Endogenous Honeybee Gut Microbiota Metabolize the Pesticide Clothianidin
Abstract
1. Introduction
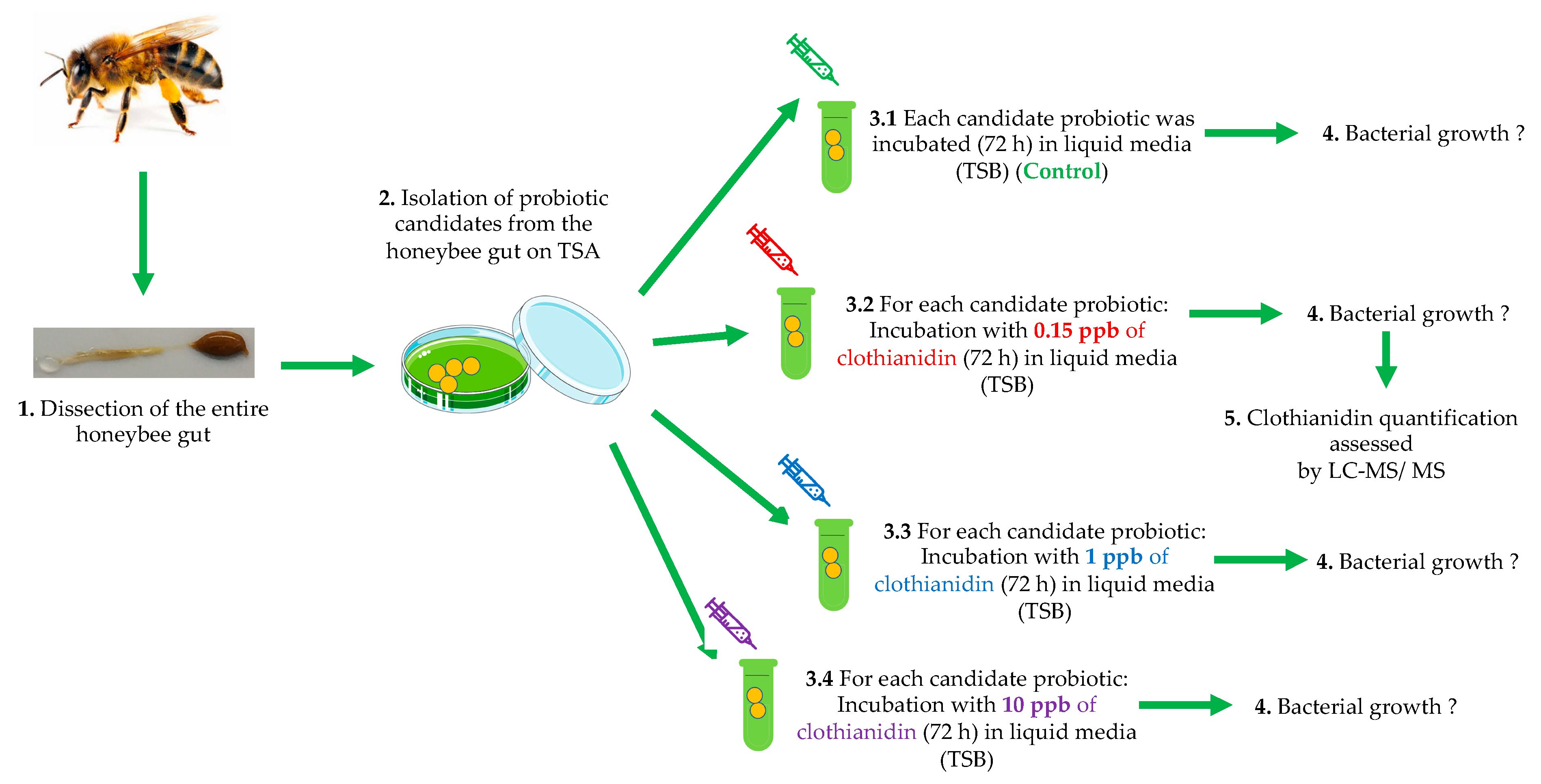
2. Material and Methods
2.1. Isolation of Probiotic Candidates from Honeybee Gut Microbiota
2.2. In Vitro Tests
2.2.1. Growth of Honeybee Gut Probiotic Candidates
2.2.2. Clothianidin Degradation by Honeybee Gut Probiotic Candidates
3. Results
3.1. Isolation of Probiotic Candidates from Honeybee Gut Microbiota
3.2. In Vitro Tests
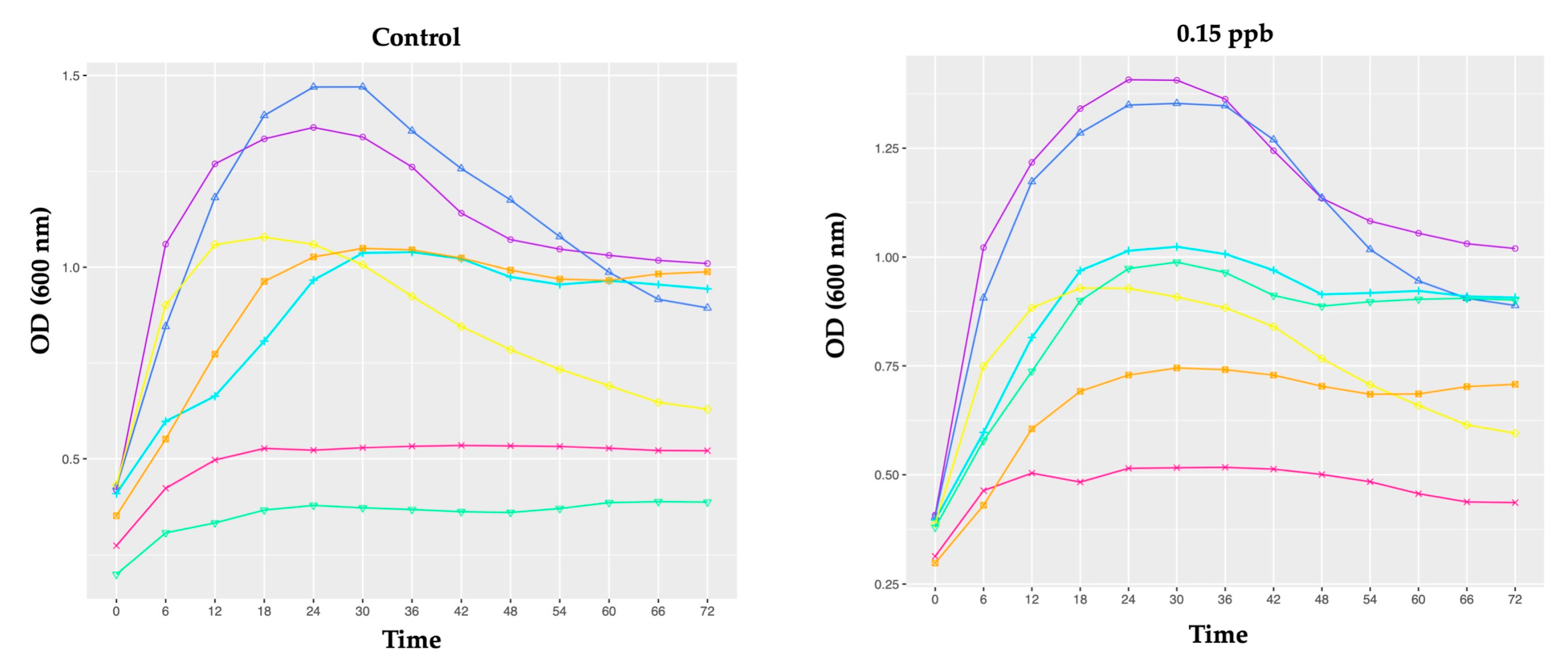
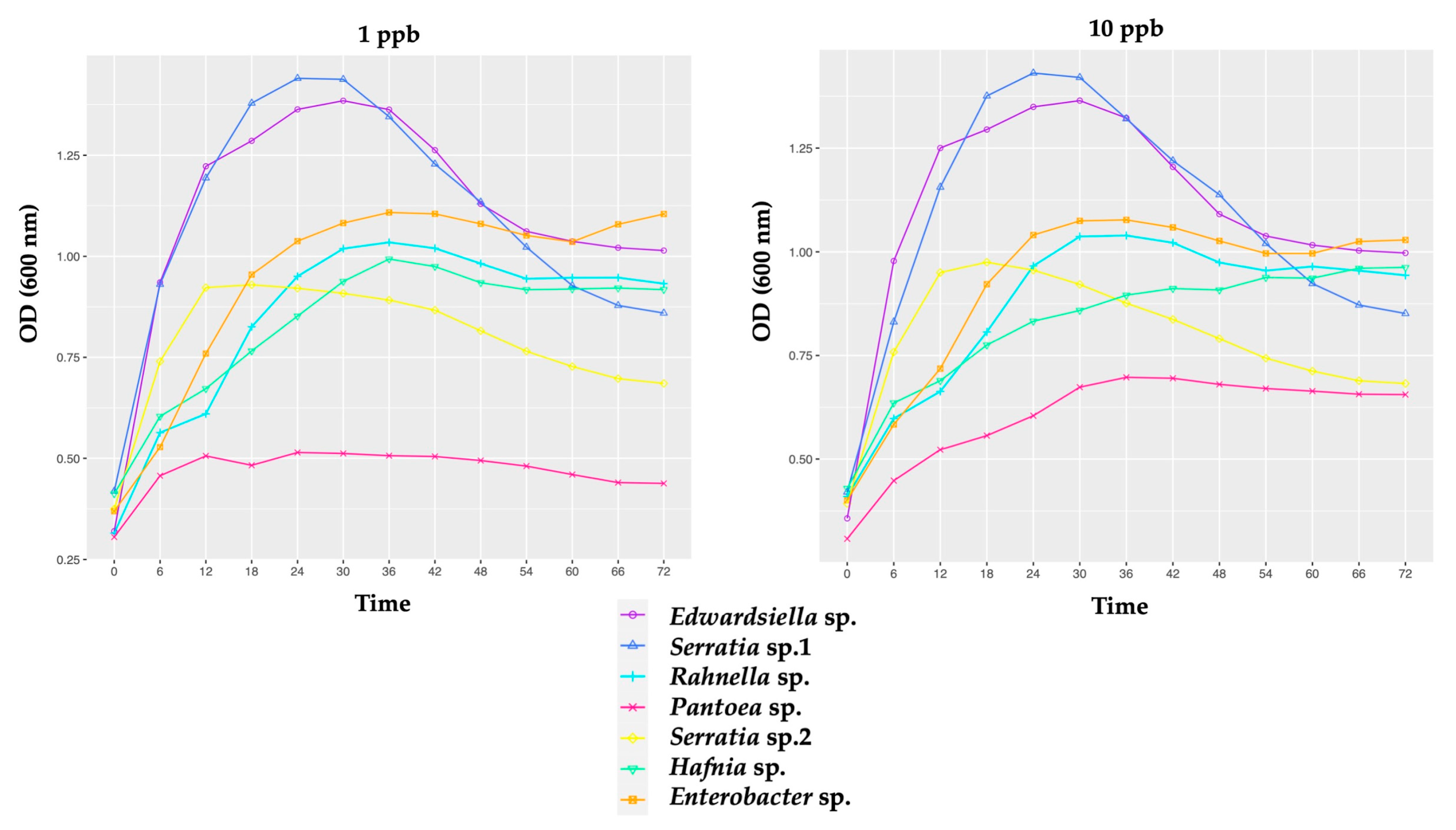
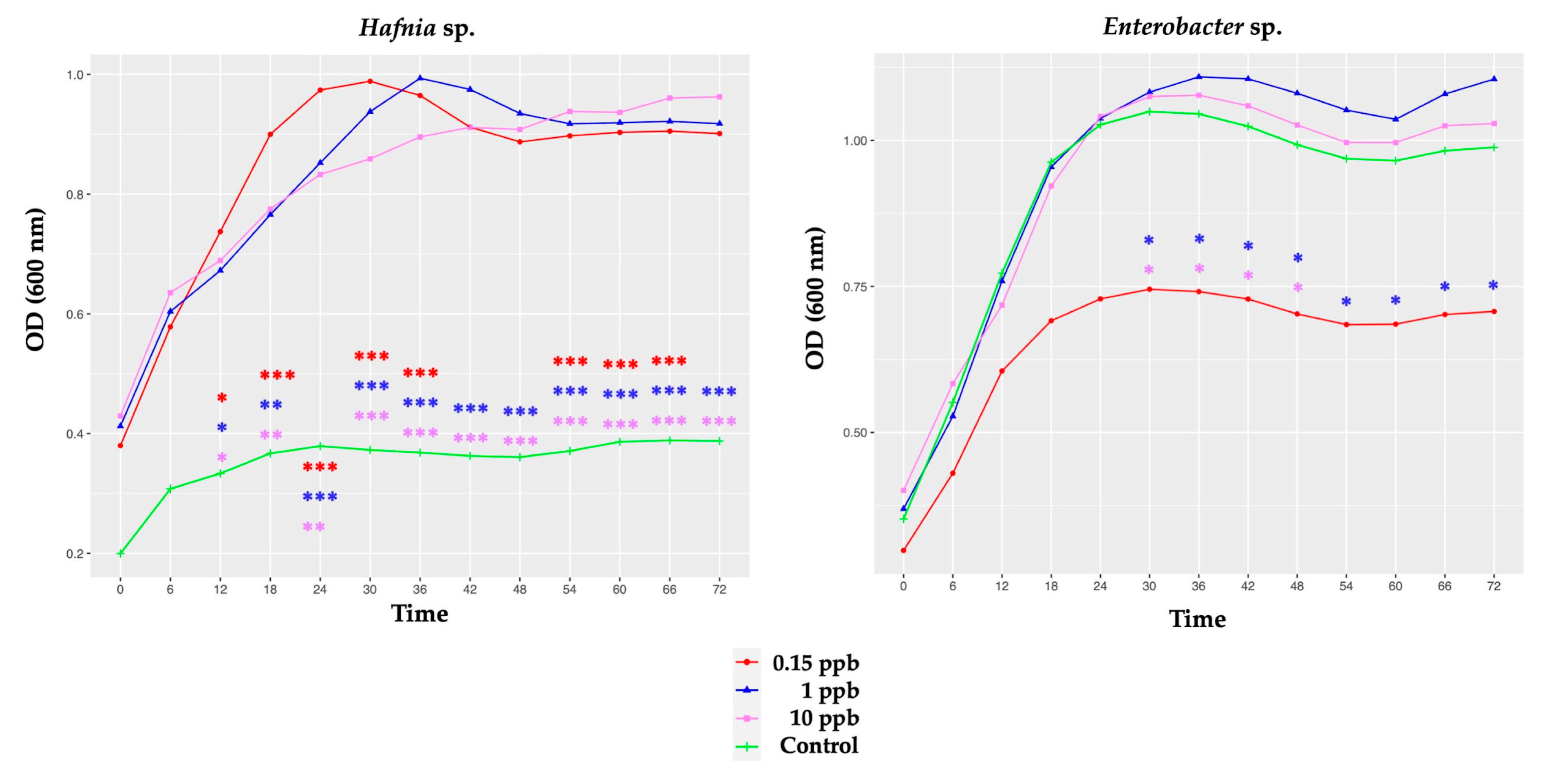
3.2.1. Growth of Honeybee Gut Probiotic Candidates
3.2.2. Clothianidin Degradation by Honeybee Gut Probiotic Candidates
4. Discussion
4.1. Growth of Honeybee Gut Probiotic Candidates
4.2. Clothianidin Degradation by Honeybee Gut Probiotic Candidates
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alberoni, D.; Gaggìa, F.; Baffoni, L.; Di Gioia, D. Beneficial Microorganisms for Honey Bees: Problems and Progresses. Appl. Microbiol. Biotechnol. 2016, 100, 9469–9482. [Google Scholar] [CrossRef]
- Huang, Y.; Xiao, L.; Li, F.; Xiao, M.; Lin, D.; Long, X.; Wu, Z. Microbial Degradation of Pesticide Residues and an Emphasis on the Degradation of Cypermethrin and 3-Phenoxy Benzoic Acid: A Review. Molecules 2018, 23, 2313. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Cabirol, A.; Devaud, J.-M.; Barron, A.B.; Lihoreau, M. Why Bees Are So Vulnerable to Environmental Stressors. Trends Ecol. Evol. 2017, 32, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Bartomeus, I.; Potts, S.G.; Steffan-Dewenter, I.; Vaissière, B.E.; Woyciechowski, M.; Krewenka, K.M.; Tscheulin, T.; Roberts, S.P.M.; Szentgyörgyi, H.; Westphal, C.; et al. Contribution of Insect Pollinators to Crop Yield and Quality Varies with Agricultural Intensification. PeerJ 2014, 2, e328. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, M. The Silent Beehive: How the Decline of Honey Bee Populations Shifted the Environmental Protection Agency’s Pesticide Policy towards Pollinators. Ecol. Law Q. 2017, 44, 311. [Google Scholar]
- Marshman, J.; Blay-Palmer, A.; Landman, K. Anthropocene Crisis: Climate Change, Pollinators, and Food Security. Environments 2019, 6, 22. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee Declines Driven by Combined Stress from Parasites, Pesticides, and Lack of Flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Dussaubat, C.; Maisonnasse, A.; Crauser, D.; Tchamitchian, S.; Bonnet, M.; Cousin, M.; Kretzschmar, A.; Brunet, J.-L.; Le Conte, Y. Combined Neonicotinoid Pesticide and Parasite Stress Alter Honeybee Queens’ Physiology and Survival. Sci. Rep. 2016, 6, 31430. [Google Scholar] [CrossRef]
