Synergy of Cellulase Systems between Acetivibrio thermocellus and Thermoclostridium stercorarium in Consolidated-Bioprocessing for Cellulosic Ethanol
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Culturing Conditions
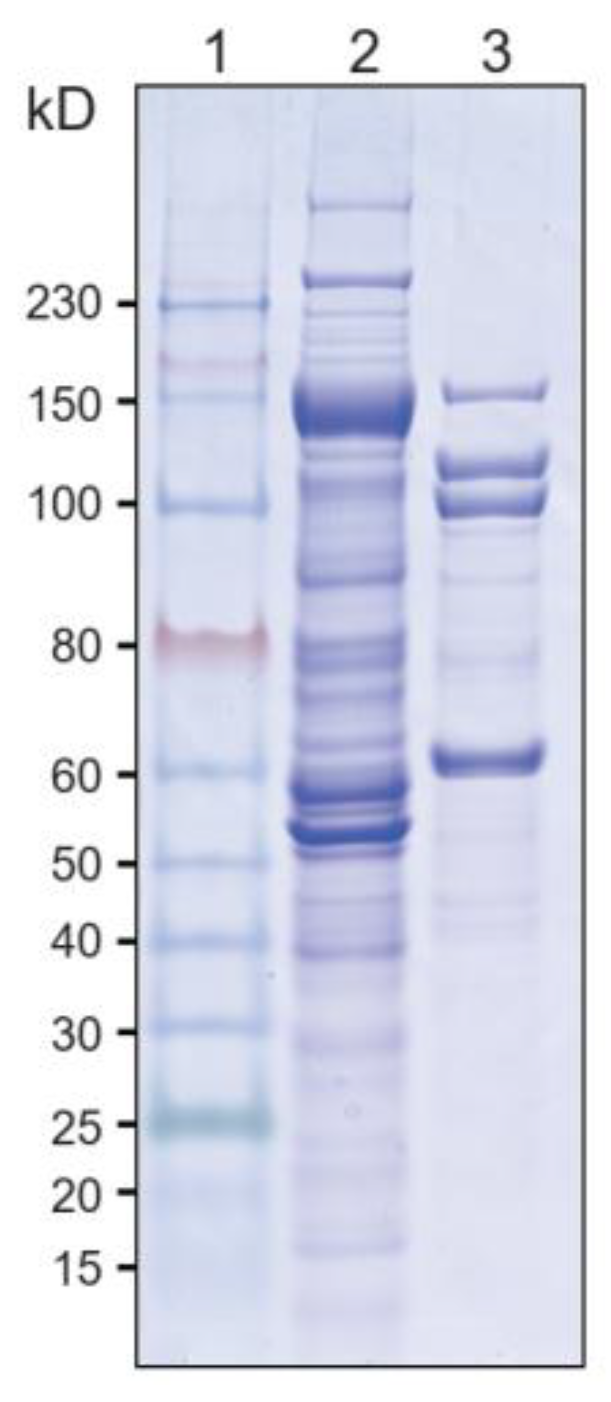
2.2. Cellulase Systems Isolation
2.3. Enzyme Activity Measurement
2.4. Synergistic Interaction Assay
2.5. Fermentation and Analytical Methods
3. Results
3.1. Comparison of Components of Cellulase Systems between A. thermocellus and T. stercorarium
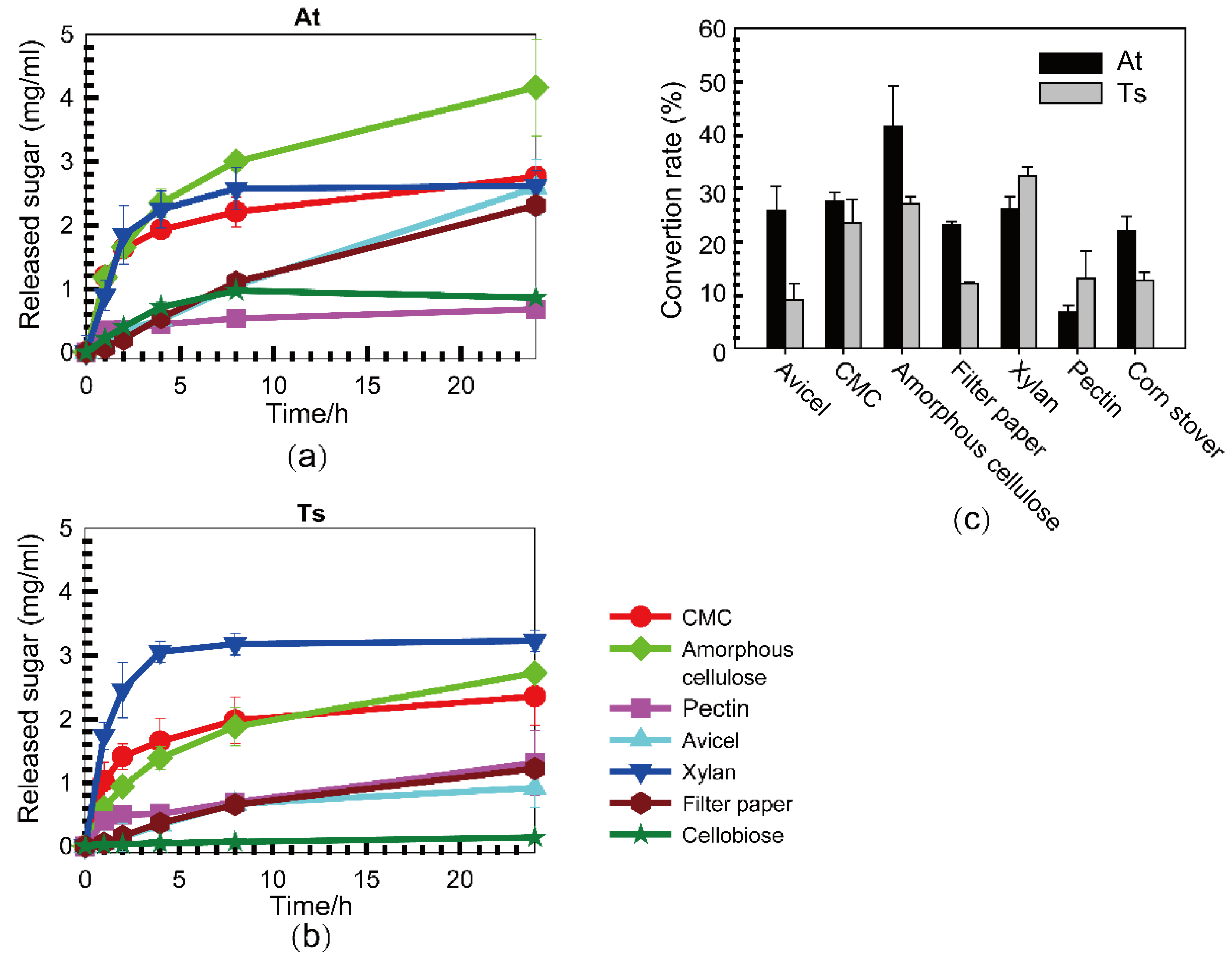
3.2. Substrate Specificities of Cellulase Systems from A. thermocellus and T. stercorarium
3.3. The Synergistic Effect of Cellulase Systems between A. thermocellus and T. stercorarium
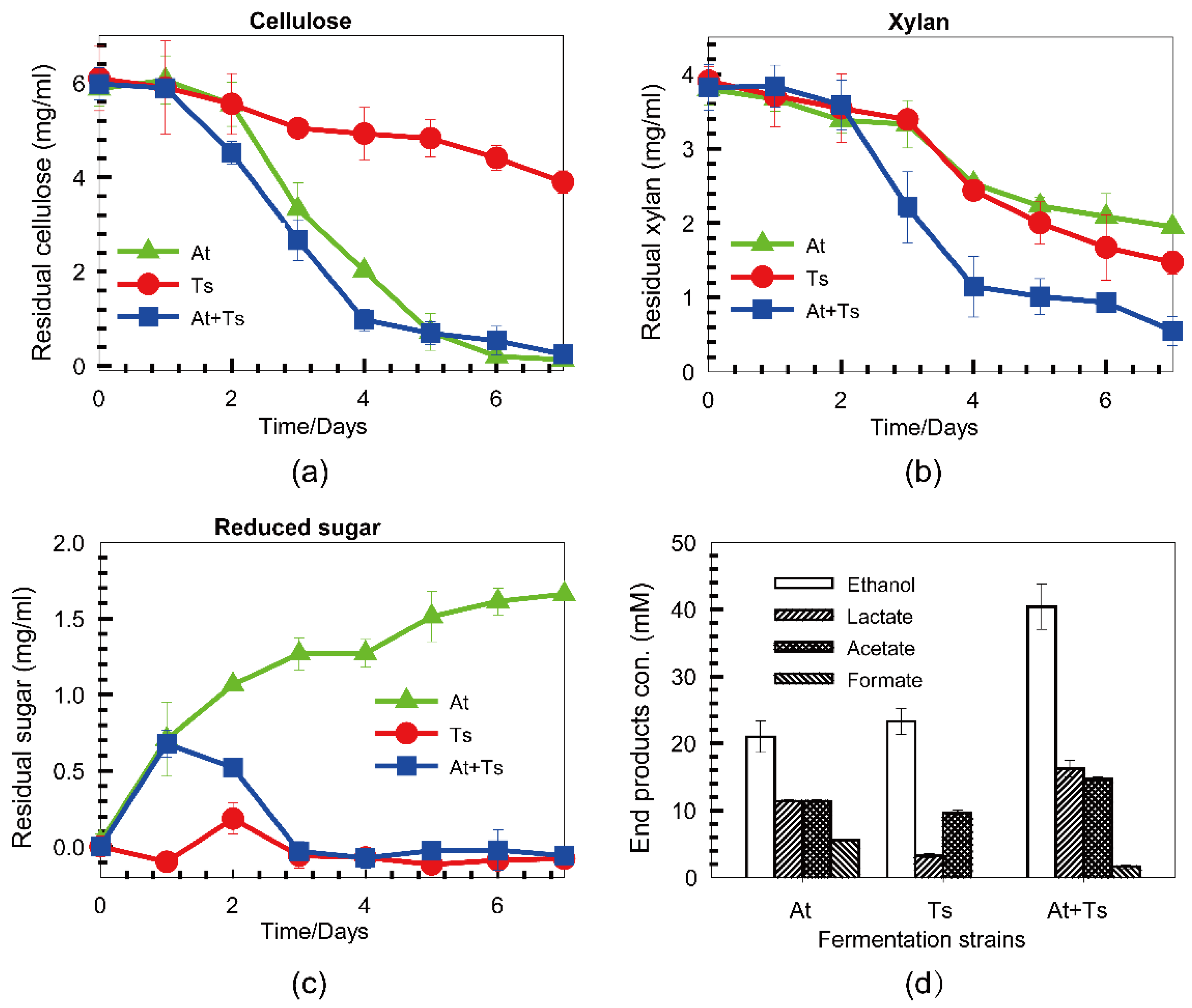
3.4. Co-Fermentation of A. thermocellus and T. stercorarium with a Mixture of Cellulose and Xylan
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Lynd, L.R. The grand challenge of cellulosic biofuels. Nat. Biotechnol. 2017, 35, 912–915. [Google Scholar] [CrossRef] [PubMed]
- Chandel, A.K.; Garlapati, V.K.; Singh, A.K.; Antunes, F.A.F.; da Silva, S.S. The path forward for lignocellulose biorefineries: Bottlenecks, solutions, and perspective on commercialization. Bioresour Technol. 2018, 264, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Antunes, F.A.F.; Chandel, A.K.; Teran-Hilares, R.; Ingle, A.P.; Rai, M.; Dos Santos Milessi, T.S.; da Silva, S.S.; Dos Santos, J.C. Overcoming challenges in lignocellulosic biomass pretreatment for second-generation (2G) sugar production: Emerging role of nano, biotechnological and promising approaches. Biotechnology 2019, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Sanford, K.; Chotani, G.; Danielson, N.; Zahn, J.A. Scaling up of renewable chemicals. Curr. Opin. Biotechnol. 2016, 38, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Pratto, B.; de Souza, R.B.; Sousa, R., Jr.; da Cruz, A.J. Enzymatic Hydrolysis of Pretreated Sugarcane Straw: Kinetic Study and Semi-Mechanistic Modeling. Appl. Biochem. Biotechnol. 2016, 178, 1430–1444. [Google Scholar] [CrossRef]
- Hans, M.; Lugani, Y.; Chandel, A.K.; Rai, R.; Kumar, S. Production of first- and second-generation ethanol for use in alcohol-based hand sanitizers and disinfectants in India. Biomass. Convers. Biorefin. 2021, 11, 18. [Google Scholar] [CrossRef]
