Induction of Broad β-lactam Resistance in Achromobacter ruhlandii by Exposure to Ticarcillin Is Primarily Linked to Substitutions in Murein Peptide Ligase Mpl
Abstract
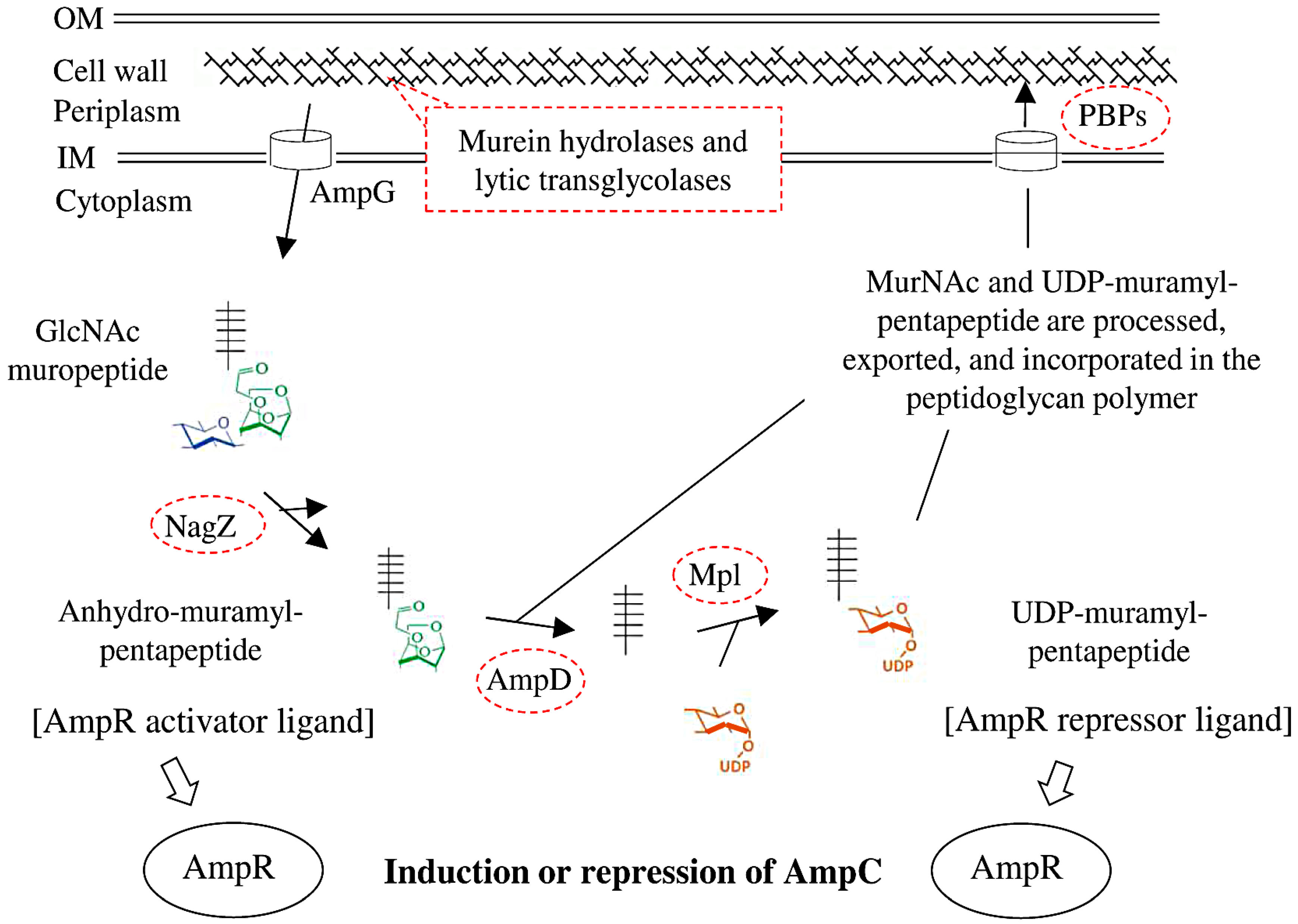
:1. Introduction
2. Materials and Methods
2.1. Isolates
2.2. Generation of Mutants
2.3. DNA Sequencing and Analysis
2.4. AmpD and Mpl Substitutions in Previously Published Genomes
2.5. Antimicrobial Susceptibility Testing
3. Results
3.1. Isolates and Mutations
3.2. β-lactam Susceptibility
3.3. AmpD and Mpl Substitutions in Previously Published Genomes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Sullivan, B.P.; Freedman, S.D. Cystic fibrosis. Lancet 2009, 373, 1891–1904. [Google Scholar] [CrossRef]
- Chmiel, J.F.; Berger, M.; Konstan, M.W. The Role of Inflammation in the Pathophysiology of CF Lung Disease. Clin. Rev. Allergy Immunol. 2002, 23, 5–27. [Google Scholar] [CrossRef]
- Emerson, J.; McNamara, S.; Buccat, A.M.; Worrell, K.; Burns, J.L. Changes in cystic fibrosis sputum microbiology in the United States between 1995 and 2008. Pediatr. Pulmonol. 2010, 45, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Amoureux, L.; Bador, J.; Siebor, E.; Taillefumier, N.; Fanton, A.; Neuwirth, C. Epidemiology and resistance of Achromobacter xylosoxidans from cystic fibrosis patients in Dijon, Burgundy: First French data. J. Cyst. Fibros. 2013, 12, 170–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridderberg, W.; Bendstrup, E.; Olesen, H.V.; Jensen-Fangel, S.; Nørskov-Lauritsen, N. Marked increase in incidence of Achromobacter xylosoxidans infections caused by sporadic acquisition from the environment. J. Cyst. Fibros. 2011, 10, 466–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Daele, S.; Verhelst, R.; Claeys, G.; Verschraegen, G.; Franckx, H.; Van Simaey, L.; de Ganck, C.; De Baets, F.; Vaneechoutte, M. Shared Genotypes of Achromobacter xylosoxidans Strains Isolated from Patients at a Cystic Fibrosis Rehabilitation Center. J. Clin. Microbiol. 2005, 43, 2998–3002. [Google Scholar] [CrossRef] [Green Version]
- Lambiase, A.; Catania, M.R.; Del Pezzo, M.; Rossano, F.; Terlizzi, V.; Sepe, A.; Raia, V. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur. J. Clin. Microbiol. 2011, 30, 973–980. [Google Scholar] [CrossRef] [Green Version]
- Gade, S.S.; Nørskov-Lauritsen, N.; Ridderberg, W. Prevalence and species distribution of Achromobacter sp. cultured from cystic fibrosis patients attending the Aarhus centre in Denmark. J. Med. Microbiol. 2017, 66, 686–689. [Google Scholar] [CrossRef]
- Gabrielaite, M.; Bartell, J.A.; Nørskov-Lauritsen, N.; Pressler, T.; Nielsen, F.C.; Johansen, H.K.; Marvig, R.L. Transmission and Antibiotic Resistance of Achromobacter in Cystic Fibrosis. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Bador, J.; Amoureux, L.; Duez, J.-M.; Drabowicz, A.; Siebor, E.; Llanes, C.; Neuwirth, C. First Description of an RND-Type Multidrug Efflux Pump in Achromobacter xylosoxidans, AxyABM. Antimicrob. Agents Chemother. 2011, 55, 4912–4914. [Google Scholar] [CrossRef] [Green Version]
