Abstract
The Ectothiorhodospiraceae family represents purple sulfur bacteria of the Gammaproteobacteria found primarily in alkaline soda lakes of moderate to extremely high salinity. The main microscopically visible characteristic separating them from the Chromatiaceae is the excretion of the intermediate elemental sulfur formed during oxidation of sulfide prior to complete oxidation to sulfate rather than storing it in the periplasm. We present a comparative study of 38 genomes of all species of phototrophic Ectothiorhodospiraceae. We also include a comparison with those chemotrophic bacteria that have been assigned to the family previously and critically reevaluate this assignment. The data demonstrate the separation of Halorhodospira species in a major phylogenetic branch distant from other Ectothiorhodospiraceae and support their separation into a new family, for which the name Halorhodospiraceae fam. nov. is proposed. In addition, the green-colored, bacteriochlorophyll-containing species Halorhodospira halochloris and Halorhodospira abdelmalekii were transferred to the new genus Halochlorospira gen. nov. of this family. The data also enable classification of several so far unclassified isolates and support the separation of Ectothiorhodospira shaposhnikovii and Ect. vacuolata as well as Ect. mobilis and Ect. marismortui as distinct species.
1. Introduction
The phototrophic purple sulfur bacteria are Gammaproteobacteria that use sulfide and other reduced sulfur sources as photosynthetic electron donors and oxidize these to sulfate as the final oxidation product. The genus Ectothiorhodospira originally was included in the Chromatiaceae family [1]. Phenotypic differences and the distinction of Ectothiorhodospira from Chromatiaceae by oligonucleotide patterns of 16S rRNA molecules [2,3] led to their separation in a distinct family, the Ectothiorhodospiraceae [4].
The two families of purple sulfur bacteria can be distinguished by the most obvious differences in the oxidation of sulfide which are microscopically visible, the formation of elemental sulfur globules inside the cells in Chromatiaceae and outside the cells in Ectothiorhodospiracae [5,6,7,8]. Other characteristic properties that distinguish species of both families are different internal membrane systems, which are in the form of vesicles in Chromatiaceae and in the form of membrane stacks in Ectothiorhodospiraceae [5,6,7], and by a number of chemotaxonomic properties, including the quinone, lipid and fatty acid composition [9,10] and lipopolysaccharide structures [11,12,13]. Sequence analysis of the 16S rRNA gene from available type strains of Ectothiorhodospiraceae had demonstrated the clear divergence of two groups within this family which had been recognized as two different genera: slightly to moderately halophilic species of Ectothiorhodospira and extremely halophilic species, which were transferred to the new genus Halorhodospira [14].
Further comprehensive phylogenetic studies, including large numbers of phototrophic purple bacteria of Alpha-, Beta- and Gammaproteobacteria, comparing sequences of 16S rRNA genes and bchXYZ genes [15], pufLM genes [16] and the glycine/sarcosine methyltransferase essential for glycine betaine biosynthesis [17] all supported the recognition of three major groups of purple sulfur bacteria, the Chromatiaceae, the Ectothiorhodospiraceae (except Halorhodospira species) and the extremely halophilic Halorhodospira species.
In this communication we compare the genome information of a large number of available strains and species of the Ectothiorhodospiraceae, highlight some properties related to sulfur metabolism, electron transport and photosynthesis and discuss the phylogenetic and taxonomic status of these bacteria. The data are in support of a distinction of the Halorhodospira species from Ectothiorhodospiraceae and Chromatiaceae on a family level. The name Halorhodospiraceae, fam. nov. is proposed for this new family. Ectothiorhodospiraceae and Halorhodospiraceae families are defined. In addition, the separation of Halo-rhodospira halochloris and Halorhodospira abdelmalekii into the new genus Halochlorospira gen. nov. as Halochlorospira halochloris and Halochlorospira abdelmalekii is proposed.
According to genome information, including ANI, most of the chemotrophic genera, which have been assigned in recent years in the literature and in databases to the Ectothio-rhodospiraceae, should be excluded from this family due to their large phylogenetic distance and significantly different phenotypic properties.