- Lynd, L.R.; Laser, M.S.; Bransby, D.; Dale, B.E.; Davison, B.; Hamilton, R.; Himmel, M.; Keller, M.; McMillan, J.D.; Sheehan, J.; et al. How biotech can transform biofuels. Nat. Biotechnol. 2008, 26, 169–172. [Google Scholar] [CrossRef]
- Olson, D.G.; McBride, J.E.; Shaw, A.J.; Lynd, L.R. Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 2012, 23, 396–405. [Google Scholar] [CrossRef]
- Parisutham, V.; Kim, T.H.; Lee, S.K. Feasibilities of consolidated bioprocessing microbes: From pretreatment to biofuel production. Bioresour. Technol. 2014, 161, 431–440. [Google Scholar] [CrossRef]
- Den Haan, R.; van Rensburg, E.; Rose, S.H.; Gorgens, J.F.; van Zyl, W.H. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing. Curr. Opin. Biotechnol. 2015, 33, 32–38. [Google Scholar] [CrossRef]
- Singh, N.; Mathur, A.S.; Tuli, D.K.; Gupta, R.P.; Barrow, C.J.; Puri, M. Cellulosic ethanol production via consolidated bioprocessing by a novel thermophilic anaerobic bacterium isolated from a Himalayan hot spring. Biotechnol. Biofuels 2017, 10, 73. [Google Scholar] [CrossRef]
- Xu, C.; Qin, Y.; Li, Y.; Ji, Y.; Huang, J.; Song, H.; Xu, J. Factors influencing cellulosome activity in Consolidated Bioprocessing of cellulosic ethanol. Bioresour. Technol. 2010, 101, 9560–9569. [Google Scholar] [CrossRef]
- Olson, D.G.; Horl, M.; Fuhrer, T.; Cui, J.; Zhou, J.; Maloney, M.I.; Amador-Noguez, D.; Tian, L.; Sauer, U.; Lynd, L.R. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab. Eng. 2017, 39, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, S.; Li, R.; Hong, W.; Xiao, Y.; Feng, Y.; Cui, Q.; Liu, Y.J. Efficient whole-cell-catalyzing cellulose saccharification using engineered Clostridium thermocellum. Biotechnol. Biofuels 2017, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.G.; Maloney, M.; Lanahan, A.A.; Hon, S.; Hauser, L.J.; Lynd, L.R. Identifying promoters for gene expression in Clostridium thermocellum. Metab. Eng. Commun. 2015, 2, 23–29. [Google Scholar] [CrossRef]
- Artzi, L.; Bayer, E.A.; Morais, S. Cellulosomes: Bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 2017, 15, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Ravachol, J.; Borne, R.; Meynial-Salles, I.; Soucaille, P.; Pages, S.; Tardif, C.; Fierobe, H.P. Combining free and aggregated cellulolytic systems in the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Biotechnol. Biofuels 2015, 8, 114. [Google Scholar] [CrossRef]
