Selection and Optimization of High-Yielding DNA Isolation Protocol for Quantitative Analyses of Methanogenic Archaea
Abstract
:1. Introduction
2. Materials and Methods
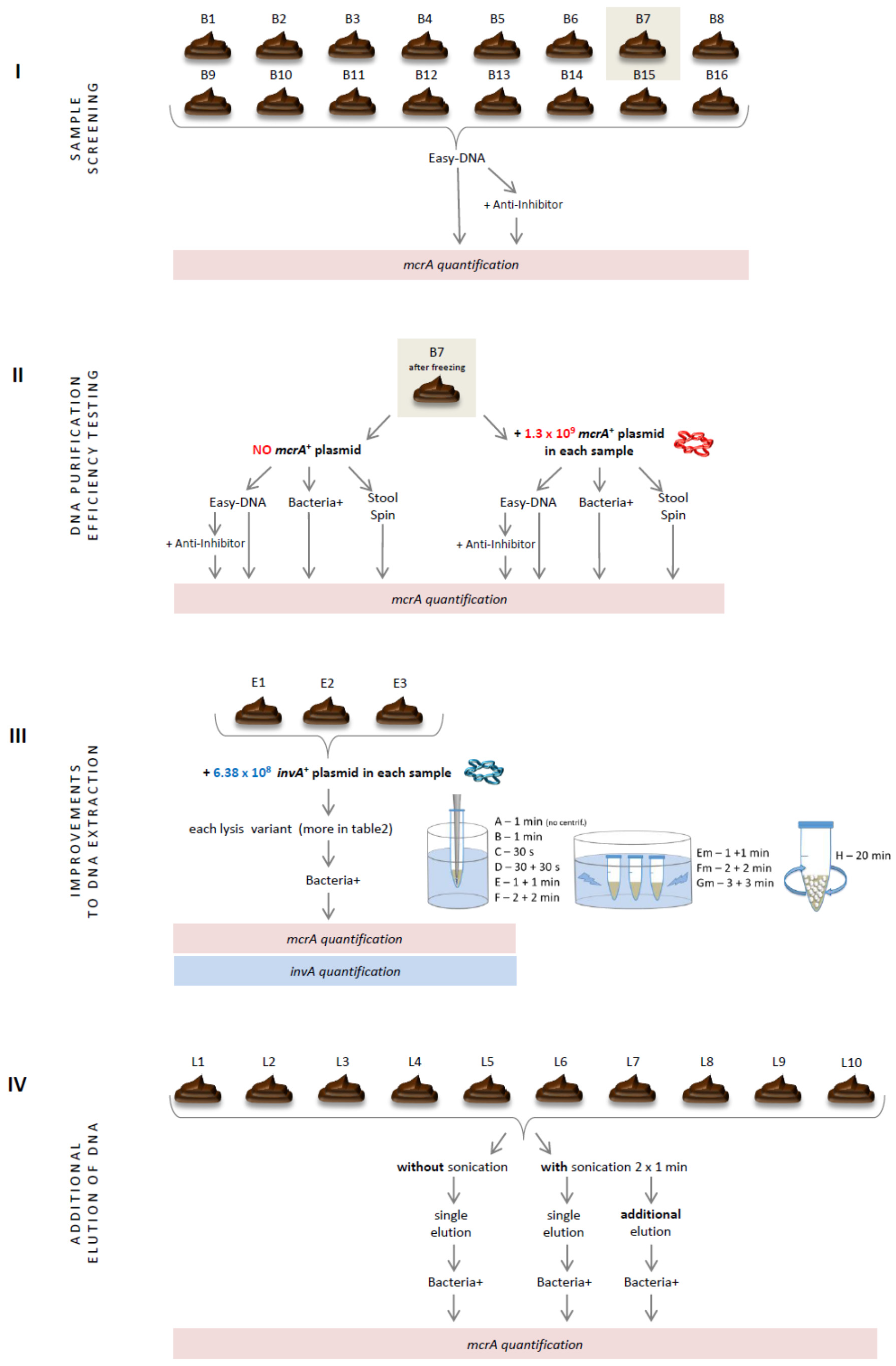
2.1. Sample Screening
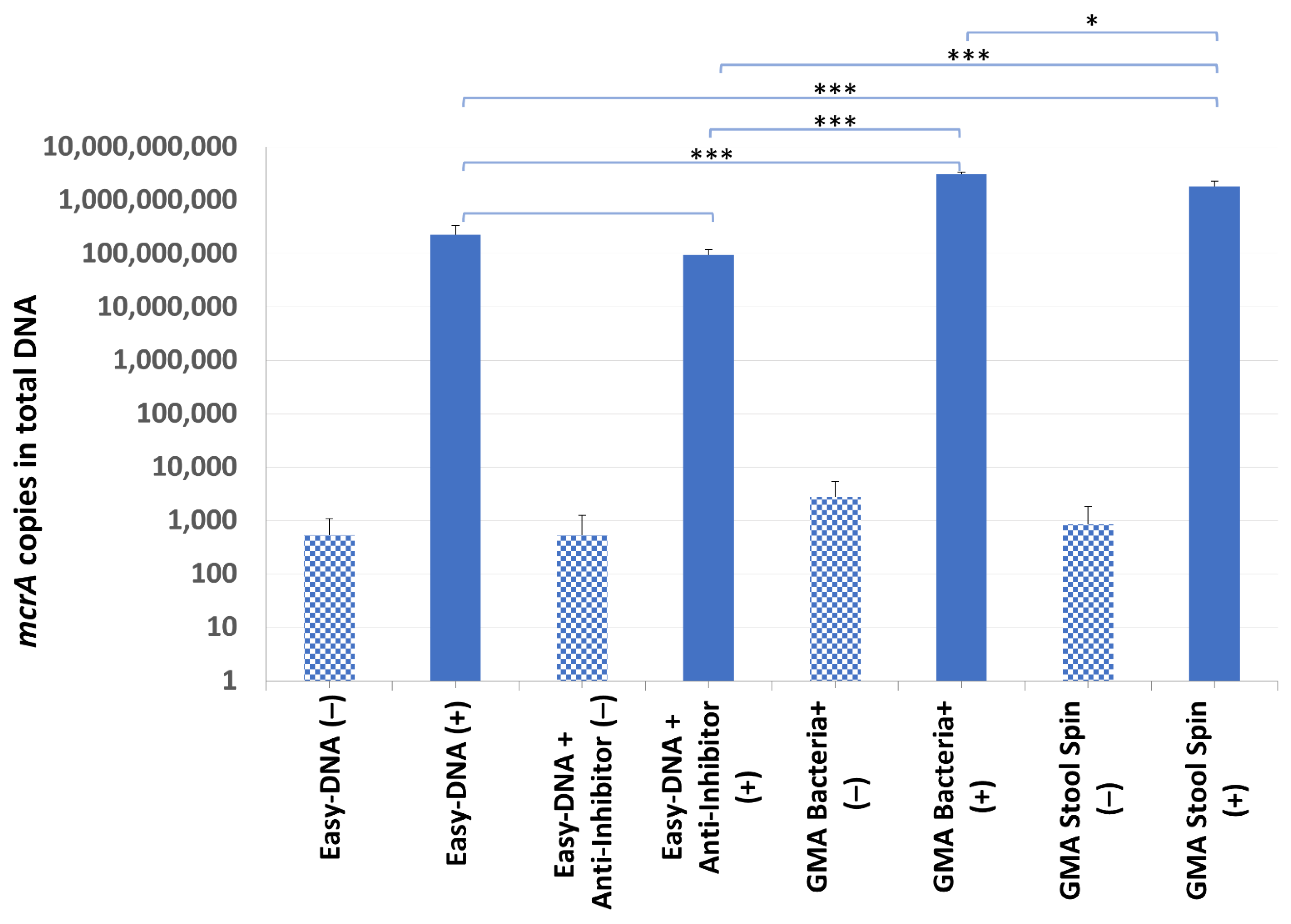
2.2. DNA Purification Efficiency Testing
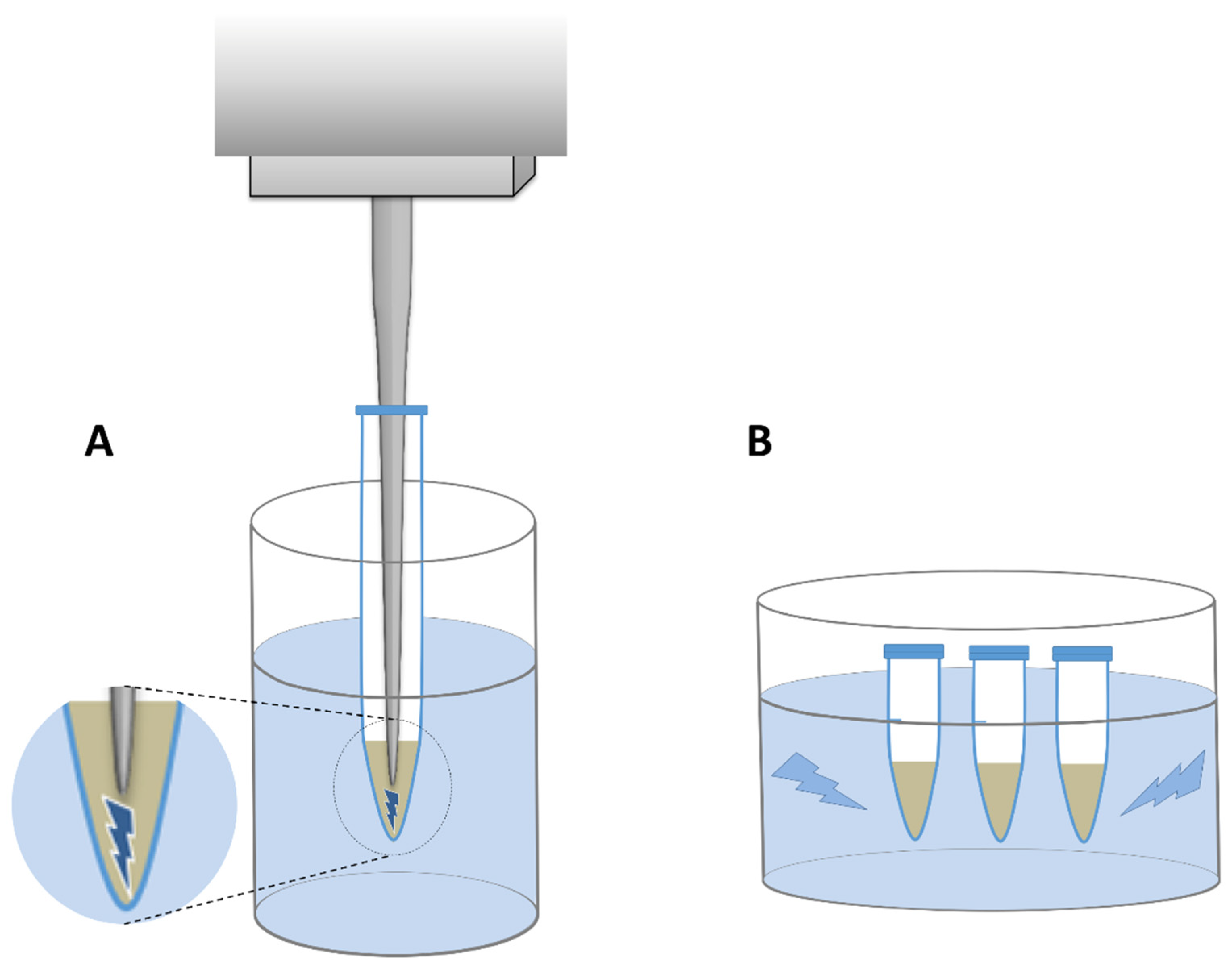