2. Material and Methods
2.1. Genome Sequences Used in this Study
Several genome sequences from phototrophic Ectothiorhodospiraceae and Halo-rhodospiraceae species have been published before [17,18,19,20,21,22,23] and a number of additional genome sequences have been established during this study (Table 1). The genome sequences from other strains of phototrophic bacteria used in this study are the following: Allochromatium humboldtianum DSM 21881 (JABZEO000000000), Allochromatium vinosum DSM 180 (CP001896), Chromatium okenii DSM 169 (NRRQ01000000), Chromatium okenii LaCa (PPGH01000000), Halochromatium glycolicum DSM 11080 (NRSJ01000000), Halochromatium salexigens DSM 4395 (NHSF01000000), Marichromatium bheemlicum DSM 18632 (JAAXKX000000000), and Marichromatium gracile DSM 203 (SMDC01000000). Those from chemotrophic species are: Acidiferrobacter thiooxydans m-1 (PSYR01000000), Alkalilimnicola ehrlichii MLHE-1 (CP000453), Alkalispirillum mobile DSM 12769 (RCDA01000000), Aquisalimonas asiatica CGMCC 1.6291 (FOEG01000000), Arhodomonas aquaeolei DSM 8974 (ARGF00000000), Halofilum ochraceum XJ16 (LVEG02000000), Halopeptonella vilamensis DSM 21056 (VMKO01000000), Inmirania thermothiophila DSM 100275 (RJVI01000000), Nitrococcus mobilis Nb-231 (AAOF00000000), Oceanococcus atlanticus 22II-S10r2 (AQQV00000000), Spiribacter salinus M19-40 (CP005963), Thioalbus denitrificans DSM 26407 (QPJY01000000), Thioalkalivibrio sulfidophilus HL-EbGr7 (CP001339), Thioalkalivibrio versutus AL2 (MVAR00000000), Acidihalobacter prosperus DSM 5130 (JQSG00000000), Thiogranum longum DSM 19610 (SMFX01000000), Thiohalomonas denitrificans HLD2 (FMWD01000000) and Thiohalospira halophila DSM 15071 (FOMJ01000000).

Table 1.
Comparison of genome features of the Ectothiorhodospiraceae genomes in this study. The genomes are colored by groups as described in the text.
2.2. Genomic DNA Extraction and Sequencing
For the strains sequenced in this study, genomic DNA was prepared from frozen cells, using the GeneJET DNA purification kit (Thermo Fisher Scientific, Waltham, MA, USA). The quantity and purity of DNA were determined using Qubit and NanoDrop instruments and showed absorbance 260/280 ratios between 1.67 and 2.12. The DNA libraries were prepared with the Nextera DNA flex library prep kit (Illumina, Inc., San Diego, CA, USA). All genomes were sequenced using 500 μL of a 1.8pM library with an Illumina MiniSeq instrument, using paired-end sequencing (2 × 150 bp). Quality control of the reads was performed using FASTQC in BaseSpace (Illumina, version 1.0.0), using a kmer size of 5 and contamination filtering. The data for each was assembled de novo using Unicycler [24] in PATRIC [25]. The genome sequences were annotated using RAST (Rapid Annotations using Subsystem Technology; version 2.0; [26]).
2.3. Whole Genome Comparison
Average percentage nucleotide identity (ANIb) between the whole genomes was calculated using JSpecies [27]. JSpecies uses a pairwise genome comparison algorithm to measure the probability of genomes belonging to the same species, with an arbitrary species cutoff of 95%. The whole genome-based phylogenetic tree was generated within PATRIC [25], using the CodonTree pipeline which uses PGFams as homology groups. Among these selected genomes, 141 PGFams were found using the CodonTree analysis, and the aligned proteins and coding DNA from single-copy genes were used for RAxML analysis [28,29]. 100 rounds of bootstrapping were performed using the ‘Rapid bootstrapping option. The resulting Newick file was used in iTOL for tree visualization [30]. Average amino acid identity (AAI) values were calculated from the proteome comparison in PATRIC. Only bi-directional hits were used for this analysis and Hlr. halophila SL1T and Hlr. halochloris DSM1059T (BN9850) as the reference strains. Pairwise 16S rRNA comparisons were performed using LALIGN (EMBL-EBI), using the genome-derived 16S rRNA sequences.
