Detection of mcr-1 Gene in Undefined Vibrio Species Isolated from Clams
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area, Sample Collection and Microbial Isolation
2.2. Phenotypic Characterisation of Isolates
2.2.1. Antimicrobial Susceptibility Test
2.2.2. Biochemical Characterisation
2.2.3. Lipid Profile
2.3. Genotypic Characterisation of Isolates
2.3.1. DNA Extraction
2.3.2. Amplification of the 16S rRNA Gene and Housekeeping Genes, Search for Antimicrobial Resistance and Virulence Genes on the Isolates
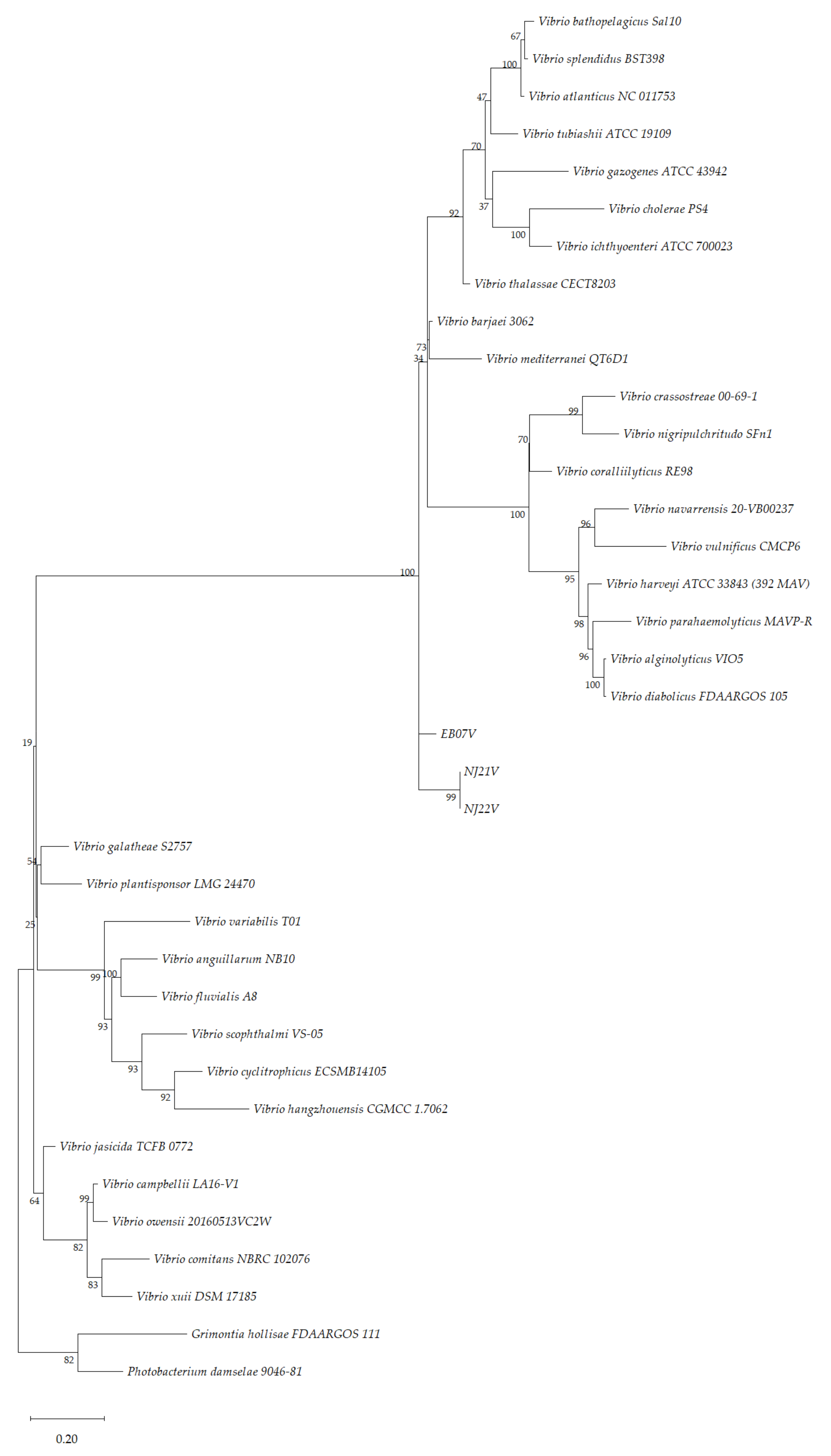
2.4. Phylogenetic Analysis
2.5. Evaluation of mcr-1 Mobility
2.5.1. Plasmid Extraction and Bacteria Transformation
2.5.2. Plasmid Curing
3. Results and Discussion
3.1. Enumeration of Vibrionaceae and Coliforms Isolates
3.2. Antimicrobial Resistance of the Isolates
3.3. Biochemical Analysis of the Isolates
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Farmer, J.J. The Family Vibrionaceae. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, F. Environmental vibrios: «A walk on the wild side». Environ. Microbiol. Rep. 2017, 9, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Gennari, M.; Ghidini, V.; Caburlotto, G.; Lleo, M.M. Virulence genes and pathogenicity islands in environmental Vibrio strains nonpathogenic to humans. FEMS Microbiol. Ecol. 2012, 82, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Trinanes, J.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Non-Cholera Vibrios: The Microbial Barometer of Climate Change. Trends Microbiol. 2017, 25, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Faruque, M.S.; Nair, G.B. Epidemiology. In The Biology of Vibrios; Thompson, F.L., Austin, B., Swings, J., Eds.; ASM Press: Washington, DC, USA, 2006; pp. 385–400. [Google Scholar]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primers 2018, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ina-Salwany, M.Y.; Al-Saari, N.; Mohamad, A.; Mursidi, F.A.; Mohd-Aris, A.; Amal, M.N.A.; Kasai, H.; Mino, S.; Sawabe, T.; Zamri-Saad, M. Vibriosis in Fish: A Review on Disease Development and Prevention. J. Aquat. Anim. Health 2019, 31, 3–22. [Google Scholar] [CrossRef] [PubMed]