- Li, J.H.; Evans, J.D.; Li, W.F.; Zhao, Y.Z.; DeGrandi-Hoffman, G.; Huang, S.K.; Li, Z.G.; Hamilton, M.; Chen, Y.P. New Evidence Showing That the Destruction of Gut Bacteria by Antibiotic Treatment Could Increase the Honey Bee’s Vulnerability to Nosema Infection. PLoS ONE 2017, 12, e0187505. [Google Scholar] [CrossRef]
- López, J.H.; Krainer, S.; Engert, A.; Schuehly, W.; Riessberger-Gallé, U.; Crailsheim, K. Sublethal Pesticide Doses Negatively Affect Survival and the Cellular Responses in American Foulbrood-Infected Honeybee Larvae. Sci. Rep. 2017, 7, 40853. [Google Scholar] [CrossRef]
- Hristov, P.; Shumkova, R.; Palova, N.; Neov, B. Factors Associated with Honey Bee Colony Losses: A Mini-Review. Vet. Sci. China 2020, 7, 166. [Google Scholar] [CrossRef]
- Lim, S.; Yunusbaev, U.; Ilyasov, R.; Lee, H.S.; Kwon, H.W. Abdominal Contact of Fluvalinate Induces Olfactory Deficit in Apis mellifera. Pestic. Biochem. Physiol. 2020, 164, 221–227. [Google Scholar] [CrossRef]
- Ilyasov, R.; Sooho, L.İ.M.; Lee, M.L.; Kwon, H.W.; Nïkolenko, A. Effect of Miticides Amitraz and Fluvalinate on Reproduction and Productivity of Honey Bee Apis mellifera. Uludağ Arıcılık Derg. 2021, 21, 21–30. [Google Scholar] [CrossRef]
- Hladik, M.L.; Main, A.R.; Goulson, D. Environmental Risks and Challenges Associated with Neonicotinoid Insecticides. Environ. Sci. Technol. 2018, 52, 3329–3335. [Google Scholar] [CrossRef]
- Jeschke, P.; Nauen, R. Neonicotinoids—From Zero to Hero in Insecticide Chemistry. Pest Manag. Sci. Former. Pestic. Sci. 2008, 64, 1084–1098. [Google Scholar] [CrossRef]
- Jeschke, P.; Nauen, R.; Schindler, M.; Elbert, A. Overview of the Status and Global Strategy for Neonicotinoids. J. Agric. Food Chem. 2011, 59, 2897–2908. [Google Scholar] [CrossRef]
- Bass, C.; Denholm, I.; Williamson, M.S.; Nauen, R. The Global Status of Insect Resistance to Neonicotinoid Insecticides. Pestic. Biochem. Physiol. 2015, 121, 78–87. [Google Scholar] [CrossRef]
- Casida, J.E. Neonicotinoids and Other Insect Nicotinic Receptor Competitive Modulators: Progress and Prospects. Annu. Rev. Entomol. 2018, 63, 125–144. [Google Scholar] [CrossRef]