The alignment for the 16S rRNA comparisons was performed using Clustal Omega [31] which uses seeded guide trees and Hidden Markov Model (HMM) profiles to generate multiple sequence alignments. The phylogenetic tree was calculated by the neighbour-joining (NJ) method [32] in JALVIEW [33] and a Newick file was generated. The Jalview NJ method uses the BLOSUM62 substitution matrix to compute a sum of scores for the residue (base) pairs at each aligned position. iTOL was used to draw the phylogenetic trees expressed in the Newick phylogenetic tree format [30]. Due to the lack of complete 16S rRNA sequences from whole genomes, Genbank 16S rRNA sequences were used instead for Ect. salini (FM244738.1), Ect. imhoffii, Arhodomonas aquaeoli (M26631.2) and Nitrococcus mobilis (NR_104912.1), for the 16S rRNA phylogenetic tree.
For the synteny analysis, comparative genome regions were generated using global PATRIC PGFam families to determine a set of genes that match a focus gene. The gene set is compared to the focus gene using BLAST and sorted by BLAST scores within PATRIC [25]. The Compare Region Viewer in PATRIC displays the focus gene along with the other genes in the same family and their flanking regions in their genomes. The bchB gene was used as a focus gene to analyze synteny of the photosynthetic and bacteriochlorophyll gene cluster.
3. Results and Discussion
With the availability of large numbers of genome sequences, the average nucleotide identity (ANI) of genomes became an established measure to compare the similarity/relatedness of bacteria and is an accepted alternative to the classical DNA–DNA hybridization method, which only provided reliable results if performed by experts familiar with this method. It has been suggested to recognize bacteria with ANI >95% as belonging to the same species [27], while bacteria with ANI <90% would be recognized in most cases as separate species. Those with values between 90 and 95% identity may be argued either way depending on other properties. As ANI is apparently more precise in the differentiation of closely related bacteria compared to the 16S rRNA gene sequences [27], we have analyzed ANI from all phototrophic Ectothiorhodospiraceae species with available genome sequences with representatives of the genera Ectothiorhodospira, Ectothiorhodosinus, Thio-rhodospira and Halorhodospira. In addition, representative chemotrophic Ectothiorhodospiraceae with available genome sequences were included.
As the species of Halorhodospira are clearly distinct from other Ectothiorhodospiraceae genera and the separation of Halorhodospira species on a family level is proposed, in the following, the term Ectothiorhodospiraceae will be used in the strict sense with the exclusion of Halorhodospira species and species of Halorhodospira will be treated separately.
3.1. Genomes of Ectothiorhodospiraceae
The Ectothiorhodospiraceae (excluding Halorhodospira) currently contain 10 named phototrophic species. The genome sequences of Trs. sibirica ATCC 700588T [18], Ect. strain PHS-1 [19], Ect. haloalkaliphila ATCC 51935T [20] and Ect. strain BSL-9 [21] have been previously published. In addition, genome sequence information for Ect. magna DSM 22250T, Ect. marina DSM 241T, Ect. marismortui DSM 4180T, Ect. mobilis DSM 237T, Ect. shaposhnikovii DSM 243T and Ectothiorhodosinus mongolicus DSM 15479T was presented in earlier publications [15,16,17]. Here we include additional genome sequences of type strains of Ect. vacuolata DSM 2111T and Ect. variabilis DSM 21381T and additional isolates (DG9, A-7R, A-7Y, B14B, WN21Y, WN21R, WN2R, BN9902 = C, BN9100 = YC6.1 and BN9905 = ScotB) and use this information for a comparison of species and strains (Table 1). Based on comparison of genome length and its G + C content (mol%), several groups of strains/species can be clearly distinguished (Table 1). The group containing Ect. mobilis DSM 237 and both Ect. marismortui strains (DSM 4180 and DG9) has a smaller genome size (2.62–2.80 Mbp) and higher percentage of 68.2–68.4 mol% G + C as compared to the majority of the genomes that have genome sizes from 3.20 Mbp to 3.79 Mbp (with the exception of Ect. spec. strain PHS-1 with 2.94 Mbp) and a G + C content of 62.3 to 63.7 mol%. Ectothiorhodosinus mongolicus, Thiorhodospira sibirica and Ect. magna apparently are more distinct and also have a lower G + C content (Table 1).
The ANI comparison of the phototrophic Ectothiorhodospiraceae (Table 2) clearly identifies three groups of species, with ANI values >80%: (i) including the type strains of Ect. vacuolata, Ect. shaposhnikovii and Ect. magna (colored orange in Table 2), with Ect. magna being clearly more distant to the others with ANI values between 83–87%; (ii) including the type strains of Ect. haloalkaliphila, Ect. variabilis and Ect. marina (colored yellow in Table 2); (iii) including the type strains of Ect. mobilis and Ect. marismortui (colored green in Table 2). More distant to these groups are Ectothiorhodosinus mongolicus and Thiorhodospira sibirica, with ANI values of <70% relative to the others.

