Dis. 2016, 7, 573–585. [Google Scholar] [CrossRef]
- Tirloni, L.; Braz, G.; Nunes, R.D.; Gandara, A.C.P.; Vieira, L.R.; Assumpcao, T.C.; Sabadin, G.A.; da Silva, R.M.; Guizzo, M.G.; Machado, J.A.; et al. A physiologic overview of the organ-specific transcriptome of the cattle tick Rhipicephalus microplus. Sci. Rep. 2020, 10, 18296. [Google Scholar] [CrossRef]
- Oleaga, A.; Carnero-Morán, A.; Valero, M.L.; Pérez-Sánchez, R. Proteomics informed by transcriptomics for a qualitative and quantitative analysis of the sialoproteome of adult Ornithodoros moubata ticks. Parasite Vector 2021, 14, 396. [Google Scholar] [CrossRef]
- Chmelar, J.; Kotal, J.; Karim, S.; Kopacek, P.; Francischetti, I.M.B.; Pedra, J.H.F.; Kotsyfakis, M. Sialomes and mialomes: A systems-biology view of tick tissues and tick-host interactions. Trends Parasitol. 2016, 32, 242–254. [Google Scholar] [CrossRef] [Green Version]
- De la Fuente, J.; Contreras, M. Tick vaccines: Current status and future directions. Expert Rev. Vaccines 2015, 14, 1367–1376. [Google Scholar] [CrossRef] [Green Version]
- Merino, O.; Alberdi, P.; de la Lastra, J.M.P.; de la Fuente, J. Tick vaccines and the control of tick-borne pathogens. Front. Cell. Infect. Microbiol. 2013, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Perez-Sanchez, R.; Manzano-Roman, R.; Obolo-Mvoulouga, P.; Oleaga, A. In silico selection of functionally important proteins from the mialome of Ornithodoros erraticus ticks and assessment of their protective efficacy as vaccine targets. Parasit Vectors 2019, 12, 508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Fuente, J.; Estrada-Pena, A. Why new vaccines for the control of ectoparasite vectors have not been registered and commercialized? Vaccines 2019, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- Murfin, K.E.; Fikrig, E. Tick bioactive molecules as novel therapeutics: Beyond vaccine targets. Front. Cell. Infect. Microbiol. 2017, 7, 222. [Google Scholar] [CrossRef] [Green Version]
- Chmelar, J.; Kotal, J.; Kovarikova, A.; Kotsyfakis, M. The use of tick salivary proteins as novel therapeutics. Front. Physiol. 2019, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Franck, C.; Foster, S.R.; Johansen-Leete, J.; Chowdhury, S.; Cielesh, M.; Bhusal, R.P.; Mackay, J.P.; Larance, M.; Stone, M.J.; Payne, R.J. Semisynthesis of an evasin from tick saliva reveals a critical role of tyrosine sulfation for chemokine binding and inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 12657–12664. [Google Scholar] [CrossRef] [PubMed]
- Vial, L. Biological and ecological characteristics of soft ticks (Ixodida: Argasidae) and their impact for predicting tick and associated disease distribution. Parasite 2009, 16, 191–202. [Google Scholar] [CrossRef]
- Elbir, H.; Raoult, D.; Drancourt, M. Relapsing fever borreliae in Africa. Am. J. Trop. Med. Hyg. 2013, 89, 288–292. [Google Scholar] [CrossRef] [Green Version]
- Penrith, M.L.; Bastos, A.D.; Etter, E.M.C.; Beltran-Alcrudo, D. Epidemiology of African swine fever in Africa today: Sylvatic cycle versus socio-economic imperatives. Transbound Emerg. Dis. 2019, 66, 672–686. [Google Scholar] [CrossRef]
- Buysse, M.; Duhayon, M.; Cantet, F.; Bonazzi, M.; Duron, O. Vector competence of the African argasid tick Ornithodoros moubata for the Q fever agent Coxiella burnetii. PLoS Negl. Trop. Dis. 2021, 15, e0009008. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Uzcategui, N.Y.; Gould, E.A.; Nuttall, P.A. Ixodid and argasid tick species and west nile virus. Emerg. Infect. Dis. 2004, 10, 653–657. [Google Scholar] [CrossRef]