- CDC. Chapter 6-Isolation and Identification of Vibrio cholerae. In Laboratory Methods for the Diagnosis of Epidemic Dysentery and Cholera; CDC: Atlanta, Georgia, 1999. [Google Scholar]
- Alcaide, E.; Blasco, M.-D.; Esteve, C. Occurrence of drug-resistant bacteria in two European eel farms. Appl. Environ. Microbiol. 2005, 71, 3348–3350. [Google Scholar] [CrossRef]
- Pedersen, K.; Skall, H.F.; Lassen-Nielsen, A.M.; Nielsen, T.F.; Henriksen, N.H.; Olesen, N.J. Surveillance of health status on eight marine rainbow trout, Oncorhynchus mykiss (Walbaum), farms in Denmark in 2006. J. Fish. Dis. 2008, 31, 659–667. [Google Scholar] [CrossRef]
- Oliveira, J.; Reygaert, W.C. Gram Negative Bacteria; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Hertz, F.B.; Nielsen, J.B.; Schønning, K.; Littauer, P.; Knudsen, J.D.; Løbner-Olesen, A.; Frimodt-Møller, N. Population structure of Drug-Susceptible, -Resistant and ESBL-producing Escherichia coli from Community-Acquired Urinary Tract Infections. BMC Microbiol. 2016, 16, 63. [Google Scholar] [CrossRef]
- Kontopidou, F.; Giamarellou, H.; Katerelos, P.; Maragos, A.; Kioumis, I.; Trikka-Graphakos, E.; Valakis, C.; Maltezou, H.C. Infections caused by carbapenem-resistant Klebsiella pneumoniae among patients in intensive care units in Greece: A multi-centre study on clinical outcome and therapeutic options. CMI 2014, 20, O117–O123. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.I.; Eltayeb, I.B. Self-medication practices with antibiotics and antimalarials among Sudanese undergraduate university students. Ann. Pharmacother. 2007, 41, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Ruuskanen, M.; Muurinen, J.; Meierjohan, A.; Pärnänen, K.; Tamminen, M.; Lyra, C.; Kronberg, L.; Virta, M. Fertilizing with animal manure disseminates antibiotic resistance genes to the farm environment. J. Environ. Qual. 2016, 45, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Harris, P.N.A. Colistin resistance: A major breach in our last line of defence. Lancet Infect. Dis. 2016, 16, 132–133. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. AMR Rev. 2016. Available online: http://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 22 December 2021).