- Bador, J.; Amoureux, L.; Blanc, E.; Neuwirth, C. Innate Aminoglycoside Resistance of Achromobacter xylosoxidans Is Due to AxyXY-OprZ, an RND-Type Multidrug Efflux Pump. Antimicrob. Agents Chemother. 2012, 57, 603–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolston, K.; Messer, M. The in-vitro susceptibility of Alcatigenes deni-trificans subsp.xylosoxidans to40 anti-microbial agents. J. Antimicrob. Chemother. 1990, 26, 857–859. [Google Scholar] [CrossRef] [PubMed]
- Almuzara, M.; Limansky, A.; Ballerini, V.; Galanternik, L.; Famiglietti, A.; Vay, C. In vitro susceptibility of Achromobacter spp. isolates: Comparison of disk diffusion, Etest and agar dilution methods. Int. J. Antimicrob. Agents 2010, 35, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.M.; Penstoft, L.N.; Nørskov-Lauritsen, N. Motility, Biofilm Formation and Antimicrobial Efflux of Sessile and Planktonic Cells of Achromobacter xylosoxidans. Pathogens 2019, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Doi, Y.; Poirel, L.; Paterson, D.L.; Nordmann, P. Characterization of a Naturally Occurring Class D β-Lactamase from Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 2008, 52, 1952–1956. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.W.; Fisher, J.F.; Mobashery, S. Bacterial cell-wall recycling. Ann. N. Y. Acad. Sci. 2012, 1277, 54–75. [Google Scholar] [CrossRef] [Green Version]
- Tamma, P.D.; Doi, Y.; A Bonomo, R.; Johnson, J.K.; Simner, P.J. Antibacterial Resistance Leadership Group A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. 2019, 69, 1446–1455. [Google Scholar] [CrossRef] [Green Version]
- Calvopiña, K.; Avison, M.B. Disruption of mpl Activates β-Lactamase Production in Stenotrophomonas maltophilia and Pseudomonas aeruginosa Clinical Isolates. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Tsutsumi, Y.; Tomita, H.; Tanimoto, K. Identification of Novel Genes Responsible for Overexpression ofampCin Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2013, 57, 5987–5993. [Google Scholar] [CrossRef] [Green Version]
- Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Mutation-Driven Evolution of Pseudomonas aeruginosa in the Presence of either Ceftazidime or Ceftazidime-Avibactam. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xia, L.; Chen, J.; Lian, Y.; Dandekar, A.A.; Xu, F.; Wang, M. Resistance elicited by sub-lethal concentrations of ampicillin is partially mediated by quorum sensing in Pseudomonas aeruginosa. Environ. Int. 2021, 156, 106619. [Google Scholar] [CrossRef] [PubMed]
- Park, J.T.; Uehara, T. How Bacteria Consume Their Own Exoskeletons (Turnover and Recycling of Cell Wall Peptidoglycan). Microbiol. Mol. Biol. Rev. 2008, 72, 211–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spilker, T.; Vandamme, P.; Lipuma, J.J. A Multilocus Sequence Typing Scheme Implies Population Structure and Reveals Several Putative Novel Achromobacter Species. J. Clin. Microbiol. 2012, 50, 3010–3015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Seemann, T. Snippy: Rapid Haploid Variant Calling and Core Genome Alignment. GitHub. Available online: https://github.com/tseemann/snippy (accessed on 10 February 2020).
