Abstract
Streptomyces sp. N11-34 is a producer of bicyclic peptides named nyuzenamides A and B. We elucidated its taxonomic position and surveyed its nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) gene clusters by whole genome analysis. Streptomyces sp. N11-34 showed 16S rRNA gene sequence similarities of 99.9% and 99.8% to Streptomyces hygroscopicus NBRC 13472T and Streptomyces demainii NRRL B-1478T, respectively. Although these members formed a clade in a phylogenetic tree based on 16S rRNA gene sequences, the clade split into two closely related subclades in multilocus sequence analysis (MLSA). One included Streptomyces sp. N11-34, S. demainii NRRL B-1478T, S. hygroscopicus NBRC 100766, S. hygroscopicus NBRC 16556 and S. hygroscopicus TP-A0867 and the other comprised S. hygroscopicus NBRC 13472T and S. hygroscopicus NBRC 12859. These phylogenetic relationships were supported by phylogenomic analysis. Although Streptomyces sp. N11-34 was classified to S. hygroscopicus at the species level based on MLSA evolutionary distances and DNA–DNA relatedness, these distances and relatedness of members between the two subclades were comparatively far (0.004–0.006) and low (75.4–76.4%), respectively. Streptomyces sp. N11-34 possessed six NRPS, seven PKS and four hybrid PKS/NRPS gene clusters in the genome. Among the seventeen, ten were identified to be biosynthetic gene clusters (BGCs) of nyuzenamide, echoside, coelichelin, geldanamycin, mediomycin, nigericin, azalomycin, spore pigment, alchivemycin and totopotensamide, whereas the remaining seven were orphan in our bioinformatic analysis. All seventeen are conserved in S. hygroscopicus NBRC 100766, S. hygroscopicus NBRC 16556 and S. hygroscopicus TP-A0867. In contrast, S. hygroscopicus NBRC 13472T and S. hygroscopicus NBRC 12859 lacked the BGCs of alchivemycin, totopotensamide, a nonribosomal peptide and a hybrid polyketide/nonribosomal peptide compound. This difference was in a good accordance with the abovementioned phylogenetic relationship. Based on phenotypic differences in addition to phylogenetic relationship, DNA–DNA relatedness and BGCs, strains of S. hygroscopicus should be reclassified to two subspecies: S. hygroscopicus subsp. hygroscopicus and a new subspecies, for which we proposed S. hygroscopicus subsp. sporocinereus subsp. nov. The type strain is NBRC 100766T (=ATCC 43692T = DSM 41460T = INMI 32T = JCM 9093T = NRRL B-16376T = VKM Ac-312T). S. demainii was classified in this subspecies.
1. Introduction
Nonribosomal peptides and polyketides are the two largest families in the secondary metabolites of actinomycetes. These compounds are structurally diverse and often exhibit pharmaceutically useful biological activities. Half to two thirds of the secondary metabolite–biosynthetic gene clusters (smBGCs) in each actinomycetal genome are nonribosomal peptide synthetase (NRPS), polyketide synthase (PKS) and hybrid PKS/NRPS gene clusters, while each strain, such as that of the genus Streptomyces, harbors dozen of smBGCs [1]. NRPS and PKS pathways share a similar biosynthetic mechanism. Backbones of these products are synthesized by incorporation of building blocks, such as amino acid or acyl-CoA, respectively, into the growing chains. Biosyntheses by NRPS and type-I PKS pathways are catalyzed by large modular enzymes with multiple domains, according to the co-linearity rule of assembly line fashion. A minimum NRPS module consists of an adenylation (A) domain for selecting the incoming amino acid, a condensation (C) domain for condensing the building block with the peptidyl intermediate from the previous module and a thiolation (T) domain for carrying the growing polypeptide chain. Similarly, a minimal PKS module consists of an acyltransferase (AT) domain for selecting incoming acyl-CoAs, a ketosynthase (KS) domain for condensing the new building block with the acyl intermediate from the previous module and an acyl carrier protein (ACP) domain for carrying the growing polyketide chain. Individual modules are responsible for the incorporation of either one amino acid or acyl-CoA as a building block into the chain [2,3]. Optional domains may be present in each module, which methylate or epimerize incorporated amino acid residues in nonribosomal peptides or reduce a keto group in polyketide chains. Thus, we can predict backbones of the products based on module numbers, domain organization and the substrates of A and AT domains in each gene cluster by bioinformatic analysis [3,4]. Hence, PKS and NRPS gene clusters are often investigated to access the potential of each strain to produce diverse secondary metabolites [5,6,7,8].