- Hussain, S.; Hartley, C.J.; Shettigar, M.; Pandey, G. Bacterial Biodegradation of Neonicotinoid Pesticides in Soil and Water Systems. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Pang, S.; Lin, Z.; Zhang, W.; Mishra, S.; Bhatt, P.; Chen, S. Insights Into the Microbial Degradation and Biochemical Mechanisms of Neonicotinoids. Front. Microbiol. 2020, 11, 868. [Google Scholar] [CrossRef]
- Parte, S.G.; Kharat, A.S. Aerobic Degradation of Clothianidin to 2-Chloro-Methyl Thiazole and Methyl 3-(Thiazole-Yl) Methyl Guanidine Produced by Pseudomonas Stutzeri Smk. J. Environ. Public Health 2019, 2019, 4807913. [Google Scholar] [CrossRef]
- Iancu, V.-I.; Petre, J.; Galaon, T.; Radu, G.L. Occurrence of Neonicotinoid Residues in Danube River and Tributaries. Rev. Chim. 2019, 70, 313–318. [Google Scholar] [CrossRef]
- Mancini, F.; Woodcock, B.A.; Isaac, N.J.B. Agrochemicals in the Wild: Identifying Links between Pesticide Use and Declines of Non-Target Organisms. Curr. Opin. Environ. Sci. Health 2019, 11, 53–58. [Google Scholar] [CrossRef]
- Desneux, N.; Decourtye, A.; Delpuech, J.-M. The Sublethal Effects of Pesticides on Beneficial Arthropods. Annu. Rev. Entomol. 2007, 52, 81–106. [Google Scholar] [CrossRef]
- Imran, M. Neonicotinoid Insecticides: A Threat to Pollinators. In Trends in Integrated Insect Pest Management; Soundararajan, R.P., Narayanasamy, C., Eds.; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar]
- Palmer, M.J.; Moffat, C.; Saranzewa, N.; Harvey, J.; Wright, G.A.; Connolly, C.N. Cholinergic Pesticides Cause Mushroom Body Neuronal Inactivation in Honeybees. Nat. Commun. 2013, 4, 1634. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Qiao, N.-H.; Diao, Q.-Y.; Jing, Z.; Vukanti, R.; Dai, P.-L.; Ge, Y. Thiacloprid Exposure Perturbs the Gut Microbiota and Reduces the Survival Status in Honeybees. J. Hazard. Mater. 2019, 389, 121818. [Google Scholar] [CrossRef]
- Brandt, A.; Gorenflo, A.; Siede, R.; Meixner, M.; Büchler, R. The Neonicotinoids Thiacloprid, Imidacloprid, and Clothianidin Affect the Immunocompetence of Honey Bees (Apis mellifera L.). J. Insect Physiol. 2016, 86, 40–47. [Google Scholar] [CrossRef]
- Pettis, J.S.; vanEngelsdorp, D.; Johnson, J.; Dively, G. Pesticide Exposure in Honey Bees Results in Increased Levels of the Gut Pathogen Nosema. Naturwissenschaften 2012, 99, 153–158. [Google Scholar] [CrossRef]