Table 2.
Average percentage nucleotide identity (ANI) between pairs of genomes. ANI values >80% are colored and values above the species cutoff (>95%) are presented in bold.
The whole genome-based phylogenetic tree (Figure 1), constructed using unique PGFams as homology groups, supports the ANI analysis and confirms the three groups of strains and species as separate clades in the tree. It also includes representative genomes of chemotrophic species that had previously been considered as belonging to Ectothio-rhodospiraceae but are phylogenetically at a distance, which suggests their grouping in a separate new family. The only chemotrophic bacteria that are especially closely related to the phototrophic Ectothiorhodospiraceae species, in particular to Thiorhodospira sibirica, are species of the two groups of Thioalkalivibrio (Figure 1). Of the two distinct groups, one includes, among others, the type species Thioalkalivibrio versutus and Thioalkalivibrio halo-philus, and the other, among others, Thioalkalivibrio sulfidophilus and Thioalkalivibrio denitrificans [34].
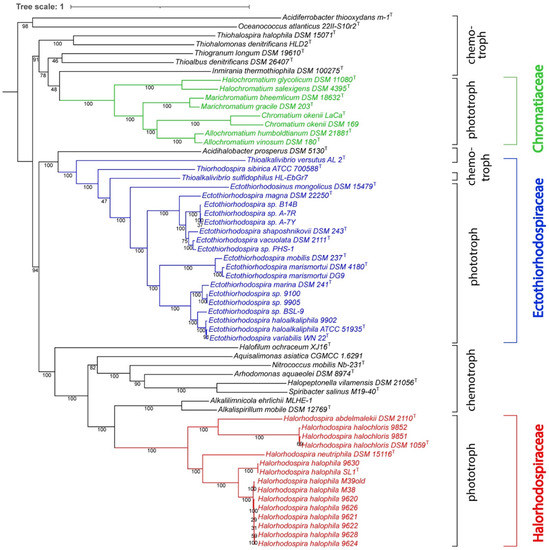
Figure 1.
Whole genome-based phylogenetic tree of the Ectothiorhodospiraceae. The support values for the phylogenetic tree are generated using 100 rounds of the ‘Rapid bootstrapping’ option of RaxML. The tree was rooted at midpoint and the branch length tree scale is defined as the mean number of substitutions per site, which is an average across both nucleotide and amino acid changes. Oceanococcus and Acidiferrobacter were found not to belong to any of the three families but were included in the tree as outgroups.
In addition to the whole genome-based tree, we also constructed a 16S rRNA-based phylogenetic tree that includes a comparison of all sequenced phototrophic Ectothio-rhodospiraceae (Figure 2), including Ect. salini and Ect. imhoffii, for which there is no genome sequence available. Included for comparison were 16S rRNAs of the chemotrophic species previously assigned under Ectothiorhodospiraceae. The results of the 16S rRNA tree are consistent with the whole genome-based tree and ANI comparisons.
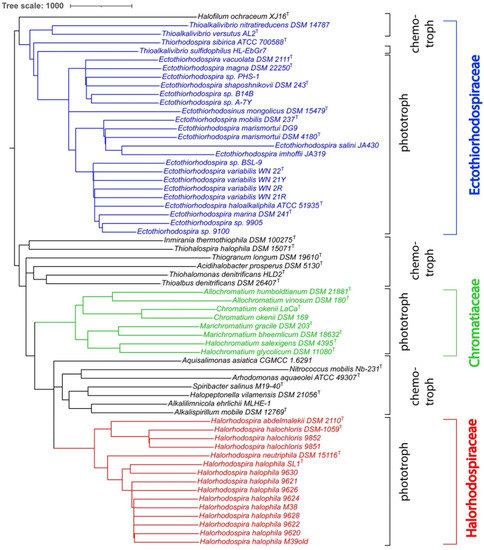
Figure 2.