- Francischetti, I.M.; Meng, Z.; Mans, B.J.; Gudderra, N.; Hall, M.; Veenstra, T.D.; Pham, V.M.; Kotsyfakis, M.; Ribeiro, J.M. An insight into the salivary transcriptome and proteome of the soft tick and vector of epizootic bovine abortion, Ornithodoros coriaceus. J. Proteom. 2008, 71, 493–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francischetti, I.M.; Mans, B.J.; Meng, Z.; Gudderra, N.; Veenstra, T.D.; Pham, V.M.; Ribeiro, J.M. An insight into the sialome of the soft tick, Ornithodorus parkeri. Insect Biochem. Mol. Biol 2008, 38, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Roman, R.; Perez-Sanchez, R. Functional annotation and analysis of the Ornithodoros moubata midgut genes differentially expressed after blood feeding. Ticks Tick Borne Dis. 2017, 8, 693–708. [Google Scholar] [CrossRef] [PubMed]
- Landulfo, G.A.; Patane, J.S.L.; Silva, D.; de-Azevedo, I.L.M.J.; Mendonca, R.Z.; Simons, S.M.; Carvalho, E.; Barros-Battesti, D.M. Gut transcriptome analysis on females of Ornithodoros mimon (Acari: Argasidae) and phylogenetic inference of ticks. Rev. Bras. Parasitol. Vet. 2017, 26, 185–204. [Google Scholar] [CrossRef] [Green Version]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Roman, R.; Perez-Sanchez, R. De novo assembly and analysis of midgut transcriptome of the argasid tick Ornithodoros erraticus and identification of genes differentially expressed after blood feeding. Ticks Tick Borne Dis. 2018, 9, 1537–1554. [Google Scholar] [CrossRef]
- Araujo, R.N.; Silva, N.C.S.; Mendes-Sousa, A.; Paim, R.; Costa, G.C.A.; Dias, L.R.; Oliveira, K.; Sant’Anna, M.R.V.; Gontijo, N.F.; Pereira, M.H.; et al. RNA-seq analysis of the salivary glands and midgut of the Argasid tick Ornithodoros rostratus. Sci. Rep. 2019, 9, 6764. [Google Scholar] [CrossRef]
- Ribeiro, J.M.; Labruna, M.B.; Mans, B.J.; Maruyama, S.R.; Francischetti, I.M.; Barizon, G.C.; de Miranda Santos, I.K. The sialotranscriptome of Antricola delacruzi female ticks is compatible with non-hematophagous behavior and an alternative source of food. Insect Biochem. Mol. Biol. 2012, 42, 332–342. [Google Scholar] [CrossRef] [Green Version]
- Bourret, T.J.; Boyle, W.K.; Zalud, A.K.; Valenzuela, J.G.; Oliveira, F.; Lopez, J.E. The relapsing fever spirochete Borrelia turicatae persists in the highly oxidative environment of its soft-bodied tick vector. Cell. Microbiol. 2019, 21, e12987. [Google Scholar] [CrossRef] [Green Version]
- Oleaga, A.; Escudero-Poblacion, A.; Camafeita, E.; Perez-Sanchez, R. A proteomic approach to the identification of salivary proteins from the argasid ticks Ornithodoros moubata and Ornithodoros erraticus. Insect Biochem. Mol. Biol. 2007, 37, 1149–1159. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Martin, V.; Manzano-Roman, R.; Valero, L.; Oleaga, A.; Encinas-Grandes, A.; Perez-Sanchez, R. An insight into the proteome of the saliva of the argasid tick Ornithodoros moubata reveals important differences in saliva protein composition between the sexes. J. Proteom. 2013, 80, 216–235. [Google Scholar] [CrossRef] [Green Version]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Roman, R.; Perez-Sanchez, R. Midgut proteome of an argasid tick, Ornithodoros erraticus: A comparison between unfed and engorged females. Parasit. Vectors 2015, 8, 525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Roman, R.; Perez-Sanchez, R. A proteomic insight into the midgut proteome of Ornithodoros moubata females reveals novel information on blood digestion in argasid ticks. Parasit. Vectors 2017, 10, 366. [Google Scholar] [CrossRef] [Green Version]