We recently isolated Streptomyces sp. N11-34 from deep sea water and found two novel compounds designated nyuzenamides A and B from the strain. Nyuzenamides are bicyclic peptides (Figure 1) with antifungal and cytotoxic activity [9]. Although these compounds seem to be synthesized through an NRPS pathway, the biosynthetic gene cluster (BGC) has not yet been elucidated. In the present study, we investigated the taxonomic position of Streptomyces sp. N11-34 and analyzed NRPS and PKS gene clusters to identify the nyuzenamide-BGC and reveal hidden potential to produce other compounds. Consequently, we classified Streptomyces sp. N11-34 to S. hygroscopicus.
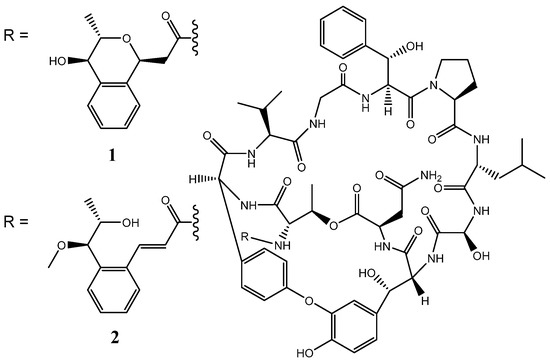
Figure 1.
Chemical structures of nyuzenamides A (1) and B (2).
S.hygroscopicus is known to include strains significant in industrial and biotechnological applications. As various bioactive secondary metabolites have been discovered from the members, it is expected as a source for searching novel bioactive compounds in pharmaceutical industries. This species once included four subspecies with validly published names. However, as they were reclassified to independent species, S. hygroscopicus includes no subspecies at present [10]. In the present study, we compared S. hygroscopicus N11-34 with its taxonomic neighbors, and consequently revealed that members of S. hygroscopicus can be classified into two groups. Thus, we here propose a new subspecies of S. hygroscopicus.
2. Materials and Methods
Streptomyces sp. N11-34 was isolated in the previous study [9]. This strain has been deposited to and available from the NBRC Culture Collection as NBRC 113678. EzBioCloud [11] was used to search for taxonomic neighbors based on 16S rRNA gene sequences. Multilocus sequence analysis (MLSA) was conducted using DNA sequences of five housekeeping genes—atpD, gyrB, recA, rpoB and trpB—as established in the genus Streptomyces [12]. The accession numbers of gene sequences used for MLSA are listed in Table S1. The phylogenetic trees were reconstructed using ClustalX 2.1 [13]. Genomic DNA of Streptomyces sp. N11-34 for whole genome sequencing was prepared from cultured cells via the method of Saito and Kimura [14]. The whole genome was sequenced by the Kazusa DNA Research Institute using a single-molecule real-time (SMRT) strategy in the same manner of our previous report [7]. The assembled genome sequences were deposited to DDBJ under the accession numbers BNEK01000001–BNEK01000009. Phylogenomic tree was constructed using the TYGS webserver [15]. DNA–DNA relatedness was digitally calculated using whole genome sequences by Formula 2 of the Genome-to-Genome Distance Calculator (GGDC), an in silico method that reliably mimics conventional DNA–DNA hybridization experiments [16]. PKS and NRPS gene clusters in the genomes were surveyed using antiSMASH, which allows the rapid genome-wide identification, annotation and analysis of smBGCs in microbial genomes [4], and then manually analyzed as reported previously [6]. Whole genome sequences used for DNA–DNA relatedness calculation and NRPS and PKS gene cluster analysis are listed in Table 1.