- El Khoury, S.; Gauthier, J.; Bouslama, S.; Cheaib, B.; Giovenazzo, P.; Derome, N. Dietary Contamination with a Neonicotinoid (Clothianidin) Gradient Triggers Specific Dysbiosis Signatures of Microbiota Activity along the Honeybee (Apis mellifera) Digestive Tract. Microorganisms 2021, 9, 2283. [Google Scholar] [CrossRef]
- Zhu, L.; Qi, S.; Xue, X.; Niu, X.; Wu, L. Nitenpyram Disturbs Gut Microbiota and Influences Metabolic Homeostasis and Immunity in Honey Bee (Apis mellifera L.). Environ. Pollut. 2019, 258, 113671. [Google Scholar] [CrossRef]
- Alburaki, M.; Boutin, S.; Mercier, P.-L.; Loublier, Y.; Chagnon, M.; Derome, N. Neonicotinoid-Coated Zea Mays Seeds Indirectly Affect Honeybee Performance and Pathogen Susceptibility in Field Trials. PLoS ONE 2015, 10, e0125790. [Google Scholar] [CrossRef] [PubMed]
- Alburaki, M.; Chen, D.; Skinner, J.A.; Meikle, W.G.; Tarpy, D.R.; Adamczyk, J.; Stewart, S.D. Honey Bee Survival and Pathogen Prevalence: From the Perspective of Landscape and Exposure to Pesticides. Insects 2018, 9, 65. [Google Scholar] [CrossRef]
- Morfin, N.; Goodwin, P.H.; Guzman-Novoa, E. Interaction of field realistic doses of clothianidin and Varroa destructor parasitism on adult honey bee (Apis mellifera L.) health and neural gene expression, and antagonistic effects on differentially expressed genes. PLoS ONE 2020, 15, e0229030. [Google Scholar] [CrossRef] [PubMed]
- Tosi, S.; Nieh, J.C.; Sgolastra, F.; Cabbri, R.; Medrzycki, P. Neonicotinoid Pesticides and Nutritional Stress Synergistically Reduce Survival in Honey Bees. Proc. R. Soc. B Biol. Sci. 2017, 284, 20171711. [Google Scholar] [CrossRef] [PubMed]
- Sadd, B.M.; Barribeau, S.M.; Bloch, G.; de Graaf, D.C.; Dearden, P.; Elsik, C.G.; Gadau, J.; Grimmelikhuijzen, C.J.P.; Hasselmann, M.; Lozier, J.D.; et al. The Genomes of Two Key Bumblebee Species with Primitive Eusocial Organization. Genome Biol. 2015, 16, 76. [Google Scholar] [CrossRef]
- Cremer, S.; Armitage, S.A.O.; Schmid-Hempel, P. Social Immunity. Curr. Biol. 2007, 17, R693–R702. [Google Scholar] [CrossRef]
- Berenbaum, M.R.; Johnson, R.M. Xenobiotic Detoxification Pathways in Honey Bees. Curr. Opin. Insect Sci 2015, 10, 51–58. [Google Scholar] [CrossRef]
- Mao, W.; Schuler, M.A.; Berenbaum, M.R. Honey Constituents up-Regulate Detoxification and Immunity Genes in the Western Honey Bee Apis mellifera. Proc. Natl. Acad. Sci. USA 2013, 110, 8842–8846. [Google Scholar] [CrossRef]