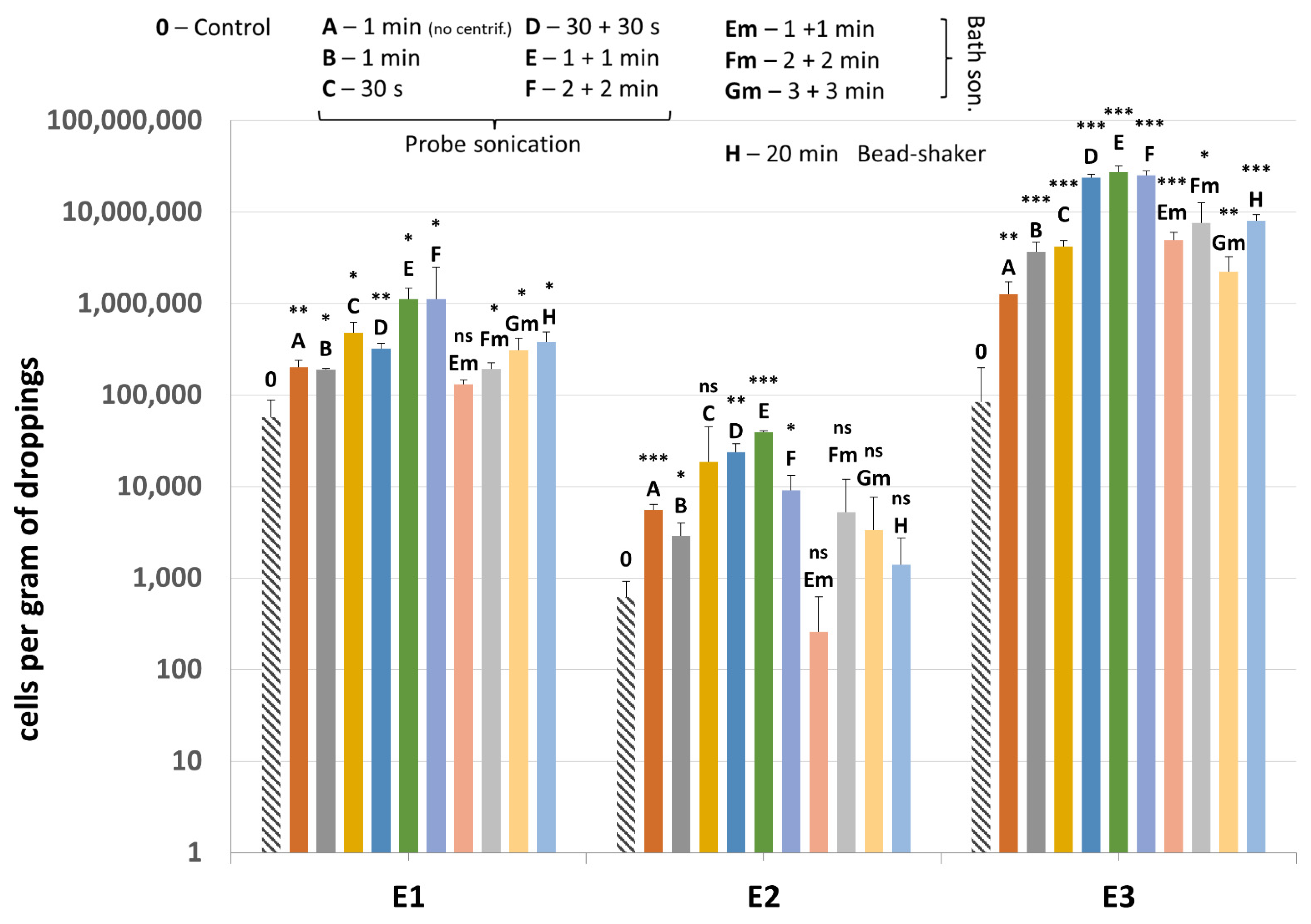
2.3. Improvements to DNA Extraction
2.4. Additional Elution of DNA
3. Results
3.1. Sample Screening
3.2. DNA Purification Efficiency Testing
3.3. Improvements to DNA Extraction
3.4. Additional Elution of DNA
- Transfer 100 mg of a fresh dropping sample (or 200 mg if the sample is rich in plant debris) into a 15 mL tube and add 350–500 µL BS suspension buffer up to the point when the mix becomes viscous, 30 µL lysozyme (10 mg/mL), and 7 µL mutanolysin (10 U/µL). Incubate at 37 °C for 15 min and then at 50 °C for 25 min.
- Add 500 µL LS lysis buffer and 35 µL proteinase K (20 mg/mL). Incubate at 50 °C for 60 min. Mix by hand sporadically.
- Centrifuge at 5500×g for 10 min at room temperature.
- Transfer the supernatant (1st fraction) into a 2 mL tube, add 5 µL RNase A (10 mg/mL), and leave at room temperature. Add 350 µL LS buffer to the remaining pellet and resuspend it with pipette.
- Sonicate the resuspended pellet for 1 min with constant pulse at 40% amplitude in a probe sonicator.
- Centrifuge at 5500×g for 10 min at room temperature.