Table 1.
Whole genome sequences of Streptomyces sp. N11-34 and its taxonomic neighbors.
3. Results
3.1. Taxonomic Positions of Streptomyces sp. N11-34
Streptomyces sp. N11-34 showed 16S rRNA gene sequence similarities of 99.9% (1448/1449) and 99.8% (1446/1449) to Streptomyces hygroscopicus NBRC 13472T and Streptomyces demainii NRRL B-1478T, respectively. In a phylogenetic tree based on 16S rRNA gene sequences, Streptomyces sp. N11-34 formed a clade with these members (Figure S1). As S. hygroscopicus has two heterotypic synonyms, Streptomyces endus and Streptomyces sporocinereus [17], we included their type strains in the tree. S. hygroscopicus TP-A0867 is an alchivemycin producer [18]. S. hygroscopicus NBRC 16556 is a strain for which we reported the whole genome sequence [19].
As 16S rRNA gene sequence analysis is known to be low in the resolution, we next conducted MLSA and phylogenomic analysis. MLSA is often used for elucidating phylogenetic relationships with higher resolutions [12], whereas phylogenomic analysis can clarify whole genome sequence-based phylogenies [15]. In the MLSA-based phylogenetic tree, Streptomyces sp. N11-34 formed a clade with members of S. hygroscopicus and S. demainii. However, the topology within the clade was different from that in the 16S rRNA gene sequence-based phylogenetic tree. The clade clearly split into two subclades with the bootstrap values of 100%: one comprises only the type strains of S. hygroscopicus and S. endus whereas the other is composed of the remaining members, including Streptomyces sp. N11-34 and the type strains of S. sporocinereus and S. demainii (Figure S2). Similarly, the members within the S. hygroscopicus clades split into two in the phylogenomic tree (Figure 2).
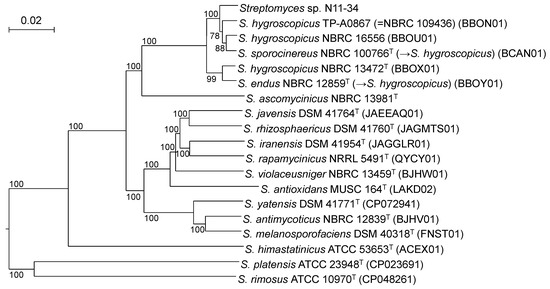
Figure 2.
Phylogenomic tree constructed by the TYGS server. Confidence limits above 50% are at branching points. The codes in parentheses show accession numbers of whole genome sequences or WGS Projects. S. albus NBRC 13014T (BBQG01) was used as an outgroup (not shown).
DNA–DNA relatedness value of 70% is established as the cut-off for species delineations in bacteria systematics [20]. In the genus Streptomyces, 0.007 in MLSA evolutionary distance is recognized to correspond to the cut-off [12]. Among Streptomyces sp. N11-34 and the phylogenetic neighbors, the DNA–DNA relatedness and MLSA evolutionary distances ranged 75.4–90.1% and 0.000–0.006, respectively (Table 2). This suggests that these members represent the same species. Hence, Streptomyces sp. N11-34 is classified to S. hygroscopicus. The seven strains have been phylogenetically grouped into two, one is 1–5 and the other is 6–7, as stated above. Within the group 1–5, DNA–DNA relatedness and MLSA evolutionary distances are 85.0–91.8% and 0.000–0.003, respectively. Similarly, these values are 90.1% and 0.000 within the other 6–7, respectively. In contrast, values between the two groups are 75.4–76.4% and 0.004–0.006, respectively. As the threshold for subspecies demarcation is reported to be 79–80% in the DNA–DNA relatedness in bacteria [21], members between groups 1–5 and 6–7 are discriminated at subspecies level.