- Paye, J.M.; Guseva, A.; Hammer, S.K.; Gjersing, E.; Davis, M.F.; Davison, B.H.; Olstad, J.; Donohoe, B.S.; Nguyen, T.Y.; Wyman, C.E. Biological lignocellulose solubilization: Comparative evaluation of biocatalysts and enhancement via cotreatment. Biotechnol. Biofuels 2016, 9, 8. [Google Scholar] [CrossRef]
- Broeker, J.; Mechelke, M.; Baudrexl, M.; Mennerich, D.; Hornburg, D.; Mann, M.; Schwarz, W.H.; Liebl, W.; Zverlov, V.V. The hemicellulose-degrading enzyme system of the thermophilic bacterium Clostridium stercorarium: Comparative characterisation and addition of new hemicellulolytic glycoside hydrolases. Biotechnol. Biofuels 2018, 11, 229. [Google Scholar] [CrossRef]
- Fardeau, M.L.; Ollivier, B.; Garcia, J.L.; Patel, B.K.C. Transfer of Thermobacteroides leptospartum and Clostridium thermolacticum as Clostridium stercorarium subsp leptospartum subsp nov., comb. nov and C. stercorarium subsp thermolacticum subsp nov., comb. nov. Int. J. Syst. Evo. Micr. 2001, 51, 1127–1131. [Google Scholar] [CrossRef][Green Version]
- Poehlein, A.; Zverlov, V.V.; Daniel, R.; Schwarz, W.H.; Liebl, W. Complete Genome Sequence of Clostridium stercorarium subsp. stercorarium Strain DSM 8532, a Thermophilic Degrader of Plant Cell Wall Fibers. Genome Announc. 2013, 1, e0007313. [Google Scholar] [CrossRef] [PubMed]
- Fontes, C.M.; Gilbert, H.J. Cellulosomes: Highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 2010, 79, 655–681. [Google Scholar] [CrossRef]
- Artzi, L.; Morag, E.; Barak, Y.; Lamed, R.; Bayer, E.A. Clostridium clariflavum: Key Cellulosome Players Are Revealed by Proteomic Analysis. Microbiology 2015, 6, e00411–e00415. [Google Scholar] [CrossRef]
- Herlet, J.; Schwarz, W.H.; Zverlov, V.V.; Liebl, W.; Kornberger, P. Addition of beta-galactosidase boosts the xyloglucan degradation capability of endoglucanase Cel9D from Clostridium thermocellum. Biotechnol. Biofuels 2018, 11, 238. [Google Scholar] [CrossRef]