- Carboneras, B.; Villaseñor, J.; Fernandez-Morales, F.J. Modelling Aerobic Biodegradation of Atrazine and 2,4-Dichlorophenoxy Acetic Acid by Mixed-Cultures. Bioresour. Technol. 2017, 243, 1044–1050. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Fu, J.; Hu, S.; Qu, J. Degradation of Acetochlor and Beneficial Effect of Phosphate-Solubilizing Bacillus Sp. ACD-9 on Maize Seedlings. 3 Biotech 2020, 10, 67. [Google Scholar] [CrossRef]
- Kosolapov, D.B.; Kuschk, P.; Vainshtein, M.B.; Vatsourina, A.V.; Wiessner, A.; Kästner, M.; Müller, R.A. Microbial Processes of Heavy Metal Removal from Carbon-Deficient Effluents in Constructed Wetlands. Eng. Life Sci. 2004, 4, 403–411. [Google Scholar] [CrossRef]
- Mulligan, R.A.; Tomco, P.L.; Howard, M.W.; Schempp, T.T.; Stewart, D.J.; Stacey, P.M.; Ball, D.B.; Tjeerdema, R.S. Aerobic versus Anaerobic Microbial Degradation of Clothianidin under Simulated California Rice Field Conditions. J. Agric. Food Chem. 2016, 64, 7059–7067. [Google Scholar] [CrossRef]
- Mori, T.; Wang, J.; Tanaka, Y.; Nagai, K.; Kawagishi, H.; Hirai, H. Bioremediation of the Neonicotinoid Insecticide Clothianidin by the White-Rot Fungus Phanerochaete sordida. J. Hazard. Mater. 2017, 321, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, K.M.; Brochet, S.; Bonilla-Rosso, G.; Emery, O.; Glover, N.; Hadadi, N.; Jaron, K.S.; van der Meer, J.R.; Robinson-Rechavi, M.; Sentchilo, V.; et al. Genomic Changes Underlying Host Specialization in the Bee Gut Symbiont Lactobacillus Firm5. Mol. Ecol. 2019, 28, 2224–2237. [Google Scholar] [CrossRef] [PubMed]
- Morfin, N.; Goodwin, P.H.; Hunt, G.J.; Guzman-Novoa, E. Effects of Sublethal Doses of Clothianidin And/or V. Destructor on Honey Bee (Apis mellifera) Self-Grooming Behavior and Associated Gene Expression. Sci. Rep. 2019, 9, 5196. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, S.; Rousseau, A.; Lecoeur, A.; Cheaib, B.; Bouslama, S.; Mercier, P.-L.; Demey, V.; Castex, M.; Giovenazzo, P.; Derome, N. Deleterious Interaction between Honeybees (Apis mellifera) and Its Microsporidian Intracellular Parasite Nosema Ceranae Was Mitigated by Administrating Either Endogenous or Allochthonous Gut Microbiota Strains. Front. Ecol. Evol. 2018, 6, 58. [Google Scholar] [CrossRef]
- Royston, J.P. Shapiro-Wilk Normality Test and P-Value. Appl. Stat. 1995, 44, 547–551. [Google Scholar] [CrossRef]
- Paradis, D.; Bérail, G.; Bonmatin, J.-M.; Belzunces, L.P. Sensitive Analytical Methods for 22 Relevant Insecticides of 3 Chemical Families in Honey by GC-MS/MS and LC-MS/MS. Anal. Bioanal. Chem. 2014, 406, 621–633. [Google Scholar] [CrossRef]