Table 2.
MLSA evolutionary distances and DNA–DNA relatedness among Streptomyces sp. N11-34 and the phylogenetically close strains.
3.2. NRPS and PKS Gene Clusters of Streptomyces sp. N11-34
Streptomyces sp. N11-34 harbored six NRPS, seven PKS and four hybrid PKS/NRPS gene clusters in the genome, as listed in Table 3. We identified nrps-1 and -2 to be BGCs for echoside and coelichelin, respectively, by bioinformatic analysis. The NRPSs showed high similarities to EchA and SCO0492, responsible for echoside and coelichelin biosyntheses, respectively (Table 4). Although nrps-3 was not a reported gene cluster, we identified it to be the nyuzenamide-BGC because the domain organization well accounts for it. Predicted amino acid residues of the A domains (Thr–X–Val–Gly–Phe–Pro–Leu–Gly–Tyr–Asn) are in a good accordance with those in nyuzenamides (Thr–Hpg–Val–Gly–Hpa–Pro–Leu–Hgy–Htr–Asn). As nrps-4, -5 and -6 were also unreported gene clusters, we predicted their products to be an octapeptide derived from dX–Thr–dX–Val–dX–dVal–dAla–Val, a Thr-containing molecule and a tripeptide including Gly, respectively, based on the domain organization and predicted substrate of A domains. Among the seven PKS gene clusters in this strain, four type-I PKS (t1pks) and one type-II (t2pks) gene clusters were identified to be BGCs of geldanamycin, mediomycin, nigericin, azalomycin and spore pigment, respectively, which were supported by high sequence similarities of the NRPSs and PKSs to the reported enzymes (Table 4). The remaining two were not reported gene clusters. Domain organization of t1pks-5 partially resembled that of a butylolactol-BGC. Although the butyrolactol-BGC encodes six PKSs [22], t1pks-5 encodes only five of the six. As the module number of t1pks-5 is eight, the product was predicted to be a compound derived from an octaketide, which is similar to butyrolactol, but the alkyl chain is shorter than that of butyrolactol. Only a single module was present in t1pks-6. As it did not show similarities to BGCs of known compounds, the product could not be predicted. Among the hybrid PKS/NRPS gene clusters in this strain, pks/nrps-1 and -2 were BGCs of alchivemycin [18] and totopotensamide, respectively, whose NRPSs and PKSs correspond to the biosynthetic enzymes (Table 4). In contrast, pks/nrps-3 and -4 were orphan gene clusters. PKSs in pks/nrps-3 were AT-less and resembled to those of leinamycin. However, as the domain organization differed from that of leinamycin-BGC, the product was predicted to be a new macrolactam compound like leinamycin. According to module number and domain organization, pks/nrps-4 was predicted to synthesis octapeptide with a polyketide moiety.

Table 3.
NRPSs and PKSs in these gene clusters in the genome of Streptomyces sp. N11-34.

Table 4.
Similarities of enzymes involved in the biosynthesis of known compounds to those of Streptomyces sp. N11-34.
3.3. Distributions of the NRPS and PKS Gene Clusters in Streptomyces sp. N11-34 to the Phylogenetically Close Strains
As S. hygroscopicus NBRC 100766, NBRC 16556, TP-A0867, NBRC 12859 and NBRC 13472T are phylogenetically close to Streptomyces sp. N11-34, as described in the Section 3.1, we examined whether the seventeen PKS and NRPS gene clusters found in Streptomyces sp. N11-34 are present in the genomes of these S. hygroscopicus strains. As summarized in Table 5, all the gene clusters were present in S. hygroscopicus NBRC 100766, NBRC 16556 and TP-A0867, which are closer to Streptomyces sp. N11-34, but S. hygroscopicus NBRC 12859 and NBRC 13472T, phylogenetically discriminated from Streptomyces sp. N11-34, lacked nrps-6, pks/nrps-1 (avm), -2 (tot) and -3.