- Cui, G.Z.; Hong, W.; Zhang, J.; Li, W.L.; Feng, Y.; Liu, Y.J.; Cui, Q. Targeted gene engineering in Clostridium cellulolyticum H10 without methylation. J. Microbiol. Methods 2012, 89, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Lamounier, K.F.R.; Rodrigues, P.O.; Pasquini, D.; Dos Santos, A.S.; Baffi, M.A. Ethanol Production and Other Bioproducts by Galactomyces geotrichum from Sugarcane Bagasse Hydrolysate. Curr. Microbiol. 2020, 77, 738–745. [Google Scholar] [CrossRef]
- Lamed, R.; Kenig, R.; Setter, E. Major characteristics of the cellulolytic system of Clostrdium thermocellum coincide with those of the purified cellulosome. Enzym. Microb. Technol. 1985, 7, 37–41. [Google Scholar] [CrossRef]
- Han, S.O.; Yukawa, H.; Inui, M.; Doi, R.H. Effect of carbon source on the cellulosomal subpopulations of Clostridium cellulovorans. Microbiology 2005, 151, 1491–1497. [Google Scholar] [CrossRef][Green Version]
- Qi, M.; Jun, H.S.; Forsberg, C.W. Characterization and synergistic interactions of Fibrobacter succinogenes glycoside hydrolases. Appl. Environ. Microbiol. 2007, 73, 6098–6105. [Google Scholar] [CrossRef]
- Tokin, R.; Ipsen, J.O.; Westh, P.; Johansen, K.S. The synergy between LPMOs and cellulases in enzymatic saccharification of cellulose is both enzyme- and substrate-dependent. Biotechnol. Lett. 2020, 42, 1975–1984. [Google Scholar] [CrossRef]
- Zhang, P.; Hai, H.; Sun, D.; Yuan, W.; Liu, W.; Ding, R.; Teng, M.; Ma, L.; Tian, J.; Chen, C. A high throughput method for total alcohol determination in fermentation broths. BMC Biotechnol. 2019, 19, 30. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Hu, X.; Lu, S.; Cui, R.; Zhao, J.; Hu, X.; Song, Y. Cellulose nanocrystals produced using recyclable sulfuric acid as hydrolysis media and their wetting molecular dynamics simulation. Int. J. Biol. Macromol. 2021, 184, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wu, X.; Huang, C.; Ling, Z.; Lai, C.; Yong, Q. Natural surfactant-aided dilute sulfuric acid pretreatment of waste wheat straw to enhance enzymatic hydrolysis efficiency. Bioresour. Techno. 2021, 324, 124651. [Google Scholar] [CrossRef]
- Fierobe, H.P.; Mingardon, F.; Mechaly, A.; Bélaïch, A.; Rincon, M.T.; Pagès, S.; Lamed, R.; Tardif, C.; Bélaïch, J.-P.; Bayer, E.A. Action of designer cellulosomes on homogeneous versus complex substrates: Controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 2005, 280, 16325–16334. [Google Scholar] [CrossRef] [PubMed]