- Jeannot, K.; Bolard, A.; Plesiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Rabanal, F.; Cajal, Y. Recent advances and perspectives in the design and development of polymyxins. Nat. Prod. Rep. 2017, 34, 886–908. [Google Scholar] [CrossRef] [PubMed]
- Kempf, I.; Fleury, M.A.; Drider, D.; Bruneau, M.; Sanders, P.; Chauvin, C.; Madec, J.-Y.; Jouy, E. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int. J. Antimicrob. Agents 2013, 42, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Chung, E.S.; Na, I.Y.; Kim, H.; Shin, D.; Ko, K.S. Development of colistin resistance in pmrA-, phoP-, parR-and cprR-inactivated mutants of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2014, 69, 2966–2971. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infec. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef] [PubMed]
- Irrgang, A.; Roschanski, N.; Tenhagen, B.-A.; Grobbel, M.; Skladnikiewicz-Ziemer, T.; Thomas, K.; Roesler, U.; Kaesbohrer, A. Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010–2015. PLoS ONE 2016, 11, e0159863. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Benito, R.; Iglesias, M.R.; Quijada, N.M.; Campos, M.J.; Ugarte-Ruiz, M.; Hernández, M.; Pazos, C.; Rodríguez-Lázaro, D.; Garduño, E.; Domínguez, L.; et al. Escherichia coli ST167 carrying plasmid mobilisable mcr-1 and bla(CTX-M-15) resistance determinants isolated from a human respiratory infection. Int. J. Antimicrob. Agents. 2017, 50, 285–286. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 2016, 71, 2342–2344. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Lu, X.; Munir, A.; Abdullah, S.; Liu, Y.; Xiao, X.; Wang, Z.; Mohsin, M. Widespread prevalence and molecular epidemiology of tet(X4) and mcr-1 harboring Escherichia coli isolated from chickens in Pakistan. Sci. Total Environ. 2021, 806, 150689. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Wu, Y. China bans colistin as a feed additive for animals. Lancet Infect. Dis. 2016, 16. [Google Scholar] [CrossRef]
- McGann, P.; Snesrud, E.; Maybank, R.; Corey, B.; Ong, A.C.; Clifford, R.; Hinkle, M.; Whitman, T.; Lesho, E.; Schaecher, K.E. Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First Report of mcr-1 in the United States. Antimicrob. Agents Chemother. 2016, 60, 4420–4421. [Google Scholar] [CrossRef] [PubMed]
- Elnahriry, S.S.; Khalifa, H.O.; Soliman, A.M.; Ahmed, A.M.; Hussein, A.M.; Shimamoto, T.; Shimamoto, T. Emergence of Plasmid-Mediated Colistin Resistance Gene mcr-1 in a Clinical Escherichia coli Isolate from Egypt. Antimicrob. Agents. Chemother. 2016, 60, 3249–3250. [Google Scholar] [CrossRef]
- Khedher, M.B.; Baron, S.A.; Riziki, T.; Ruimy, R.; Raoult, D.; Diene, S.M.; Rolain, J.-M. Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: Is there another role for these enzymes? Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, N.; Nordmann, P.; Poirel, L. Moraxella Species as Potential Sources of MCR-Like Polymyxin Resistance Determinants. Antimicrob. Agents. Chemother. 2017, 61, e00129-17. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Zhang, J.; Jiang, F.; He, M.; Zeng, H.; Chen, M.; Wu, S.; Wang, J.; Ding, Y.; Wu, Q. First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 2019, 308, 108290. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Uchino, M.; Kalinovskaya, N.I.; Mikhailov, V.V. Isolation, phylogenetic analysis and screening of marine mollusc-associated bacteria for antimicrobial, hemolytic and surface activities. Microbiol. Res. 2008, 163, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Richards, G.P. Microbial purification of shellfish: A review of depuration and relaying. J. Food Prot. 1988, 51, 218–251. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L. The potential for marine bivalve shellfish to act as transmission vehicles for outbreaks of protozoan infections in humans: A review. Int. J. Food Microbiol. 2007, 120, 201–216. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. Antimicrobial Susceptibility Testing—EUCAST Disk Diffusion Method. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2021_manuals/Manual_v_9.0_EUCAST_Disk_Test_2021.pdf (accessed on 21 January 2021).
- EUCAST. Broth microdilution—EUCAST Reading Guide v 4.0. Available online: https://www.eucast.org/ast_of_bacteria/mic_determination/?no_cache=1 (accessed on 14 January 2022).
- Coico, R. Gram staining. Curr. Protoc. 2006. [Google Scholar] [CrossRef]
- Suslow, T.V.; Schroth, M.N.; Isaka, M. Application of a rapid method for Gram differentiation of plant pathogenic and saprophytic bacteria without staining. Phytopathology 1982, 72, 917–918. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Letchumanan, V.; Chan, K.-G.; Lee, L.-H. An insight of traditional plasmid curing in Vibrio species. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Law, E.U. Microbiological Criteria for Foodstuffs. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A02005R2073-20200308 (accessed on 10 August 2021).