Table 5.
Distribution of the NRPS and PKS gene clusters in Streptomyces sp. N11-34 to the phylogenetically close S. hygroscopicus strains.
4. Discussion
The description of S. hygroscopicus (Jensen 1931) Yüntsen et al. 1956 (Approved Lists 1980) was emended in 2017, and this species has two heterotypic synonyms, S. endus Anderson and Gottlieb 1952 (Approved Lists 1980) and S. sporocinereus (ex Krassilnikov 1970) Preobrazhenskaya 1986 [17]. Once, S. hygroscopicus had four subspecies with validly approved names, such as S. hygroscopicus subsp. angustmyceticus, S. hygroscopicus subsp. decoyicus, S. hygroscopicus subsp. glebosus and S. hygroscopicus subsp. ossamyceticus. However, these subspecies have been reclassified as independent species by rank up [10,12,23] or reclassified to a synonym of another species [24]. Consequently, S. hygroscopicus has no subspecies at present [10]. In the present study, Streptomyces sp. N11-34 was classified to S. hygroscopicus as well as Streptomyces sp. TP-A0867, an alchivemycin producer [18], and S. hygroscopicus NBRC 16556 [19]. We have proposed a hypothesis that strains classified to the same species harbor a similar set of NRPS and PKS gene clusters in the genomes [6,7]. Although our present study supported the hypothesis in principle, it was unexpectedly observed that S. hygroscopicus NBRC 13472T and NBRC 12859 lack four NRPS and PKS gene clusters among the seventeen clusters present in Streptomyces sp. N11-34. The lack was well correlated with the phylogenetic relationship since S. hygroscopicus NBRC 13472T and NBRC 12859 were phylogenetically discriminated from the other members examined here. As summarized in Table 6, many different features were observed between Streptomyces sp. N11-34, S. demainii DSM 41600T, S. hygroscopicus NBRC 107666, NBRC 16556 and TP-A 0867 (group A) and S. hygroscopicus NBRC 13472T and NBRC 12859 (group B). Although whole genome sequence of S. demainii DSM 41600T has not been published, genome sizes are larger in the group A. Housekeeping gene sequences differ between the two groups. Members of the group B lack four smBGCs in their genome. The distinctive phenotypic characteristics between the groups A and B are given in the previous reports as follows: spore wall ornamentations are warty or rugose in the group A whereas those are smooth in the group B [25]; although members of the group A utilize D-fructose as a sole carbon source for growth, those of the group B do not [25,26]; and maltose utilization is stronger in members of the group A than of the group B [12]. Taken together, it is considered that members in the group A are a new subspecies of S. hygroscopicus, for which we propose Streptomyces hygroscopicus subsp. sporocinereus subsp. nov.

Table 6.
Genotypic differentiation between the two groups.
5. Descriptions of Streptomyces hygroscopicus and Its Subspecies
5.1. Description of Streptomyces hygroscopicus subsp. sporocinereus subsp. nov.
Streptomyces hygroscopicus subsp. sporocinereus (spo.ro.ci.ne’re.us. Gr. n. spora seed; L. adj. cinereus ash-colored; N.L. masc. adj. sporocinereus ash-colored spores).
The description is as given for Streptomyces sporocinereus (ex Krassilnikov 1970) Preobrazhenskaya 1986 [25,27]. This subspecies is also discriminated from Streptomyces hygroscopicus subsp. hygroscopicus by the genomic feature shown in Table 5. The genome size ranges from 9.9–10.4 Mb. The type strain is NBRC 100766T (=ATCC 43692T = DSM 41460T = INMI 32T = JCM 9093T = NRRL B-16376T = VKM Ac-312T). Accession numbers of the 16S rRNA gene and whole genome sequences in the type strain are AB249933 and BCAN01000001–BCAN01000217, respectively.