- Aymé, L.; Hébert, A.; Henrissat, B.; Lombard, V.; Franche, N.; Perret, S.; Jourdier, E.; Heiss-Blanquet, S. Characterization of three bacterial glycoside hydrolase family 9 endoglucanases with different modular architectures isolated from a compost metagenome. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129848. [Google Scholar] [CrossRef]
- Dash, S.; Olson, D.G.; Joshua Chan, S.H.; Amador-Noguez, D.; Lynd, L.R.; Maranas, C.D. Thermodynamic analysis of the pathway for ethanol production from cellobiose in Clostridium thermocellum. Metab. Eng. 2019, 55, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.G.; Sparling, R.; Lynd, L.R. Ethanol production by engineered thermophiles. Curr. Opin. Biotechnol. 2015, 33, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Beckham, G.T.; Matthews, J.F.; Peters, B.; Bomble, Y.J.; Himmel, M.E.; Crowley, M.F. Molecular-Level Origins of Biomass Recalcitrance: Decrystallization Free Energies for Four Common Cellulose Polymorphs. J. Phys. Chem. B 2011, 115, 4118–4127. [Google Scholar] [CrossRef]
- Shaw, A.J.; Podkaminer, K.K.; Desai, S.G.; Bardsley, J.S.; Rogers, S.R.; Thorne, P.G.; Hogsett, D.A.; Lynd, L.R. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. USA 2008, 105, 13769–13774. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.P.; Eley, K.L.; Martin, S.; Tuffin, M.I.; Burton, S.G.; Cowan, D.A. Thermophilic ethanologenesis: Future prospects for second-generation bioethanol production. Trends. Biotechnol. 2009, 27, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, S.; Ogawa, S.; Nishida, A.; Kobayashi, Y.; Kurosawa, T.; Karita, S. Cellulosomes localise on the surface of membrane vesicles from the cellulolytic bacterium Clostridium thermocellum. FEMS Microbiol. Lett. 2019, 366, 114. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Mathur, A.S.; Gupta, R.P.; Barrow, C.J.; Tuli, D.K.; Puri, M. Enzyme systems of thermophilic anaerobic bacteria for lignocellulosic biomass conversion. Int. J. Biol. Macromol. 2021, 168, 572–590. [Google Scholar] [CrossRef] [PubMed]