16S rRNA-derived phylogenetic tree for Ectothiorhodospiraceae species. The phylogenetic tree was calculated by the neighbour-joining (NJ) method [32] in Jalview [33]. The Jalview NJ method uses the BLOSUM62 substitution matrix to compute a sum of scores for the residue (base) pairs at each aligned position. The length of the branches is proportional to the number of nucleotide substitutions per site. iTOL was used to draw the phylogenetic trees expressed in the Newick phylogenetic tree format [30].
These data show that the type strains of Ect. variabilis DSM 21381T (WN 22T) and Ect. haloalkaliphila ATCC 51935T are highly similar. With an ANI of 98.3%, which is above the suggested 95% species cutoff, they may be regarded belonging to the same species. Both strains originate from soda lakes of the Wadi el-Natrun in Egypt [14,35,36]. Additional isolates from the Wadi el-Natrun (BN9902, WN21Y, WN21R, WN2R) are also highly similar to this species (Table 1 [36]). Despite some earlier results [14,36], 16S rRNA sequences support this high similarity, with 97.2% similarity between the two type strains, but may not necessarily demark species identity. This issue will have to be resolved in more detailed studies.
The results also support the recognition of the following closely related but distinct species. Ect. marismortui DSM 4180T is close to Ect. mobilis DSM 237T and Ect. shaposhnikovii DSM 243T close to Ect. vacuolata DSM 2111T. Previously, the taxonomic status of Ect. marismortui DSM 4180T and Ect. vacuolata DSM 2111T were disputed. They were recognized as distinct species by Imhoff [6] and Imhoff and Süling [14], though it was suggested by Ventura et al. [37,38] to combine Ect. vacuolata with Ect. shaposhnikovii and Ect. marismortui with Ect. mobilis, primarily based on data on DNA–DNA hybridization and ribotyping. The results of ribotyping, however, did not clearly support this conclusion. They revealed identity between two strains of Ect. mobilis (DSM 237T and DSM 240) but clear differences of these to Ect. marismortui DSM 4180T. They also revealed the identity of two strains of Ect. vacuolata (DSM 2111T and B3) with clear differences to Ect. shaposhnikovii DSM 243T [37]. In addition, the values of DNA–DNA hybridization were only 80–84% [37]. In contradiction to this suggestion and based on genetic studies of Rubisco and nitrogenase genes, Tourova et al. [39] recognized the four species as distinct species. The presented data on ANI and whole genome-based comparisons are in good agreement with considerations by Imhoff [6] and Tourova et al. [39], recognizing Ect. vacuolata DSM 2111T and Ect. shaposhnikovii DSM 243T as distinct species with only 90% ANI and also recognizing Ect. marismortui DSM 4180T and Ect. mobilis DSM 237T as distinct species with ANI values of 93%. Therefore, ANI supports the distinction of these four bacteria at the species level and also confirms the close relationships of the two couples of species.
The results also suggest the assignment of several new isolates to one or the other of the established species. Of two strains that were isolated as arsenite-oxidizing bacteria [40], strain BSL-9 originating from Big Soda Lake represents a distinct new species closely related to Ect. haloalkaliphila and Ect. marina, while strain PHS-1 originating from Mono Lake is a strain of Ect. vacuolata (Figure 1 and Table 2). Strain DG9 is an additional isolate of Ect. marismortui from Berikei Sulfur Springs in Dagestan (Russia, [41]). Strains BN9100 (=YC6.1), an isolate from Solar Lake by H. Biebl (Braunschweig, Germany) and BN9905 (=ScotB), isolated from the seashore near Inverary (Scotland) by one of us (JFI), are new isolates of Ect. marina. Strains A-7R, A-7Y and B14B form a separate clade on the whole genome phylogenetic tree and have ANI values <90% with respect to any of the other strains, but high ANI values amongst themselves, and therefore represent a new species related to Ect. shaposhnikovii and Ect. vacuolata (Figure 1 and Table 2).
The present data support the distinction of Ectothiorhodosinus mongolicus [42] and also Thiorhodospira sibirica [43] in genera separate from Ectothiorhodospira. Ect. magna [44], though distantly related to other species of the genus, is placed inside a group of species together with Ect. shaposhnikovii and Ect. vacuolata.
3.2. Genomes of Halorhodospira Species
Four Halorhodospira species are currently recognized: Hlr. halophila [45,46], Hlr. halochloris [47], Hlr. abdelmalekii [48] and Hlr. neutriphila [49]. The genome sequence of Hlr. halophila SL1T was previously determined [22] and information on genome sequences of Hlr. halophila BN9626, BN9620, BN9630, Hlr. neutriphila DSM 15116T, Hlr. halochloris DSM 1059T and Hlr. abdelmalekii DSM 2110T were reported earlier [15,16,17].