Streptomyces demainii Goodfellow et al. 2008 is included in this subspecies. Streptomyces sporocinereus (ex Krassilnikov 1970) Preobrazhenskaya 1986 is a basonym of this subspecies.
5.2. Emended Description of Streptomyces hygroscopicus subsp. hygroscopicus (Jensen 1931) Yüntsen et al. 1956 (Approved Lists 1980) emend. Komaki et al. 2017
The description is as given for Streptomyces hygroscopicus subsp. hygroscopicus (Jensen 1931) Yüntsen et al. 1956 (Approved Lists 1980) emend. Komaki et al. 2017 [17] with the following modifications. The genome size of the type strain is 9.5 Mb. Streptomyces sporocinereus (ex Krassilnikov 1970) Preobrazhenskaya 1986 is not included in this subspecies. Streptomyces endus Anderson and Gottlieb 1952 (Approved Lists 1980) is a member of this subspecies.
5.3. Emended Description of Streptomyces hygroscopicus (Jensen 1931) Yüntsen et al. 1956 (Approved Lists 1980)
The description is as given for Streptomyces hygroscopicus subsp. hygroscopicus (Jensen 1931) Yüntsen et al. 1956 (Approved Lists 1980) emend. Komaki et al. 2017 [17] with the following modifications. Spore wall ornamentation is smooth, warty or rugose. Utilization of D-fructose is different between its subspecies. Genome sizes range from 9.5–10.4 Mb. Accession numbers of 16S rRNA gene and whole genome sequences in the type strain are AB184428 and BBOX01000001–BBOX01000680, respectively. Streptomyces endus Anderson and Gottlieb 1952 (Approved Lists 1980), Streptomyces demainii Goodfellow et al. 2008 and Streptomyces sporocinereus (ex Krassilnikov 1970) Preobrazhenskaya 1986 are later heterotypic synonyms of this species [12,17].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10020349/s1, Figure S1: Phylogenetic tree based on 16S rRNA gene sequences, Figure S2: Phylogenetic tree based on MLSA, Table S1: Accession numbers of gene sequences used for MLSA.
Author Contributions
Conceptualization, H.K. and Y.I.; methodology, T.T.; investigation, H.K.; resources, Y.I.; data curation, H.K.; writing—original draft preparation, H.K.; writing—review and editing, Y.I. and T.T.; supervision, Y.I.; project administration, T.T.; funding acquisition, T.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by a commissioned project from the Japan Patent Office.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The whole genome shotgun project of Streptomyces sp. N11-34 has been deposited at DDBJ under the accession number BNEK00000000. Accession numbers of the BioProject and the BioSample are PRJDB9821 and SAMD00228011, respectively.
Acknowledgments
We are grateful to Shinpei Ino and Takahiro Matsuyama for genome DNA preparation and Aya Uohara for depositing the whole genome sequence to DDBJ, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Owen, J.G.; Reddy, B.V.; Ternei, M.A.; Brady, S.F. Chemical-biogeographic survey of secondary metabolism in soil. Proc. Natl. Acad. Sci. USA 2014, 111, 3757–3762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzer, D.; Marahiel, M.A. Multimodular biocatalysts for natural product assembly. Naturwissenschaften 2001, 88, 93–101. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Oguchi, A.; Tamura, T.; Hamada, M.; Ichikawa, N. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters in the genus Acrocarpospora. J. Gen. Appl. Microbiol. 2021, 66, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef] [Green Version]
- Komaki, H.; Tamura, T. Differences at species level and in repertoires of secondary metabolite biosynthetic gene clusters among Streptomyces coelicolor A3(2) and type strains of S. coelicolor and its taxonomic neighbors. Appl. Microbiol. 2021, 1, 573–585. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Suzuki, K.; Fujita, N. Genome-based analysis of type-I polyketide synthase and nonribosomal peptide synthetase gene clusters in a novel strain taxonomically close to the genus Salinispora. J. Antibiot. 2015, 68, 767–770. [Google Scholar] [CrossRef]