- Eibinger, M.; Ganner, T.; Plank, H.; Nidetzky, B. A Biological Nanomachine at Work: Watching the Cellulosome Degrade Crystalline Cellulose. ACS Cent. Sci. 2020, 6, 739–746. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Kont, R.; Beckham, G.T.; Crowley, M.F.; Gudmundsson, M.; Sandgren, M.; Stahlberg, J.; Valjamae, P.; Knott, B.C. The dissociation mechanism of processive cellulases. Proc. Natl. Acad. Sci. USA 2019, 116, 23061–23067. [Google Scholar] [CrossRef]
- Gilmore, S.P.; Lillington, S.P.; Haitjema, C.H.; de Groot, R.; O’Malley, M.A. Designing chimeric enzymes inspired by fungal cellulosomes. Synth. Syst. Biotechnol. 2020, 5, 23–32. [Google Scholar] [CrossRef]
- Zajki-Zechmeister, K.; Kaira, G.S.; Eibinger, M.; Seelich, K.; Nidetzky, B. Processive Enzymes Kept on a Leash: How Cellulase Activity in Multienzyme Complexes Directs Nanoscale Deconstruction of Cellulose. ACS Catal. 2021, 11, 13530–13542. [Google Scholar] [CrossRef]
- Yarbrough, J.M.; Zhang, R.; Mittal, A.; Vander Wall, T.; Bomble, Y.J.; Decker, S.R.; Himmel, M.E.; Ciesielski, P.N. Multifunctional Cellulolytic Enzymes Outperform Processive Fungal Cellulases for Coproduction of Nanocellulose and Biofuels. ACS Nano. 2017, 11, 3101–3109. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Minton, N.P.; Zhang, Y.; Li, Q.; Liu, J.; Jiang, Y.; Yang, S. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab. Eng. 2017, 39, 38–48. [Google Scholar] [CrossRef]
- Wen, Z.; Ledesma-Amaro, R.; Lin, J.; Jiang, Y.; Yang, S. Improved n-Butanol Production from Clostridium cellulovorans by Integrated Metabolic and Evolutionary Engineering. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Argyros, D.A.; Tripathi, S.A.; Barrett, T.F.; Rogers, S.R.; Feinberg, L.F.; Olson, D.G.; Foden, J.M.; Miller, B.B.; Lynd, L.R.; Hogsett, D.A.; et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl. Environ. Microbiol. 2011, 77, 8288–8294. [Google Scholar] [CrossRef]
- Xu, L.; Tschirner, U. Immobilized anaerobic fermentation for bio-fuel production by Clostridium co-culture. Bioprocess. Biosyst. Eng. 2014, 37, 1551–1559. [Google Scholar] [CrossRef]
- Mori, Y. Characterization of a Symbiotic Coculture of Clostridium thermohydrosulfuricum YM3 and Clostridium thermocellum YM4. Appl. Environ. Microbiol. 1990, 56, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, B.; Zhang, Z.; Yan, J.; Liu, P.; Song, C.; Shabbir, S.; Zhu, Q.; Yang, S.; Peng, N.; et al. Cellulosic ethanol production by consortia of Scheffersomyces stipitis and engineered Zymomonas mobilis. Biotechnol. Biofuels 2021, 14, 221. [Google Scholar] [CrossRef] [PubMed]