- Sonenshine, D.E.; Roe, R.M. Biology of Ticks, 2nd ed.; Oxford University Press: New York, NY, USA, 2014. [Google Scholar]
- Hajdusek, O.; Sima, R.; Ayllon, N.; Jalovecka, M.; Perner, J.; de la Fuente, J.; Kopacek, P. Interaction of the tick immune system with transmitted pathogens. Front. Cell. Infect. Microbiol. 2013, 3, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudenko, N.; Golovchenko, M.; Edwards, M.J.; Grubhoffer, L. Differential expression of Ixodes ricinus tick genes induced by blood feeding or Borrelia burgdorferi infection. J. Med. Entomol. 2005, 42, 36–41. [Google Scholar] [CrossRef]
- Mahmood, S.; Sima, R.; Urbanova, V.; Trentelman, J.J.A.; Krezdorn, N.; Winter, P.; Kopacek, P.; Hovius, J.W.; Hajdusek, O. Identification of tick Ixodes ricinus midgut genes differentially expressed during the transmission of Borrelia afzelii spirochetes using a transcriptomic approach. Front. Immunol. 2021, 11, 3731. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, Z.; Torina, A.; Mitra, R.; Alongi, A.; Scimeca, S.; Kocan, K.M.; Galindo, R.C.; Almazan, C.; Blouin, E.F.; Villar, M.; et al. Subolesin expression in response to pathogen infection in ticks. BMC Immunol. 2010, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Mercado-Curiel, R.F.; Palmer, G.H.; Guerrero, F.D.; Brayton, K.A. Temporal characterisation of the organ-specific Rhipicephalus microplus transcriptional response to Anaplasma marginale infection. Int. J. Parasitol. 2011, 41, 851–860. [Google Scholar] [CrossRef] [Green Version]
- Ayllon, N.; Villar, M.; Galindo, R.C.; Kocan, K.M.; Sima, R.; Lopez, J.A.; Vazquez, J.; Alberdi, P.; Cabezas-Cruz, A.; Kopacek, P.; et al. Systems biology of tissue-specific response to Anaplasma phagocytophilum reveals differentiated apoptosis in the tick vector Ixodes scapularis. PLoS Genet. 2015, 11, e1005120. [Google Scholar] [CrossRef] [Green Version]
- Martins, L.A.; Galletti, M.; Ribeiro, J.M.; Fujita, A.; Costa, F.B.; Labruna, M.B.; Daffre, S.; Fogaca, A.C. The distinct transcriptional response of the midgut of Amblyomma sculptum and Amblyomma aureolatum ticks to Rickettsia rickettsii correlates to their differences in susceptibility to infection. Front. Cell. Infect. Microbiol. 2017, 7, 129. [Google Scholar] [CrossRef]
- Schwan, E.V.; Hutton, D.; Shields, K.J.; Townson, S. Artificial feeding and successful reproduction in Ornithodoros moubata moubata (Murray, 1877) (Acarina: Argasidae). Exp. Appl. Acarol. 1991, 13, 107–115. [Google Scholar] [CrossRef]
- Larsson, C.; Lundqvist, J.; van Rooijen, N.; Bergstrom, S. A novel animal model of Borrelia recurrentis louse-borne relapsing fever borreliosis using immunodeficient mice. PLoS Negl. Trop. Dis. 2009, 3, e522. [Google Scholar] [CrossRef] [PubMed]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 2019, 8, giz100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Tautz, D.; Domazet-Loso, T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011, 12, 692–702. [Google Scholar] [CrossRef]