We have now added genome sequences of additional strains of Hlr. halochloris and Hlr. halophila, which originate from the Wadi el-Natrun in Egypt [35,50] and from Mongolian soda lakes (Gorlenko, personal communication) (Table 1). The ANI, whole genome-based and also 16S rRNA-based phylogenetic tree comparison of the Halorhodospira species (Table 3, Figure 1 and Figure 2) show a clear distinction of five different species.

Table 3.
ANI, average percentage nucleotide identity between pairs of genomes. ANI values >90% are colored and values above the species cutoff (>95%) are presented in bold.
The first group consists of Hlr. halophila SL1T and Hlr. halophila BN9630. The close relationship of strain BN9630 to the Hlr. halophila type strain was demonstrated previously by 16S rRNA sequence analyses and fatty acid analyses [9,14] and is now supported by near identity in terms of the ANI (>99%) (Table 3). The 16S rRNA sequences delineated from the genome sequence actually are only two bases different between the two strains.
A second group is formed by strain BN9626 and a number of other isolates (BN9620, BN9621, BN9622, BN9624, BN9628, M38 and M39old), which differed from strain SL1T with respect to fatty acid composition [9] and revealed only 85–86% ANI identity to Hlr. halophila SL1T (Table 3). This indicates that these strains could be recognized as strains of a distinct species. The average amino acid identity (AAI) is 89.3% for 2414 orthologues of strains SL1T and BN9626, which is consistent with the ANI. The 16S rRNA identity (obtained from the genome sequences) between Hlr. halophila BN9626 and SL1T with 18 differences out of 1537 bp is 98.8%, which is within a range allowing species distinction according to [51] (see systematic conclusions, below). More detailed studies are required to clarify the situation.
Third, according to the whole genome phylogenetic tree (Figure 1) Hlr. neutriphila DSM 15166T [49] is clearly distinct from Hlr. halophila. The ANI to strains of Hlr. halophila is 77.4–77.6%, indicating considerably greater divergence. The 16S rRNA obtained from the genome sequence also averages 54 bases different from that of strains SL1T and BN9626 (96.5% identity), which places Hlr. neutriphila on a separate clade on the 16S rRNA tree (Figure 2). The AAI for Hlr. neutriphila and strains SL1T and BN9626 is 72.9% (1730 orthologs). The G + C content of the genome sequence from Hlr. neutriphila is 72 mol% and thus significantly higher than all other Halorhodospira species (Table 1).
Further, according to the genome-based phylogenetic tree, the green-colored, bacteriochlorophyll b containing species Hlr. halochloris and Hlr. abdelmalekii form a major branch distinct from the groups of red-colored species, which have bacteriochlorophyll a (Figure 1). The complete genome sequence of Hlr. halochloris DSM 1059T (=BN9850T) (APO17372) was established by [23] and we have added a second genome sequence of this strain (NRRM00000000). The genome sequence supposedly determined from the Hlr. halochloris strain AT (=DSM 1059T) by the authors of [52] turned out to be almost identical to the one from Ect. haloalkaliphila ATCC 51935T (AJUE00000000) and is not from Hlr. halochloris. It could originate from a culture contaminant or a mislabeled culture.
The genomes of the three Hlr. halochloris strains (BN9850T, BN9851 and BN9852) have a significantly lower G + C content (55.8–56.1 mol%) compared to Hlr. abdelmalekii (62.9 mol%) and other Halorhodospira species (>67.9 mol%; Table 1). The genome sequence of Hlr. abdelmalekii BN9840T (= DSM 2110T) is significantly different from Hlr. halochloris (ANI of 72.5%), Hlr. halophila and Hlr. neutriphila (ANI of approximately 73–74%). There are 65 differences (95.8% identity out of 1551 bp) in the 16S rRNA between Hlr. abdelmalekii and Hlr. halochloris, as obtained from the genome sequences. The 16S rRNA of the green-colored species, Hlr. halochloris and Hlr. abdelmalekii, on average has 87 differences to the red-colored species. The AAI between the two green-colored species is 71.6% (1856 orthologs) and that for the two green-colored species and the three red-colored species is 68.6%.