- Transfer the supernatant (2nd fraction) into 2 mL tube and mix with the 1st fraction. Add 300 µL LS buffer to the remaining pellet and resuspend it with pipette.
- Repeat steps 5–6.
- Transfer the supernatant (3rd fraction) into 2 mL tube and mix with the other two fractions. Leave at room temperature. for 10 min.
- Centrifuge at 12,000×g for 5 min at room temperature.
- Prepare the purification column by adding 800 µL K1 balancing buffer.
- Transfer total supernatant onto purification column. Wait until it flows through the column.
- Add 1.5 mL K2 washing buffer.
- Repeat washing with 1.5 mL K2 washing buffer.
- Add 250 µL K3 elution buffer (this will allow one to discard the dead volume of buffer from the column).
- Place the column into the precipitation tube.
- Add 1 mL K3 buffer. DNA that flows out of the column becomes the 1st eluate. Close the tube.
- Transfer the column into another precipitation tube.
- Add 1 mL K3 buffer (this will result in elution of any residual DNA into the 2nd eluate).
- Add 800 µL PM precipitation buffer to each of the eluates.
- Centrifuge at 10,000×g for 10 min.
- Discard the supernatant.
- Wash the pellets with 500 µL 70% ethanol.
- Centrifuge at 10,000×g for 3 min.
- Discard the supernatant.
- Air dry the pellets for 5 min.
- Suspend the pellets from 1st and 2nd elution with 50 µL Tris buffer (10 mM Tris HCl, pH 8.5).
- Store the DNA solutions at −20 °C.
4. Discussion
4.1. Cell Lysis
4.2. DNA Purification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cisek, A.A.; Binek, M. Chicken intestinal microbiota function with a special emphasis on the role of probiotic bacteria. Pol. J. Vet. Sci. 2014, 17, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Klingl, A. S-layer and cytoplasmic membrane—Exceptions from the typical archaeal cell wall with a focus on double membranes. Front. Microbiol. 2014, 5, 624. [Google Scholar] [CrossRef] [Green Version]
- Saengkerdsub, S.; Anderson, R.C.; Wilkinson, H.H.; Kim, W.K.; Nisbet, D.J.; Ricke, S.C. Identification and quantification of methanogenic archaea in adult chicken ceca. Appl. Environ. Microbiol. 2007, 73, 353–356. [Google Scholar] [CrossRef] [Green Version]
- Steenbakkers, P.J.M.; Geerts, W.J.; Ayman-Oz, N.A.; Keltjens, J.T. Identification of pseudomurein cell wall binding domains. Mol. Microbiol. 2006, 62, 1618–1630. [Google Scholar] [CrossRef] [Green Version]
- Scupham, A.J.; Jones, J.A.; Wesley, I.V. Comparison of DNA extraction methods for analysis of turkey cecal microbiota. J. Appl. Microbiol. 2007, 102, 401–409. [Google Scholar] [CrossRef]
- Science Primer. Available online: http://scienceprimer.com/copy-number-calculator-for-realtime-pcr (accessed on 1 October 2021).
- Steinberg, L.M.; Regan, J.M. Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl. Environ. Microbiol. 2008, 74, 6663–6671. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Mustapha, A. Multiplex TaqMan® detection of pathogenic and multi-drug resistant Salmonella. Int. J. Food Microbiol. 2013, 166, 213–218. [Google Scholar] [CrossRef]