- Mans, B.J.; de Castro, M.H.; Pienaar, R.; de Klerk, D.; Gaven, P.; Genu, S.; Latif, A.A. Ancestral reconstruction of tick lineages. Ticks Tick Borne Dis. 2016, 7, 509–535. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.L.; Cheng, T.Y.; Yang, H.; Liao, Z.H. De novo assembly and analysis of midgut transcriptome of Haemaphysalis flava and identification of genes involved in blood digestion, feeding and defending from pathogens. Infect. Genet. Evol. 2016, 38, 62–72. [Google Scholar] [CrossRef]
- Heekin, A.M.; Guerrero, F.D.; Bendele, K.G.; Saldivar, L.; Scoles, G.A.; Dowd, S.E.; Gondro, C.; Nene, V.; Djikeng, A.; Brayton, K.A. Gut transcriptome of replete adult female cattle ticks, Rhipicephalus (Boophilus) microplus, feeding upon a Babesia bovis-infected bovine host. Parasitol. Res. 2013, 112, 3075–3090. [Google Scholar] [CrossRef]
- Villar, M.; Popara, M.; Ayllon, N.; de Mera, I.G.F.; Mateos-Hernandez, L.; Galindo, R.C.; Manrique, M.; Tobes, R.; de la Fuente, J. A systems biology approach to the characterization of stress response in Dermacentor reticulatus tick unfed larvae. PLoS ONE 2014, 9, e89564. [Google Scholar] [CrossRef] [PubMed]
- Cramaro, W.J.; Revets, D.; Hunewald, O.E.; Sinner, R.; Reye, A.L.; Muller, C.P. Integration of Ixodes ricinus genome sequencing with transcriptome and proteome annotation of the naive midgut. BMC Genom. 2015, 16, 871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akov, S. Blood digestion in ticks. In Physiology of Ticks: Current Themes in Tropical Science; Obenchain, F.D., Galun, R., Eds.; Elsevier: Amsterdam, The Netherlands, 1982. [Google Scholar]
- Grandjean, O.; Aeschlimann, A. Contribution to the study of digestion in ticks: Histology and fine structure of the midgut ephithelium of “Ornithodorus moubata”, Murray (Ixodoidea, Argasidae). Acta Trop. 1973, 30, 193–212. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.N. The Biology of Mosquitoes, Volume 1: Development, Nutrition and Reproduction; Chapman & Hall: London, UK, 1992. [Google Scholar]
- Charrier, N.P.; Couton, M.; Voordouw, M.J.; Rais, O.; Durand-Hermouet, A.; Hervet, C.; Plantard, O.; Rispe, C. Whole body transcriptomes and new insights into the biology of the tick Ixodes ricinus. Parasit. Vectors 2018, 11, 364. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, R.P.; Hutet, E.; Lancelot, R.; Paboeuf, F.; Duhayon, M.; Boinas, F.; de Leon, A.A.P.; Filatov, S.; Le Potier, M.F.; Vial, L. Differential vector competence of Ornithodoros soft ticks for African swine fever virus: What if it involves more than just crossing organic barriers in ticks? Parasit. Vectors 2020, 13, 618. [Google Scholar] [CrossRef]
- Bensaoud, C.; Hackenberg, M.; Kotsyfakis, M. Noncoding RNAs in parasite-vector-host interactions. Trends Parasitol. 2019, 35, 715–724. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; Mather, T.N.; Ribeiro, J.M.C. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem. Biophys. Res. Commun. 2003, 305, 869–875. [Google Scholar] [CrossRef] [Green Version]
- Hatta, T.; Kazama, K.; Miyoshi, T.; Umemiya, R.; Liao, M.; Inoue, N.; Xuan, X.; Tsuji, N.; Fujisaki, K. Identification and characterisation of a leucine aminopeptidase from the hard tick Haemaphysalis longicornis. Int. J. Parasitol. 2006, 36, 1123–1132. [Google Scholar] [CrossRef]