- Gerba, C.P.; Goyal, S.M.; LaBelle, R.L.; Cech, I.; Bodgan, G.F. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am. J. Public Health. 1979, 69, 1116–1119. [Google Scholar] [CrossRef]
- Formiga-Cruz, M.; Allard, A.K.; Conden-Hansson, A.-C.; Henshilwood, K.; Hernroth, B.E.; Jofre, J.; Lees, D.N.; Lucena, F.; Papapetropoulou, M.; Rangdale, R.E. Evaluation of potential indicators of viral contamination in shellfish and their applicability to diverse geographical areas. Appl. Environ. Microbiol. 2003, 69, 1556–1563. [Google Scholar] [CrossRef]
- Høi, L.; Dalsgaard, I.; Dalsgaard, A. Improved isolation of Vibrio vulnificus from seawater and sediment with cellobiose-colistin agar. Appl. Environ. Microbiol. 1998, 64, 1721–1724. [Google Scholar] [CrossRef]
- Mondal, M.; Nag, D.; Koley, H.; Saha, D.R.; Chatterjee, N.S. The Vibrio cholerae extracellular chitinase ChiA2 is important for survival and pathogenesis in the host intestine. PLoS ONE 2014, 9, e103119. [Google Scholar] [CrossRef] [PubMed]
- Zobell, C.E.; Rittenberg, S.C. The occurrence and characteristics of chtinoclastic bacteria in the sea. J. Bacteriol. 1938, 35, 275–287. [Google Scholar] [CrossRef]
- Wortman, A.T.; Somerville, C.C.; Colwell, R.R. Chitinase determinants of Vibrio vulnificus: Gene cloning and applications of a chitinase probe. Appl. Environ. Microbiol. 1986, 52, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Soto-GiI, R.W.; Zyskind, J.W. Chitin, Chitosan, and Related Enzymes; Zikakis, J., Ed.; Academic Press Inc.: New York, NY, USA, 1984. [Google Scholar]
- Hirono, I.; Yamashita, M.; Aoki, T. Molecular cloning of chitinase genes from Vibrio anguillarum and V. parahaemolyticus. J. Appl. Microbiol. 1998, 84, 1175–1178. [Google Scholar] [CrossRef]
- Huq, A.; Small, E.B.; West, P.A.; Huq, M.I.; Rahman, R.; Colwell, R.R. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 1983, 45, 275–283. [Google Scholar] [CrossRef]
- Pruzzo, C.; Vezzulli, L.; Colwell, R.R. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 2008, 10, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Kimmerer, W.J.; Gartside, E.; Orsi, J.J. Predation by an introduced clam as the likely cause of substantial declines in zooplankton of San Francisco Bay. Mar. Ecol. Prog. Ser. 1994, 113, 81–93. [Google Scholar] [CrossRef]
- Kimmerer, W.J.; Lougee, L. Bivalve grazing causes substantial mortality to an estuarine copepod population. J. Exp. Mar. Biol. Ecol. 2015, 473, 53–63. [Google Scholar] [CrossRef]
- Pace, M.L.; Findlay, S.E.G.; Fischer, D. Effects of an invasive bivalve on the zooplankton community of the Hudson River. Freshw. Biol. 1998, 39, 103–116. [Google Scholar] [CrossRef]
- Sun, J.; Fang, L.-X.; Wu, Z.; Deng, H.; Yang, R.-S.; Li, X.-P.; Li, S.-M.; Liao, X.-P.; Feng, Y.; Liu, Y.-H. Genetic analysis of the IncX4 plasmids: Implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 2017, 7, 424. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Qin, W.; Lin, J.; Fang, S.; Qiu, J. Antibacterial Mechanisms of Polymyxin and Bacterial Resistance. Biomed. Res. Int. 2015, 2015, 679109. [Google Scholar] [CrossRef] [PubMed]
- Bakour, S.; Olaitan, A.O.; Ammari, H.; Touati, A.; Saoudi, S.; Saoudi, K.; Rolain, J.M. Emergence of Colistin- and Carbapenem-Resistant Acinetobacter baumannii ST2 Clinical Isolate in Algeria: First Case Report. Microb. Drug. Resist. 2015, 21, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Eguchi, T. Biosynthesis of archaeal membrane lipids: Digeranylgeranylglycerophospholipid reductase of the thermoacidophilic archaeon Thermoplasma acidophilum. J. Biochem. 2006, 139, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Hadjadj, L.; Rolain, J.M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents. 2016, 48, 583–591. [Google Scholar] [CrossRef]