- Ridderberg, W.; Handberg, K.J.; Nørskov-Lauritsen, N. Prevalence of hypermutator isolates of Achromobacter spp. from cystic fibrosis patients. Int. J. Med Microbiol. 2020, 310, 151393. [Google Scholar] [CrossRef]
| Isolate | Species (Original Isolation) Induction | Non-Synonymous Mutations in Known Genes (Gene Symbols) | Non-Synonymous Mutations in Hypothetical Genes | Synonymous Mutations | Total |
|---|---|---|---|---|---|
| CF01_mut1 | A. ruhlandii (2009, Aarhus) Ticarcillin 1 | 1 (ampD) | 0 | 2 | 3 |
| CF01_mut2 | 1 (mpl) | 0 | 0 | 1 | |
| CF01_mut3 | 1 (mpl) | 0 | 2 | 3 | |
| CF01_2019 | A. ruhlandii (2019, Aarhus) Host factors 2 | 38 (ampD, radA, gabR_4, cysS, thadh, Ion, kefF_1, fan1, est, argP_5, bigR, rpsI, yscL, algD_2, patA, sasA_10, rpsB, rstB_3, regA, oprB, sdgD_2, fusA_2, acsA_1, iclR_4, pcaJ_1, clsB_1, sasA_12, btrW, dnaA, amiD_2, lptF, dctD, livF_10, rstA, murG, ywrD_4) | 14 | 14 | 66 |
| CF02_mut1 | A. ruhlandii (2014, Copenhagen) Ticarcillin 1 | 1 (mpl) | 0 | 1 | 2 |
| CF02_mut2 | 2 (mpl, ABC-trans) | 0 | 1 | 3 | |
| CF02_mut3 | 1 (mpl) | 0 | 0 | 1 | |
| CCUG_mut1 | A. ruhlandii (1998, soil, type strain; CCUG 38886T) Ticarcillin 1 | 4 (mpl, dltA, mutL, clcD) | 4 | 2 | 10 |
| CCUG_mut2 | 2 (mpl, ihfB) | 2 | 2 | 6 | |
| CCUG_mut3 | 2 (mpl, ihfB) | 2 | 1 | 5 | |
| CF03_mut1 | A. xylosoxidans (2009, Aarhus) Ticarcillin 1 | 3 (mpl, rspF, nosY) | 3 | 9 | 15 |
| CF03_mut2 | 1 (mpl) | 3 | 9 | 13 | |
| CF03_mut3 | 1 (mpl) | 2 | 6 | 9 |
| Isolate | Gene | Type | Coding Region Change | Amino Acid Change |
|---|---|---|---|---|
| CF01_mut1 | ampD | SNV | c.503A>C | p.Asp168Ala |
| CF01_mut2 | mpl | SNV | c.1018A>C | p.Thr340Pro |
| CF01_mut3 | mpl | SNV | c.331A>C | p.Thr111Pro |
| CF01_2019 | ampD | SNV | c.370G>A | p.Glu124Lys |
| CF02_mut1 | mpl | Insertion | c.45dupG | p.Leu16fs |
| CF02_mut2 | mpl | Deletion | c.534delT | p.Arg180fs |
| CF02_mut3 | mpl | Deletion | c.555delT | p.Asn185fs |
| CCUG_mut1 | mpl | Insertion | c.538dupC | p.Arg180fs |
| CCUG_mut2 | mpl | SNV | c.761A>G | p.Asp254Gly |
| CCUG_mut3 | mpl | SNV | c.706T>C | p.Trp236Arg |
| CF03_mut1 | mpl | SNV | c.458G>A | p.Arg153His |
| CF03_mut2 | mpl | SNV | c.914T>A | p.Leu305Gln |
| CF03_mut3 | mpl | Insertion | c.550dupC | p.Leu184fs |
| Antimicrobial Agent | Amoxicillin-Clavulanic Acid | Aztreonam | Cefotaxime | Ceftazidime | Ceftazidime-Avibactam | Ceftolozane-Tazobactam | Ertapenem | Imipenem | Meropenem | Piperacillin-Tazobactam | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Gene | ||||||||||