- Ali, A.; Khan, S.; Ali, I.; Karim, S.; Vaz, I.D.; Termignoni, C. Probing the functional role of tick metalloproteases. Physiol. Entomol. 2015, 40, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Perner, J.; Helm, D.; Haberkant, P.; Hatalova, T.; Kropackova, S.; Ribeiro, J.M.; Kopacek, P. The central role of salivary metalloproteases in host acquired resistance to tick feeding. Front. Cell. Infect. Microbiol. 2020, 10, 563349. [Google Scholar] [CrossRef]
- Horn, M.; Nussbaumerová, M.; Sanda, M.; Kovárová, Z.; Srba, J.; Franta, Z.; Sojka, D.; Bogyo, M.; Caffrey, C.R.; Kopácek, P.; et al. Hemoglobin digestion in blood-feeding ticks: Mapping a multipeptidase pathway by functional proteomics. Chem. Biol. 2009, 16, 1053–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grandjean, O. Blood digestion in Ornithodorus moubata Murray sensu stricto Walton females (Ixodoidea: Argasidae) II. Modifications of midgut cells related to the digestive cycle and to the triggering action of mating. Ann. Parasitol. Hum. Comp. 1983, 58, 493–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grandjean, O. Blood digestion in Ornithodoros moubata Murray sensu stricto Walton (Ixodoidea: Argasidae) females I. Biochemical changes in the midgut lumen and ultrastructure of the midgut cell, related to intracellular digestion. Acarologia 1984, 25, 147–165. [Google Scholar]
- Tellam, R.L.; Wijffels, G.; Willadsen, P. Peritrophic matrix proteins. Insect Biochem. Mol. Biol. 1999, 29, 87–101. [Google Scholar] [CrossRef]
- Yang, X.; Koči, J.; Smith, A.A.; Zhuang, X.; Sharma, K.; Dutta, S.; Rana, V.S.; Kitsou, C.; Yas, O.B.; Mongodin, E.F.; et al. A novel tick protein supports integrity of gut peritrophic matrix impacting existence of gut microbiome and Lyme disease pathogens. Cell. Microbiol. 2021, 23, e13275. [Google Scholar] [CrossRef]
- Langer, R.C.; Vinetz, J.M. Plasmodium ookinete-secreted chitinase and parasite penetration of the mosquito peritrophic matrix. Trends Parasitol. 2001, 17, 269–272. [Google Scholar] [CrossRef]
- Schlein, Y.; Jacobson, R.L.; Shlomai, J. Chitinase secreted by Leishmania functions in the sandfly vector. Proc. Biol. Sci. 1991, 245, 121–126. [Google Scholar] [CrossRef]
- Graca-Souza, A.V.; Maya-Monteiro, C.; Paiva-Silva, G.O.; Braz, G.R.; Paes, M.C.; Sorgine, M.H.; Oliveira, M.F.; Oliveira, P.L. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem. Mol. Biol. 2006, 36, 322–335. [Google Scholar] [CrossRef]
- Lara, F.A.; Lins, U.; Paiva-Silva, G.; Almeida, I.C.; Braga, C.M.; Miguens, F.C.; Oliveira, P.L.; Dansa-Petretski, M. A new intracellular pathway of haem detoxification in the midgut of the cattle tick Boophilus microplus: Aggregation inside a specialized organelle, the hemosome. J. Exp. Biol. 2003, 206, 1707–1715. [Google Scholar] [CrossRef] [Green Version]
- Karim, S.; Singh, P.; Ribeiro, J.M. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS ONE 2011, 6, e28525. [Google Scholar] [CrossRef]
- Kotsyfakis, M.; Schwarz, A.; Erhart, J.; Ribeiro, J.M. Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host. Sci. Rep. 2015, 5, 9103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaworski, D.C.; Zou, Z.; Bowen, C.J.; Wasala, N.B.; Madden, R.; Wang, Y.; Kocan, K.M.; Jiang, H.; Dillwith, J.W. Pyrosequencing and characterization of immune response genes from the American dog tick, Dermacentor variabilis (L.). Insect Mol. Biol. 2010, 19, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Giachetto, P.F.; Cunha, R.C.; Nhani, A., Jr.; Garcia, M.V.; Ferro, J.A.; Andreotti, R. Gene expression in the salivary gland of Rhipicephalus (Boophilus) microplus fed on tick-susceptible and tick-resistant hosts. Front. Cell. Infect. Microbiol. 