- Mambelli, L.I.; Teixeira, S.F.; Jorge, S.D.; Kawamura, B.; Meneguelo, R.; Barbuto, J.A.M.; de Azevedo, R.A.; Ferreira, A.K. Phosphoethanolamine induces caspase-independent cell death by reducing the expression of C-RAF and inhibits tumor growth in human melanoma model. Biomed. Pharmacother. 2018, 103, 18–28. [Google Scholar] [CrossRef]
- Massad, G.; Oliver, J.D. New selective and differential medium for Vibrio cholerae and Vibrio vulnificus. Appl. Environ. Microbiol. 1987, 53, 2262–2264. [Google Scholar] [CrossRef]
- Hankins, J.V.; Madsen, J.A.; Giles, D.K.; Brodbelt, J.S.; Trent, M.S. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8722–8727. [Google Scholar] [CrossRef]
- Henderson, J.C.; Herrera, C.M.; Trent, M.S. AlmG, responsible for polymyxin resistance in pandemic Vibrio cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J. Biol. Chem. 2017, 292, 21205–21215. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Henderson, J.C.; Crofts, A.A.; Trent, M.S. Novel coordination of lipopolysaccharide modifications in Vibrio cholerae promotes CAMP resistance. Mol. Microbiol. 2017, 106, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Crofts, A.A.; Henderson, J.C.; Pingali, S.C.; Davies, B.W.; Trent, M.S. The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. mBio 2014, 5, e02283-14. [Google Scholar] [CrossRef] [PubMed]
- Bina, X.R.; Howard, M.F.; Ante, V.M.; Bina, J.E. Vibrio cholerae LeuO links the ToxR regulon to expression of lipid A remodeling genes. Infect. Immun. 2016, 84, 3161–3171. [Google Scholar] [CrossRef]
- Zhang, H.; Srinivas, S.; Xu, Y.; Wei, W.; Feng, Y. Genetic and Biochemical Mechanisms for Bacterial Lipid A Modifiers Associated with Polymyxin Resistance. Trends Biochem. Sci. 2019, 44, 973–988. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chuang, Y.C.; Chang, C.C.; Jeang, C.L.; Chang, M.C. A K+ uptake protein, TrkA, is required for serum, protamine, and polymyxin B resistance in Vibrio vulnificus. Infect. Immun. 2004, 72, 629–636. [Google Scholar] [CrossRef]
- Tantillo, G.M.; Fontanarosa, M.; Di Pinto, A.; Musti, M. Updated perspectives on emerging vibrios associated with human infections. Lett. Appl. Microbiol. 2004, 39, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Williams, D.W. Chapter Twelve—Vibrio. In Microbiology of Waterborne Diseases, 2nd ed.; Percival, S.L., Yates, M.V., Williams, D.W., Chalmers, R.M., Gray, N.F., Eds.; Academic Press: London, UK, 2014; pp. 237–248. [Google Scholar]
- Kunitsky, C.; Osterhout, G.; Sasser, M. Identificatiom of microorganisms using fatty acid methyl ester (FAME) analysis and the MIDI Sherlock® microbial identification system. In Encyclopedia of Rapid Microbiological Methods; Miller, M.J., Ed.; PDA: Bethesda, MD, USA; DHI Publishing LLC: River Grove, IL, USA,, 2006; Volume 3. [Google Scholar]