- Etxebarria, N.; Zuloaga, O.; Olivares, M.; Bartolomé, L.J.; Navarro, P. Retention-Time Locked Methods in Gas Chromatography. J. Chromatogr. A 2009, 1216, 1624–1629. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between Resistance, Tolerance and Persistence to Antibiotic Treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Bernier, S.P.; Lebeaux, D.; DeFrancesco, A.S.; Valomon, A.; Soubigou, G.; Coppée, J.-Y.; Ghigo, J.-M.; Beloin, C. Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin. PLoS Genet. 2013, 9, e1003144. [Google Scholar] [CrossRef] [PubMed]
- DeLorenzo, M.E.; Scott, G.I.; Ross, P.E. Toxicity of Pesticides to Aquatic Microorganisms: A Review. Environ. Toxicol. Chem. 2001, 20, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-C.; Wang, Y.; Zhai, S.; Ge, F.; Liu, Z.-H.; Dai, Y.-J.; Yuan, S.; Hou, J.-Y. Biodegradation of the Neonicotinoid Insecticide Thiamethoxam by the Nitrogen-Fixing and Plant-Growth-Promoting Rhizobacterium Ensifer Adhaerens Strain TMX-23. Appl. Microbiol. Biotechnol. 2013, 97, 4065–4074. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Motta, E.V.S.; Girard, C.; Riddington, I.M.; Dinser, J.A.; Moran, N.A. Imidacloprid Decreases Honey Bee Survival Rates but Does Not Affect the Gut Microbiome. Appl. Environ. Microbiol. 2018, 84, e00545-18. [Google Scholar] [CrossRef] [PubMed]
- Ramya, S.L.; Venkatesan, T.; Murthy, K.S.; Jalali, S.K.; Varghese, A. Degradation of Acephate by Enterobacter Asburiae, Bacillus Cereus and Pantoea agglomerans Isolated from Diamondback Moth Plutella xylostella (L), a Pest of Cruciferous Crops. J. Environ. Biol. 2016, 37, 611. [Google Scholar] [PubMed]
- Staley, Z.R.; Harwood, V.J.; Rohr, J.R. A Synthesis of the Effects of Pesticides on Microbial Persistence in Aquatic Ecosystems. Crit. Rev. Toxicol. 2015, 45, 813–836. [Google Scholar] [CrossRef]
- Simon-Delso, N.; Amaral-Rogers, V.; Belzunces, L.P.; Bonmatin, J.M.; Chagnon, M.; Downs, C.; Furlan, L.; Gibbons, D.W.; Giorio, C.; Girolami, V.; et al. Systemic Insecticides (neonicotinoids and Fipronil): Trends, Uses, Mode of Action and Metabolites. Environ. Sci. Pollut. Res. Int. 2015, 22, 5–34. [Google Scholar] [CrossRef]
- Muturi, E.J.; Donthu, R.K.; Fields, C.J.; Moise, I.K.; Kim, C.-H. Effect of Pesticides on Microbial Communities in Container Aquatic Habitats. Sci. Rep. 2017, 7, 44565. [Google Scholar] [CrossRef]