- Danzeisen, J.L.; Kim, H.B.; Isaacson, R.E.; Tu, Z.J.; Johnson, T.J. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS ONE 2011, 6, e27949. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, S.I.; Ricke, S.C. Microbial populations in naked neck chicken ceca raised on pasture flock fed with commercial yeast cell wall prebiotics via an Illumina MiSeq platform. PLoS ONE 2016, 11, e0151944. [Google Scholar] [CrossRef]
- Qu, A.; Brulc, J.M.; Wilson, M.K.; Law, B.F.; Theoret, J.R.; Joens, L.A.; Konkel, M.E.; Angly, F.; Dinsdale, E.A.; Edwards, R.A.; et al. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE 2008, 3, e2945. [Google Scholar] [CrossRef] [Green Version]
- Sergeant, M.J.; Constantinidou, C.; Cogan, T.A.; Bedford, M.R.; Penn, C.W.; Pallen, M.J. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS ONE 2014, 9, e91941. [Google Scholar] [CrossRef]
- Thibodeau, A.; Fravalo, P.; Yergeau, E.; Arsenault, J.; Lahaye, L.; Letellier, A. Chicken caecal microbiome modifications induced by Campylobacter jejuni colonization and by a non-antibiotic feed additive. PLoS ONE 2015, 10, e0131978. [Google Scholar] [CrossRef] [Green Version]
- Ghavami, S.B.; Sephay, A.A.; Aghdaei, H.A. Comparison study on effect of different methods on DNA extraction of Methanobrevibacter smithii. Biol. Forum. Res. Trend 2015, 7, 549–553. [Google Scholar]
- Salonen, A.; Nikkilä, J.; Jalanka-Tuovinen, J.; Immonen, O.; Rajilić-Stojanović, M.; Kekkonen, R.A.; Palva, A.; de Vos, W.M. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 2010, 81, 127–134. [Google Scholar] [CrossRef]
- Mirmohammadsadeghi, H.; Abedi, D.; Mohmoudpour, H.R.; Akbari, V. Comparison of five methods for extraction of genomic DNA from a marine Archaea, Pyrococcus furiosus. Pak. J. Med. Sci. 2013, 29, 390–394. [Google Scholar] [CrossRef]
- Altermann, E.; Schofield, L.R.; Ronimus, R.S.; Beatty, A.K.; Reilly, K. Inhibition of Rumen methanogens by a novel archaeal lytic enzyme displayed on tailored bionanoparticles. Front. Microbiol. 2018, 9, 2378. [Google Scholar] [CrossRef] [Green Version]
- Kaur, G.; Garg, T.; Rath, G.; Goyal, A.K. Drug Delivery Archaeosomes: An excellent carrier for drug and cell delivery Archaeosomes: An excellent carrier for drug and cell delivery. Drug Deliv. 2016, 23, 2497–2512. [Google Scholar] [CrossRef]
- Van Wolferen, M.; Wagner, A.; Van Der Does, C.; Albers, S.V. The archaeal Ced system imports DNA. Proc. Natl. Acad. Sci. USA 2016, 113, 2496–2501. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Greening, C.; Rattray, J.E.; Chakraborty, A.; Chuvochina, M.; Mayumi, D.; Dolfing, J.; Li, C.; Brooks, J.M.; Bernard, B.B.; et al. Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat. Commun. 2019, 10, 1816. [Google Scholar] [CrossRef] [Green Version]
- Rantakokko-Jalava, K.; Jalava, J. Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J. Clin. Microbiol. 2002, 40, 4211–4217. [Google Scholar] [CrossRef] [Green Version]
- Meimandipour, A.; Shuhaimi, M.; Soleimani, A.F.; Azhar, K.; Hair-Bejo, M.; Kabeir, B.M.; Javanmard, A.; Muhammad Anas, O.; Yazid, A.M. Selected microbial groups and short-chain fatty acids profile in a simulated chicken cecum supplemented with two strains of Lactobacillus. Poult. Sci. 2010, 89, 470–476. [Google Scholar] [CrossRef]
- The Ribosomal RNA Database. Available online: https://rrndb.umms.med.umich.edu/ (accessed on 24 November 2021).