- de Carvalho, C.C.C.R.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Fischer, M.; Whittaker, P. Evaluating the use of fatty acid profiles to identify deep-sea Vibrio isolates. Food Chem. 2010, 122, 943–950. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Planas, M.; Pintado, J. Vibrio inhibens sp. nov., a novel bacterium with inhibitory activity against Vibrio species. J. Antibiot. 2012, 65, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk, Y.; Ogura, Y.; Hayashi, T.; Urbanczyk, H. Genomic evidence that Vibrio inhibens is a heterotypic synonym of Vibrio jasicida. Int. J. Syst. Evol. Microbiol. 2016, 66, 3214–3218. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, T.; Takata, N.; Kudo, T.; Horikoshi, K. Effect of temperature and growth phase on fatty acid composition of the psychrophilic Vibrio sp. strain no. 5710. FEMS Microbiol. Lett. 1994, 119, 77–81. [Google Scholar] [CrossRef]
- Estupiñán, M.; Hernández, I.; Saitua, E.; Bilbao, M.E.; Mendibil, I.; Ferrer, J.; Alonso-Sáez, L. Novel Vibrio spp. strains producing omega-3 fatty acids isolated from coastal seawater. Mar. Drugs 2020, 18, 99. [Google Scholar] [CrossRef] [PubMed]
- Montieri, S.; Suffredini, E.; Ciccozzi, M.; Croci, L. Phylogenetic and evolutionary analysis of Vibrio parahaemolyticus and Vibrio alginolyticus isolates based on toxR gene sequence. New Microbiol. 2010, 33, 359–372. [Google Scholar] [PubMed]
- Thompson, F.; Gevers, D.; Thompson, C.; Dawyndt, P.; Naser, S.; Hoste, B.; Munn, C.; Swings, J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. App. Environ. Microbiol. 2005, 71, 5107–5115. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.C.; Thompson, F.L.; Vicente, A.C.P.; Swings, J. Phylogenetic analysis of vibrios and related species by means of atpA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2480–2484. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; Fusco, V.; Ercolini, D.; Aponte, M.; Pepe, O.; Villani, F. Lactobacillus strain diversity based on partial hsp60 gene sequences and design of PCR-restriction fragment length polymorphism assays for species identification and differentiation. Appl. Environ. Microbiol. 2008, 74, 208–215. [Google Scholar] [CrossRef]
- Nishibuchi, M. Molecular Identification. In The biology of Vibrios; Thompson, F.L., Austin, B., Swings, J., Eds.; ASM Press: Washington, DC, USA, 2006. [Google Scholar]
- Beaz-Hidalgo, R.; Doce, A.; Pascual, J.; Toranzo, A.E.; Romalde, J.L. Vibrio gallaecicus sp. nov. isolated from cultured clams in north-western Spain. Syst. Appl. Microbiol. 2009, 32, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Faury, N.; Saulnier, D.; Thompson, F.; Gay, M.; Swings, J.; Le Roux, F. Vibrio crassostreae sp. nov., isolated from the haemolymph of oysters (Crassostrea gigas). Int. J. Syst. Evol. Microbiol. 2004, 54, 2137–2140. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, F.; Goubet, A.; Thompson, F.; Faury, N.; Gay, M.; Swings, J.; Saulnier, D. Vibrio gigantis sp. nov., isolated from the haemolymph of cultured oysters (Crassostrea gigas). Int. J. Syst. Evol. Microbiol. 2005, 55, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Dubert, J.; Balboa, S.; Regueira, M.; González-Castillo, A.; Gómez-Gil, B.; Romalde, J.L. Vibrio barjaei sp. nov., a new species of the Mediterranei clade isolated in a shellfish hatchery. Syst. Appl. Microbiol. 2016, 39, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-S.; Wang, L.-T.; Chen, J.-S.; Chen, Y.-T.; Wei, S.T.-S.; Chiang, Y.-R.; Wang, P.-L.; Lee, T.-H.; Lin, S.-T.; Huang, L.; et al. Vibrio nitrifigilis sp. nov., a marine nitrogen-fixing bacterium isolated from the lagoon sediment of an islet inside an atoll. Anton. Leeuw. 2021, 114, 933–945. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Sawabe, T.; Kita-Tsukamoto, K.; Thompson, F.L. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 2007, 189, 7932–7936. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.; Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005, 43, 4178. [Google Scholar] [CrossRef]