- Russell, R.J.; Scott, C.; Jackson, C.J.; Pandey, R.; Pandey, G.; Taylor, M.C.; Coppin, C.W.; Liu, J.-W.; Oakeshott, J.G. The Evolution of New Enzyme Function: Lessons from Xenobiotic Metabolizing Bacteria versus Insecticide-Resistant Insects. Evol. Appl. 2011, 4, 225–248. [Google Scholar] [CrossRef]
- Liu, J.; Song, Y.; Tang, M.; Lu, Q.; Zhong, G. Enhanced Dissipation of Xenobiotic Agrochemicals Harnessing Soil Microbiome in the Tillage-Reduced Rice-Dominated Agroecosystem. J. Hazard. Mater. 2020, 398, 122954. [Google Scholar] [CrossRef]
- Jilani, S. Comparative Assessment of Growth and Biodegradation Potential of Soil Isolate in the Presence of Pesticides. Saudi J. Biol. Sci. 2013, 20, 257–264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miller, D.L.; Smith, E.A.; Newton, I.L.G. A Bacterial Symbiont Protects Honey Bees from Fungal Disease. mBio 2021, e0050321. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Chatterjee, S. Biodegradation of Profenofos, an Acetylcholine Esterase Inhibitor by a Psychrotolerant Strain Rahnella Sp. PFF2 and Degradation Pathway Analysis. Int. Biodeterior. Biodegrad. 2021, 158, 105169. [Google Scholar] [CrossRef]
- Pester, M.; Bittner, N.; Deevong, P.; Wagner, M.; Loy, A. A “rare Biosphere” microorganism Contributes to Sulfate Reduction in a Peatland. ISME J. 2010, 4, 1591–1602. [Google Scholar] [CrossRef]
- Zhou, G.-C.; Wang, Y.; Ma, Y.; Zhai, S.; Zhou, L.-Y.; Dai, Y.-J.; Yuan, S. The Metabolism of Neonicotinoid Insecticide Thiamethoxam by Soil Enrichment Cultures, and the Bacterial Diversity and Plant Growth-Promoting Properties of the Cultured Isolates. J. Environ. Sci. Health B 2014, 49, 381–390. [Google Scholar] [CrossRef]
- Liu, Z.; Dai, Y.; Huang, G.; Gu, Y.; Ni, J.; Wei, H.; Yuan, S. Soil Microbial Degradation of Neonicotinoid Insecticides Imidacloprid, Acetamiprid, Thiacloprid and Imidaclothiz and Its Effect on the Persistence of Bioefficacy against Horsebean Aphid Aphis Craccivora Koch after Soil Application. Pest Manag. Sci. 2011, 67, 1245–1252. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Reeves, A.M.; Anderson, T.D.; Rodrigues, R.R.; Williams, M.A. Honey Bee Gut Microbiome Is Altered by In-Hive Pesticide Exposures. Front. Microbiol. 2016, 7, 1255. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic Exposure Perturbs the Gut Microbiota and Elevates Mortality in Honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate Perturbs the Gut Microbiota of Honey Bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef]