- Pauwels, J.; Taminiau, B.; Janssens, G.P.J.; De Beenhouwer, M.; Delhalle, L.; Daube, G.; Coopman, F. Cecal drop reflects the chickens’ cecal microbiome, fecal drop does not. J. Microbiol. Methods 2015, 117, 164–170. [Google Scholar] [CrossRef] [Green Version]




| Primer/Probe Name | Sequence | Target | Reference |
|---|---|---|---|
| mcrA_F3 | CTTGAARMTCACTTCGGTGGWTC | mcrA in methanogenic archaea (271 bp) | this study |
| mcrA_R1 (=mcrA-rev) | CGTTCATBGCGTAGTTVGGRTAGT | [7] | |
| mcrA_probe2 | [6FAM]TM[+G]GHT[+T]C[+T]WYGGWTWCGA[BHQ1] * | this study | |
| Sal_F5b | GTCCAGTTTATCGTTATTACCAAAGG | invA in Salmonella Typhimurium (200 bp) | modified from [8] |
| Sal_R5b | ATCGCACCGTCAAAGGA | ||
| Sal_probe1 | [6FAM]TTC[+T]CTGGA[+T]GGTATGCCCG[BHQ1] * | this study |
| Lysis Variant | Step 1—Enzymatic Lysis | Step 2—Centrifuging 1 | Step 3—Mechanical Lysis I | Step 4—Centrifuging 2 | Step 5—Mechanical Lysis II | Step 6—Centrifuging 3 | Step 7—Purification | |
|---|---|---|---|---|---|---|---|---|
| Probe sonicator | A | yes | no | 1 min | yes | - | - | GMA Bacteria+ |
| B | yes | yes | 1 min | yes | - | - | ||
| C | yes | yes | 30 s | yes | - | - | ||
| D | yes | yes | 30 s | yes | 30 s | yes | ||
| E | yes | yes | 1 min | yes | 1 min | yes | ||
| F | yes | yes | 2 min | yes | 2 min | yes | ||
| Bath sonicator | Em | yes | yes | 1 min | yes | 1 min | yes | |
| Fm | yes | yes | 2 min | yes | 2 min | yes | ||
| Gm | yes | yes | 3 min | yes | 3 min | yes | ||
| Shaker—H | yes | yes | 20 min, constant | yes, twice | ||||
| Control—0 | yes | yes | - | - | - | - | ||
| DNA Isolation Kit 1 | Average DNA Concentration [ng/µL] | Total DNA Yield [µg] | A260/280 Ratio |
|---|---|---|---|
| Easy-DNA (−) | 483.76 ± 39.244 | 48.38 | 1.45 |
| Easy-DNA (+) | 811.99 ± 68.165 | 81.20 | 1.49 |
| Easy-DNA + Anti-Inhibitor (−) | 123.85 ± 0.141 | 12.38 | 1.7 |
| Easy-DNA + Anti-Inhibitor (+) | 288.63 ± 0.424 | 28.86 | 1.78 |
| GMA Bacteria+ (−) | 51.44 ± 0.212 | 2.57 | 1.85 |
| GMA Bacteria+ (+) | 51.42 ± 0.580 | 2.57 | 1.85 |
| GMA Stool Spin (−) | 7.9 ± 0.212 | 0.79 | 1.66 |
| GMA Stool Spin (+) | 9.68 ± 0.269 | 0.97 | 1.59 |
| Sample | A | B | C | D | E | F | Em | Fm | Gm | H |
|---|---|---|---|---|---|---|---|---|---|---|
| E1 | 3.5 | 3.3 | 8.3 | 5.6 | 19.5 | 19.5 | 2.3 | 3.3 | 5.4 | 6.7 |
| E2 | 8.9 | 4.6 | 29.7 | 38.0 | 62.9 | 14.5 | 0.4 | 8.4 | 5.3 | 2.2 |
| E3 | 15.1 | 44.0 | 50.1 | 283.0 | 323.9 | 299.2 | 58.4 | 90.3 | 26.5 | 95.3 |
| average | 9.1 | 17.3 | 29.4 | 108.9 | 135.4 | 111.1 | 20.4 | 34.0 | 12.4 | 34.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cisek, A.A.; Bąk, I.; Stefańska, I.; Binek, M. Selection and Optimization of High-Yielding DNA Isolation Protocol for Quantitative Analyses of Methanogenic Archaea. Microorganisms 2022, 10, 523. https://doi.org/10.3390/microorganisms10030523
Cisek AA, Bąk I, Stefańska I, Binek M. Selection and Optimization of High-Yielding DNA Isolation Protocol for Quantitative Analyses of Methanogenic Archaea. Microorganisms. 2022; 10(3):523. https://doi.org/10.3390/microorganisms10030523
Chicago/Turabian StyleCisek, Agata Anna, Iwona Bąk, Ilona Stefańska, and Marian Binek. 2022. "Selection and Optimization of High-Yielding DNA Isolation Protocol for Quantitative Analyses of Methanogenic Archaea" Microorganisms 10, no. 3: 523. https://doi.org/10.3390/microorganisms10030523
APA StyleCisek, A. A., Bąk, I., Stefańska, I., & Binek, M. (2022). Selection and Optimization of High-Yielding DNA Isolation Protocol for Quantitative Analyses of Methanogenic Archaea. Microorganisms, 10(3), 523. https://doi.org/10.3390/microorganisms10030523