- Hossain, M.T.; Kim, Y.-R.; Kong, I.-S. PCR–restriction fragment length polymorphism analysis using groEL gene to differentiate pathogenic Vibrio species. Diagn. Microbiol. Infect. Dis. 2014, 78, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Lane, D. 16S/23S rRNA Sequencing, Nucleic Acid Techniques; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991. [Google Scholar]
- Nhung, P.H.; Ohkusu, K.; Mishima, N.; Noda, M.; Shah, M.M.; Sun, X.; Hayashi, M.; Ezaki, T. Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagn. Microbiol. Infect. Dis. 2007, 58, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17–00672. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Monstein, H.J.; Ostholm-Balkhed, A.; Nilsson, M.V.; Nilsson, M.; Dornbusch, K.; Nilsson, L.E. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Mulvey, M.R.; Soule, G.; Boyd, D.; Demczuk, W.; Ahmed, R. Characterization of the first extended-spectrum beta-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 2003, 41, 460–462. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef]
- Pribul, B.R.; Festivo, M.L.; Souza, M.M.S.; Rodrigues, D.d.P. Characterization of quinolone resistance in Salmonella spp. isolates from food products and human samples in Brazil. Braz. J. Microbiol. 2016, 47, 196–201. [Google Scholar] [CrossRef][Green Version]
- Hammad, E.; Helal, R. PMQR determinants among clinical isolates of ESBL and Amp C producing Serratia marcescens in Mansoura University Hospitals: A 6-year study. Int. Arab. J. Antimicrob. Agents 2015, 5. [Google Scholar] [CrossRef]
- Ruwandeepika, H.; Defoirdt, T.; Bhowmick, P.; Shekar, M.; Bossier, P.; Karunasagar, I. Presence of typical and atypical virulence genes in vibrio isolates belonging to the Harveyi clade. J. Appl. Microbiol. 2010, 109, 888–899. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Pang, L.; Qi, Z.; Chen, J.; Austin, B.; Zhang, X.-H. Distribution of five vibrio virulence-related genes among Vibrio harveyi isolates. J. Gen. Appl. Microbiol. 2008, 54, 71–78. [Google Scholar] [CrossRef][Green Version]
- Saravanan, V.; Kumar, H.S.; Karunasagar, I.; Karunasagar, I. Putative virulence genes of Vibrio cholerae from seafoods and the coastal environment of Southwest India. Int. J. Food Microbiol. 2007, 119, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Bej, A.K.; Patterson, D.P.; Brasher, C.W.; Vickery, M.C.; Jones, D.D.; Kaysner, C.A. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 1999, 36, 215–225. [Google Scholar] [CrossRef]
- Fields, P.; Popovic, T.; Wachsmuth, K.; Olsvik, Ø. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemic. J. Clin. Microbiol. 1992, 30, 2118–2121. [Google Scholar] [CrossRef]
- Rivera, I.N.; Chun, J.; Huq, A.; Sack, R.B.; Colwell, R.R. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 2001, 67, 2421–2429. [Google Scholar] [CrossRef]
- Lee, S.E.; Kim, S.Y.; Kim, S.J.; Kim, H.S.; Shin, J.H.; Choi, S.H.; Chung, S.S.; Rhee, J.H. Direct identification of Vibrio vulnificus in clinical specimens by nested PCR. J. Clin. Microbiol. 1998, 36, 2887–2892. [Google Scholar] [CrossRef]
- Karaolis, D.K.; Johnson, J.A.; Bailey, C.C.; Boedeker, E.C.; Kaper, J.B.; Reeves, P.R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 1998, 95, 3134–3139. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Latif, H.M.R.; Khalil, R.H.; Saad, T.T.; El-bably, R.Y. Identification and Molecular Characterization of Yersinia ruckeri isolated from mass mortalities of cultured Nile tilapia at Kafr El-sheikh governorate. Glob. J. Fish. Aquac. Res. 2014, 1, 1–17. [Google Scholar]