2019, 9, 477. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S.; Sukumaran, B.; Bozdogan, U.; Thomas, V.; Liang, X.; DePonte, K.; Marcantonio, N.; Koski, R.A.; Anderson, J.F.; Kantor, F.; et al. A tick antioxidant facilitates the Lyme disease agent’s successful migration from the mammalian host to the arthropod vector. Cell Host Microbe 2007, 2, 7–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grubhoffer, L.; Kovar, V.; Rudenko, N. Tick lectins: Structural and functional properties. Parasitology 2004, 129, S113–S125. [Google Scholar] [CrossRef] [PubMed]
- Rego, R.O.; Kovar, V.; Kopacek, P.; Weise, C.; Man, P.; Sauman, I.; Grubhoffer, L. The tick plasma lectin, Dorin M, is a fibrinogen-related molecule. Insect Biochem. Mol. Biol. 2006, 36, 291–299. [Google Scholar] [CrossRef]
- Kovar, V.; Kopacek, P.; Grubhoffer, L. Isolation and characterization of Dorin M, a lectin from plasma of the soft tick Ornithodoros moubata. Insect Biochem. Mol. Biol. 2000, 30, 195–205. [Google Scholar] [CrossRef]
- Vechtova, P.; Sterbova, J.; Sterba, J.; Vancova, M.; Rego, R.O.M.; Selinger, M.; Strnad, M.; Golovchenko, M.; Rudenko, N.; Grubhoffer, L. A bite so sweet: The glycobiology interface of tick-host-pathogen interactions. Parasit. Vectors 2018, 11, 594. [Google Scholar] [CrossRef]
- Honig Mondekova, H.; Sima, R.; Urbanova, V.; Kovar, V.; Rego, R.O.M.; Grubhoffer, L.; Kopacek, P.; Hajdusek, O. Characterization of Ixodes ricinus fibrinogen-related proteins (Ixoderins) discloses their function in the tick innate immunity. Front. Cell. Infect. Microbiol. 2017, 7, 509. [Google Scholar] [CrossRef] [Green Version]
- Kopacek, P.; Vogt, R.; Jindrak, L.; Weise, C.; Safarik, I. Purification and characterization of the lysozyme from the gut of the soft tick Ornithodoros moubata. Insect Biochem. Mol. Biol. 1999, 29, 989–997. [Google Scholar] [CrossRef]
- Grunclova, L.; Fouquier, H.; Hypsa, V.; Kopacek, P. Lysozyme from the gut of the soft tick Ornithodoros moubata: The sequence, phylogeny and post-feeding regulation. Dev. Comp. Immunol. 2003, 27, 651–660. [Google Scholar] [CrossRef]
- Tanaka, T.; Kawano, S.; Nakao, S.; Umemiya-Shirafuji, R.; Rahman, M.M.; Boldbaatar, D.; Battur, B.; Liao, M.; Fujisaki, K. The identification and characterization of lysozyme from the hard tick Haemaphysalis longicornis. Ticks Tick Borne Dis. 2010, 1, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Johns, R.; Sonenshine, D.E.; Hynes, W.L. Response of the tick Dermacentor variabilis (Acari: Ixodidae) to hemocoelic inoculation of Borrelia burgdorferi (Spirochetales). J. Med. Entomol. 2000, 37, 265–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogaca, A.C.; da Silva, P.I., Jr.; Miranda, M.T.; Bianchi, A.G.; Miranda, A.; Ribolla, P.E.; Daffre, S. Antimicrobial activity of a bovine hemoglobin fragment in the tick Boophilus microplus. J. Biol. Chem. 1999, 274, 25330–25334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, Y.; Ogihara, K.; Taylor, D.; Yamakawa, M. Antibacterial hemoglobin fragments from the midgut of the soft tick, Ornithodoros moubata (Acari: Argasidae). J. Med. Entomol. 2003, 40, 78–81. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).