- Strobel, H.J.; Caldwell, F.C.; Dawson, K.A. Carbohydrate Transport by the Anaerobic Thermophile Clostridium thermocellum LQRI. Appl. Environ. Microbiol. 1995, 61, 4012–4015. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Li, Q.; Liu, J.; Jin, M.; Yang, S. Consolidated bioprocessing for butanol production of cellulolytic Clostridia: Development and optimization. Microb. Biotechnol. 2020, 13, 410–422. [Google Scholar] [CrossRef]
- Joseph, R.C.; Kim, N.M.; Sandoval, N.R. Recent Developments of the Synthetic Biology Toolkit for Clostridium. Front. Microbiol. 2018, 9, 154. [Google Scholar] [CrossRef]


| Family | Number | Known Activities | |
|---|---|---|---|
| At a | Ts | ||
| GH1 | 2 | 0 | exo-β-glucosidase |
| GH2 | 1(1) | 7 | β-galactosidase |
| GH3 | 2 | 5 | β-glucosidase/xylosidase |
| GH4 | 0 | 1 | α-galactosidase |
| GH5 | 10(8) | 0 | β-mannanase/xylanase/endo-β-1,4-glucanase |
| GH8 | 1(1) | 0 | endo-β-1,4-glucanase |
| GH9 | 16(15) | 1 | Processive endoglucanase/cellobiohydrolase/endo-β-1,4-glucanase |
| GH10 | 5(4) | 4 | β-1,4-xylanase/xylanase |
| GH11 | 1(1) | 1 | endo-1,4-β-xylanase |
| GH13 | 2 | 6 | 1,4-α-glucan branching enzyme/α-glycosidase |
| GH15 | 1 | 1 | |
| GH16 | 2(1) | 0 | β-1,3-1,4-glucanase/lichenase |
| GH18 | 4(1) | 2 | Chitinase |
| GH23 | 2 | 2 | |
| GH26 | 3(3) | 1 | β-mannanase/endo-β-1,4-glucanase/β-1,4-xylanase |
| GH27 | 0 | 1 | |
| GH28 | 0 | 2 | |
| GH29 | 0 | 1 | |
| GH30 | 2(2) | 0 | glucuronoxylan xylanohydrolase |
| GH31 | 0 | 1 | |
| GH35 | 0 | 1 | β-galactosidase |
| GH36 | 0 | 2 | α-galactosidase |
| GH38 | 0 | 1 | |
| GH39 | 1(1) | 1 | β-xylosidase |
| GH43 | 6(6) | 8 | α-L-arabinofuranosidase/exo-β-1,3-galactanase/β-xylosidase/arabinosidase |
| GH44 | 1(1) | 0 | |
| GH48 | 2(1) | 1 | exo-cellulase |
| GH51 | 1 | 1 | α-L-arabinofuranosidase |
| GH53 | 1(1) | 1 | endo-β-1,4-galactanase |
| GH67 | 0 | 1 | α-glucuronidase |
| GH74 | 1(1) | 0 | Xyloglucanase |
| GH78 | 0 | 1 | α-L-rhamnosidase |
| GH81 | 1(1) | 0 | β-1,3-glucanase |
| GH88 | 0 | 1 | |
| GH94 | 3 | 2 | Cellobiose/cellodextrin phosphorylase |
| GH95 | 0 | 1 | |
| GH105 | 0 | 5 | |
| GH106 | 0 | 3 | α-L-rhamnosidase |