- Cai, S.H.; Lu, Y.S.; Wu, Z.H.; Jian, J.C.; Huang, Y.C. A novel multiplex PCR method for detecting virulent strains of Vibrio alginolyticus. Aquac. Res. 2009, 41, 27–34. [Google Scholar] [CrossRef]
- Conejero, M.J.U.; Hedreyda, C.T. PCR detection of hemolysin (vhh) gene in Vibrio harveyi. J. Gen. Appl. Microbiol. 2004, 50, 137–142. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, Z.Y.; Hu, C.Q.; Chen, C.; Zhang, L.P.; Ren, C.H. Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Lett. Appl. Microbiol. 2005, 41, 202–207. [Google Scholar] [CrossRef] [PubMed]
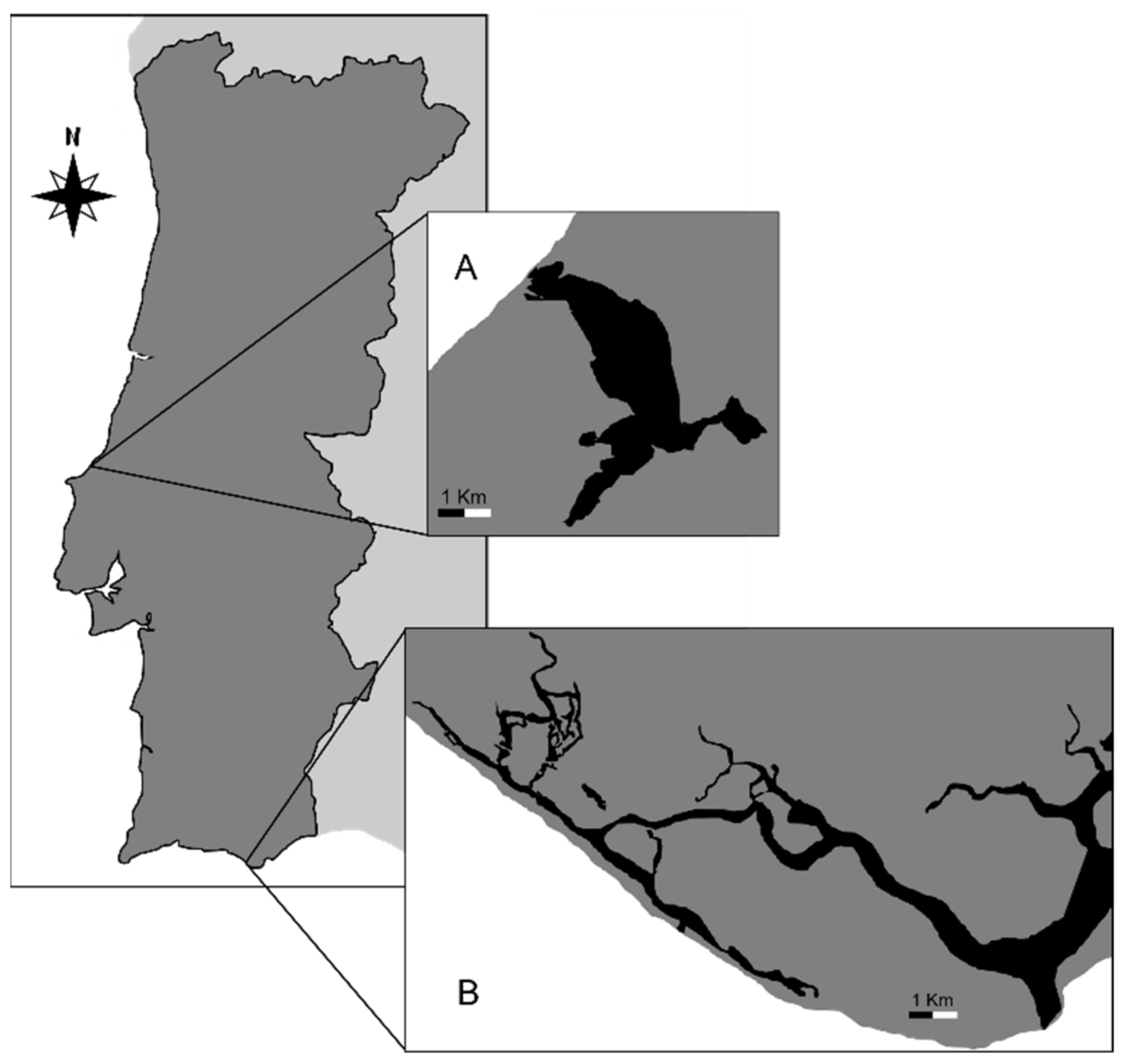
| Isolates | Óbidos Lagoon | Ria Formosa | |
|---|---|---|---|
| R. decussatus (CFU g−1) | R. phillipinarum (CFU g−1) | R. decussatus (CFU g−1) | |
| Vibrionaceae | 1.1 × 104 | 4.5 × 103 | 3.8 × 103 |
| Coliforms | 8.7 × 103 | 3.5 × 103 | 5.0 × 102 |
| Test | EB07V | NJ21V | NJ22V |
|---|---|---|---|
| Nitrate reduction | + | + | + |
| Tryptophane production | + | + | + |
| Glucose fermentation | + | − | − |
| Arginine dihydrolase | − | − | − |
| Urease | − | − | − |
| Esculin (β-glucosidase activity) | + | + | + |
| Gelatin (protease activity) | + | + | + |
| Para-NitroPhenyl-β-D-Galactopyranosidase (β-galactosidase activity) | + | + | + |
| Glucose assimilation | + | + | + |
| Arabinose assimilation | − | − | − |
| Mannose assimilation | + | + | − |
| Mannitol assimilation | + | + | + |
| N-Acetyl-Glucosamine assimilation | + | + | + |
| Maltose assimilation | + | + | + |
| Potassium gluconate assimilation | + | + | + |
| Capric acid assimilation | − | − | − |
| Adipic acid assimilation | − | NC | − |
| Malate assimilation | + | + | + |
| Trisodium citrate assimilation | + | − | − |
| Phenylacetic acid assimilation | − | − | − |
| Cytochrome oxidase | + | + | + |
| Fatty Acid | EB07V | NJ21V | NJ22V |
|---|---|---|---|
| 12:0 | 3.72 | 8.11 | 7.22 |
| 14:0 iso | 1.13 | ||
| 14:0 | 5.29 | 3.12 | 2.70 |
| 15:0 iso | 0.96 | ||
| 16:0 iso | 1.28 | 0.82 | 0.88 |
| 16:0 | 15.85 | 25.05 | 26.87 |
| 17:0 iso | 1.76 | ||
| 18:3 w6c | 14.24 | 20.78 | 12.17 |
| 18:0 | 3.60 | 2.88 | 2.43 |
| Summed Feature 3 | 30.46 | 24.53 | 26.59 |
| Summed Feature 8 | 21.15 | 12.68 | 18.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdez, C.; Costa, C.; Simões, M.; de Carvalho, C.C.C.R.; Baptista, T.; Campos, M.J. Detection of mcr-1 Gene in Undefined Vibrio Species Isolated from Clams. Microorganisms 2022, 10, 394. https://doi.org/10.3390/microorganisms10020394
Valdez C, Costa C, Simões M, de Carvalho CCCR, Baptista T, Campos MJ. Detection of mcr-1 Gene in Undefined Vibrio Species Isolated from Clams. Microorganisms. 2022; 10(2):394. https://doi.org/10.3390/microorganisms10020394
Chicago/Turabian StyleValdez, Christian, Cátia Costa, Marco Simões, Carla C. C. R. de Carvalho, Teresa Baptista, and Maria J. Campos. 2022. "Detection of mcr-1 Gene in Undefined Vibrio Species Isolated from Clams" Microorganisms 10, no. 2: 394. https://doi.org/10.3390/microorganisms10020394
APA StyleValdez, C., Costa, C., Simões, M., de Carvalho, C. C. C. R., Baptista, T., & Campos, M. J. (2022). Detection of mcr-1 Gene in Undefined Vibrio Species Isolated from Clams. Microorganisms, 10(2), 394. https://doi.org/10.3390/microorganisms10020394