| Sequence Name | Identity Percentage |
|---|---|
| Edwardsiella tarda strain ATCC 15947 | 99% |
| Serratia marcescens strain NBRC 102204 | 98% |
| Rahnella woolbedingensis strain FRB 227 | 99% |
| Pantoea agglomerans strain ATCC 27155 | 98% |
| Serratia nematodiphila strain DZ0503SBS1 | 98% |
| Hafnia paralvei strain ATCC 29927 | 99% |
| Enterobacter sp. | 90% |
| numDF | denDF | F-Value | p-Value | |
|---|---|---|---|---|
| (Intercept) | 1 | 672 | 44.59160 | <0.0001 |
| PC | 6 | 56 | 0.27428 | 0.9467 |
| Treatment | 3 | 56 | 0.56574 | 0.6399 |
| Time | 12 | 672 | 136.42373 | <0.0001 |
| PC:Treatment | 18 | 56 | 0.44272 | 0.9707 |
| PC:Time | 72 | 672 | 9.41233 | <0.0001 |
| Treatment:Time | 36 | 672 | 1.71556 | 0.0064 |
| PC:Treatment:Time | 216 | 672 | 1.38909 | 0.0011 |
| numDF | denDF | F-Value | p-Value | |
|---|---|---|---|---|
| (Intercept) | 1 | 32 | 34.75724 | <0.0001 |
| PC | 7 | 16 | 1.95330 | 0.1267 |
| Time | 2 | 32 | 4.88490 | 0.0141 |
| PC:Time | 14 | 32 | 3.18569 | 0.0033 |
| PC | T24 | T48 | T72 |
|---|---|---|---|
| Edwardsiella sp. | 61% | 100% *** | 100% *** |
| Serratia sp.1 | 1% | 100% *** | 100% *** |
| Serratia sp.2 | 48% | 88% *** | 100% *** |
| Rahnella sp. | 61% | 100% *** | 100% *** |
| Pantoea sp. | 48% | 88% ** | 100% *** |
| Hafnia sp. | 48% | 81% ** | 100% *** |
| Enterobacter sp. | 34% | 68% ** | 100% *** |
| vs. TSB + clothianidin (0.15 ppb) (control) | 0% | 0% | 0% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Khoury, S.; Giovenazzo, P.; Derome, N. Endogenous Honeybee Gut Microbiota Metabolize the Pesticide Clothianidin. Microorganisms 2022, 10, 493. https://doi.org/10.3390/microorganisms10030493
El Khoury S, Giovenazzo P, Derome N. Endogenous Honeybee Gut Microbiota Metabolize the Pesticide Clothianidin. Microorganisms. 2022; 10(3):493. https://doi.org/10.3390/microorganisms10030493
Chicago/Turabian StyleEl Khoury, Sarah, Pierre Giovenazzo, and Nicolas Derome. 2022. "Endogenous Honeybee Gut Microbiota Metabolize the Pesticide Clothianidin" Microorganisms 10, no. 3: 493. https://doi.org/10.3390/microorganisms10030493
APA StyleEl Khoury, S., Giovenazzo, P., & Derome, N. (2022). Endogenous Honeybee Gut Microbiota Metabolize the Pesticide Clothianidin. Microorganisms, 10(3), 493. https://doi.org/10.3390/microorganisms10030493