| GH112 | 0 | 1 | |
| GH115 | 0 | 1 | |
| GH124 | 1(1) | 0 | endo-β-1,4-glucanase |
| GH126 | 1 | 0 | |
| GH127 | 0 | 2 | |
| GH130 | 1 | 1 | |
| GH140 | 0 | 1 | |
| GH141 | 1 | 0 | xylanase E |
| GH154 | 0 | 2 | |
| NC | 1 | 1 | |
| Substrate | Activity in IU/mg | |
|---|---|---|
| At | Ts | |
| Avicel PH101 | 0.050 ± 0.038 | 0.021 ± 0.043 |
| Carboxymethylcellulose | 0.549 ± 0.073 | 0.472 ± 0.136 |
| Amorphous cellulose | 0.543 ± 0.054 | 0.279 ± 0.048 |
| Waterman No. 1# filter paper | 0.047 ± 0.004 | 0.036 ± 0.011 |
| Xylan from oat spelt | 0.414 ± 0.108 | 0.800 ± 0.098 |
| Pectin from citrus peel | 0.162 ± 0.010 | 0.185 ± 0.021 |
| Cellobiose | 0.101 ± 0.007 | 0.011 ± 0.001 |
| Sugar Con. (mM) | Cellulase Systems | |||
|---|---|---|---|---|
| At | Ts | At + Ts a | ||
| Avicel | Glucose | 2.13 ± 0.057 | 0.97 ± 0.010 | 2.30 ± 0.072 |
| Cellobiose | 2.51 ± 0.070 | 0.92 ± 0.015 | 3.28 ± 0.093 | |
| Corn stover | Glucose | 1.03 ± 0.056 | 0.80 ± 0.025 | 1.04 ± 0.032 |
| Xylose | 0.35 ± 0.015 | 0.91 ± 0.020 | 0.71 ± 0.071 | |
| Cellobiose | 1.92 ± 0.036 | 0.69 ± 0.026 | 2.34 ± 0.035 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Yan, Z.; Liu, N.; Zhang, X.; Xu, C. Synergy of Cellulase Systems between Acetivibrio thermocellus and Thermoclostridium stercorarium in Consolidated-Bioprocessing for Cellulosic Ethanol. Microorganisms 2022, 10, 502. https://doi.org/10.3390/microorganisms10030502
Wang N, Yan Z, Liu N, Zhang X, Xu C. Synergy of Cellulase Systems between Acetivibrio thermocellus and Thermoclostridium stercorarium in Consolidated-Bioprocessing for Cellulosic Ethanol. Microorganisms. 2022; 10(3):502. https://doi.org/10.3390/microorganisms10030502
Chicago/Turabian StyleWang, Na, Zhihua Yan, Na Liu, Xiaorong Zhang, and Chenggang Xu. 2022. "Synergy of Cellulase Systems between Acetivibrio thermocellus and Thermoclostridium stercorarium in Consolidated-Bioprocessing for Cellulosic Ethanol" Microorganisms 10, no. 3: 502. https://doi.org/10.3390/microorganisms10030502
APA StyleWang, N., Yan, Z., Liu, N., Zhang, X., & Xu, C. (2022). Synergy of Cellulase Systems between Acetivibrio thermocellus and Thermoclostridium stercorarium in Consolidated-Bioprocessing for Cellulosic Ethanol. Microorganisms, 10(3), 502. https://doi.org/10.3390/microorganisms10030502