| CF01 | ≤4 | 4 | 2 | ≤0.5 | ≤0.5 | 4 | ≤0.12 | 2 | ≤0.12 | ≤1 | |
| CF01_mut1 | ampD | 64 | >32 | >8 | 4 | ≤0.5 | >32 | 0.5 | 2 | 4 | 8 |
| CF01_mut2 | mpl | 64 | >32 | >8 | 4 | ≤0.5 | >32 | 0.5 | 2 | 4 | 8 |
| CF01_mut3 | mpl | 32 | >32 | >8 | 4 | ≤0.5 | 32 | 0.5 | 2 | 2 | 8 |
| CF01_2019 | ampD | 64 | >32 | >8 | 8 | 2 | 32 | 2 | 16 | 16 | 8 |
| CF02 | 16 | >32 | >8 | 4 | 4 | 32 | ≤0.12 | 2 | 0.25 | 2 | |
| CF02_mut1 | mpl | >64 | >32 | >8 | 8 | 4 | >32 | 1 | 2 | 4 | 32 |
| CF02_mut2 | mpl | >64 | >32 | >8 | 8 | 4 | >32 | 0.5 | 2 | 4 | 32 |
| CF02_mut3 | mpl | 64 | >32 | >8 | 8 | 4 | >32 | 0.5 | 2 | 4 | 32 |
| CCUG | 32 | >32 | >8 | 8 | 8 | 8 | ≤0.12 | 2 | ≤0.12 | 2 | |
| CCUG_mut1 | mpl | 64 | >32 | >8 | 8 | 8 | >32 | 0.5 | 2 | 4 | 16 |
| CCUG_mut2 | mpl | >64 | >32 | >8 | 16 | 16 | >32 | 1 | 4 | 4 | 32 |
| CCUG_mut3 | mpl | 64 | >32 | >8 | 16 | 8 | >32 | 0.5 | 2 | 2 | 16 |
| CF03 | 8 | >32 | >8 | 16 | 16 | >32 | ≤0.12 | 4 | 0.5 | ≤1 | |
| CF03_mut1 | mpl | >64 | >32 | >8 | 16 | 8 | >32 | 1 | 8 | 4 | 16 |
| CF03_mut2 | mpl | >64 | >32 | >8 | 16 | 16 | >32 | 1 | 8 | 8 | 16 |
| CF03_mut3 | mpl | 64 | >32 | >8 | 16 | 8 | >32 | 0.5 | 8 | 2 | 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersen, C.; Gabrielaite, M.; Nørskov-Lauritsen, N. Induction of Broad β-lactam Resistance in Achromobacter ruhlandii by Exposure to Ticarcillin Is Primarily Linked to Substitutions in Murein Peptide Ligase Mpl. Microorganisms 2022, 10, 420. https://doi.org/10.3390/microorganisms10020420
Andersen C, Gabrielaite M, Nørskov-Lauritsen N. Induction of Broad β-lactam Resistance in Achromobacter ruhlandii by Exposure to Ticarcillin Is Primarily Linked to Substitutions in Murein Peptide Ligase Mpl. Microorganisms. 2022; 10(2):420. https://doi.org/10.3390/microorganisms10020420
Chicago/Turabian StyleAndersen, Camilla, Migle Gabrielaite, and Niels Nørskov-Lauritsen. 2022. "Induction of Broad β-lactam Resistance in Achromobacter ruhlandii by Exposure to Ticarcillin Is Primarily Linked to Substitutions in Murein Peptide Ligase Mpl" Microorganisms 10, no. 2: 420. https://doi.org/10.3390/microorganisms10020420
APA StyleAndersen, C., Gabrielaite, M., & Nørskov-Lauritsen, N. (2022). Induction of Broad β-lactam Resistance in Achromobacter ruhlandii by Exposure to Ticarcillin Is Primarily Linked to Substitutions in Murein Peptide Ligase Mpl. Microorganisms, 10(2), 420. https://doi.org/10.3390/microorganisms10020420