- Karim, M.R.U.; In, Y.; Zhou, T.; Harunari, E.; Oku, N.; Igarashi, Y. Nyuzenamides A and B: Bicyclic peptides with antifungal and cytotoxic activity from a marine-derived Streptomyces sp. Org. Lett. 2021, 23, 2109–2113. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T. Reclassification of four subspecies in the genus Streptomyces to Streptomyces rubradiris sp. nov., Streptomyces asoensis sp. nov., Streptomyces fructofermentans sp. nov. and Streptomyces ossamyceticus sp. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 005078. [Google Scholar] [CrossRef]
- Yoon, S.; Ha, S.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Huang, Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. Appl. Microbiol. 2012, 35, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, H.; Miura, K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 1963, 72, 619–629. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komaki, H.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Tamura, T.; Fujita, N.; Suzuki, K.I. Genome analysis-based reclassification of Streptomyces endus and Streptomyces sporocinereus as later heterotypic synonyms of Streptomyces hygroscopicus subsp. hygroscopicus. Int. J. Syst. Evol. Microbiol. 2017, 67, 343–345. [Google Scholar] [CrossRef]
- Komaki, H.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Harunari, E.; Kodani, S.; Fujita, N.; Igarashi, Y. Draft genome sequence of Streptomyces sp. TP-A0867, an alchivemycin producer. Stand. Genomic. Sci. 2016, 11, 85. [Google Scholar] [CrossRef] [Green Version]
- Komaki, H.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Tamura, T.; Suzuki, K.; Fujita, N. Draft genome sequence of Streptomyces hygroscopicus subsp. hygroscopicus NBRC 16556. Genome. Announc. 2016, 4, e00139-16. [Google Scholar]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komaki, H.; Ichikawa, N.; Hosoyama, A.; Fujita, N.; Igarashi, Y. Draft genome sequence of Streptomyces sp. TP-A0882 reveals putative butyrolactol biosynthetic pathway. FEMS Microbiol. Lett. 2015, 362, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, Y.; Goodfellow, M. Reclassification of Streptomyces hygroscopicus strains as Streptomyces aldersoniae sp. nov., Streptomyces angustmyceticus sp. nov., comb. nov., Streptomyces ascomycinicus sp. nov., Streptomyces decoyicus sp. nov., comb. nov., Streptomyces milbemycinicus sp. nov. and Streptomyces wellingtoniae sp. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 769–775. [Google Scholar] [PubMed] [Green Version]
- Komaki, H.; Tamura, T. Reclassification of Streptomyces hygroscopicus subsp. glebosus and Streptomyces libani subsp. rufus as later heterotypic synonyms of Streptomyces platensis. Int. J. Syst. Evol. Microbiol. 2020, 70, 4398–4405. [Google Scholar]
- Kämpfer, P. Genus I. Streptomyces Waksman and Henrici 1943, 399AL emend. Witt and Stackbrandt 1990, 370 emend. Wellington, Stackebrandt, Sanders, Wolstrup and Jorgensen 1992, 159. In Bergey’s Manual of Systematic Bacteriology: The Actinobactera, Part B; Whitman, W.B., Parte, A., Goodfellow, M., Kämpfer, P., Busse, H., Eds.; Springer: New York, NY, USA, 2012; pp. 1455–1767. [Google Scholar]
- Goodfellow, M.; Kumar, Y.; Labeda, D.P.; Sembiring, L. The Streptomyces violaceusniger clade: A home for Streptomycetes with rugose ornamented spores. Antonie Van Leeuwenhoek 2007, 92, 173–199. [Google Scholar] [CrossRef]
- Gauze, G.F.; Preobrazhenskaya, T.P.; Sveshnikova, M.A.; Terekhova, L.P.; Maximova, T.S. A Guide for the Determination of Actinomycetes. Genera Streptomyces, Streptoverticillium, and Chainia; Nauka: Moscow, Russia, 1983. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).