3.3. Genome-Delineated Properties
3.3.1. Sulfur and Thiosulfate Oxidation
In the Chromatiaceae, elemental sulfur is stored in the periplasm inside a proteinaceous membrane made up of one to five sulfur globule proteins rich in alternating Gly and Tyr residues [53,54]. None of the Ectothiorhodospiraceae and the Halorhodospira species, for that matter, store sulfur intracellularly and contain sulfur globule proteins; the sulfur is transported outside the cells and must be transported back inside for further oxidation, once all the sulfide and thiosulfate are exhausted. It is only then that it is further oxidized to sulfate in the cytoplasm. Detailed studies on processes of sulfur oxidation in purple sulfur bacteria have been made by Dahl and coworkers [55,56].
Most of the Ectothiorhodospiraceae species contain the thiosulfate-oxidizing enzymes SoxAXYZB, with the apparent exception of Ect. magna and Trs. sibirica, that only contain SoxY and Z. The genes are organized in two separate operons. As is apparent from the complete genome of Ect. BSL-9 containing a single chromosome, the genes soxA and soxX are in one location and soxYZB in another. In those Halorhodospira species that use thiosulfate, the diheme SoxA and SoxX are fused into the single triheme protein, SoxXA. Hlr. abdelmalekii and Hlr. halochloris do not use thiosulfate as a growth substrate and do not contain Sox enzymes. In Chromatiaceae, genes of the thiosulfate-oxidizing enzymes are also located in two operons; however, soxB is associated with soxAX and the two gene clusters are soxAXB and soxYZ.
3.3.2. Glutathione
Glutathione is a common small molecule reductant in bacteria, which has several important functional roles [57]. It is present in most bacteria, though in some phototrophic purple bacteria it is replaced by the glutathione amide, in which the terminal glycine carboxylate is amidated, as was first shown for Marichromatium gracile [58]. In most bacteria, glutathione reductase has an Arg21 that, according to three-dimensional structural analysis, contributes to the binding of glutathione via a salt bridge to the carboxyl group of the glutathione glycine residue [59]. As found for the glutathione reductase of Marichromatium gracile and other Chromatiaceae, the Arg21 is substituted by Glu21 and in consequence glutathione, but not its amide, is repelled from the binding site [57]. Thus, glutathione reductase Glu/Arg21 can be used as a proxy for the presence or absence of glutathione amide. We have found through genome sequencing that all Chromatiaceae and Ectothiorhodospiraceae (with the exceptions of Ect. mobilis DSM 237T and both strains of Ect. marismortui, DSM 4180T and DG9, where a glutathione reductase has not been found) have Glu21 and are thus likely to have glutathione amide. However, glutathione reductases of all Halorhodospira species have Arg21 like the majority of bacteria and are likely to have the normal glutathione.
3.3.3. Carboxysome Genes
The carboxysome is an important structure in many autotrophic bacteria, such as the cyanobacteria [60] and many autotrophic proteobacteria [61]. The carboxysome forms a proteinaceous membrane that encloses Rubisco and carbonic anhydrase and its genes are associated with bicarbonate transporters. The carboxysome shields Rubisco from non-functional reactions with oxygen and provides CO2 where it is needed. Though a definite proof for the physical structures in purple sulfur bacteria is lacking, we have found genes of putative carboxysome peptides A and B and carboxysome shell proteins csoS1, csoS2 and csoS3 in many Chromatiaceae and Ectothiorhodospiraceae. They are present in seven species of Ectothiorhodospiraceae but not in in Ect. mobilis DSM 237T and both strains of Ect. marismortui (DSM 4180T and DG9), Ect. PHS-1 and Trs. Sibirica. Carboxysome genes appear to be absent from Halorhodospiraceae, though ribulose bisphosphate carboxylase, phosphoribulose kinase and enzymes involved ini the Calvin Cycle are present.
3.3.4. Nitric Oxide Reduction
As mentioned above, Ectothiorhodospira strains A-7Y, A-7R and B14B are closely related but different from Ect. shaposhnikovii DSM 243T and Ect. vacuolata DSM 2111T. These three strains have 15 unique PGFams that are not present in any of the other Ectothio-rhodospiraceae genomes and all of them contain a gene cluster for nitric oxide reductase (NOR) that seems to be missing from all the other species. The norCBQD cluster contains two NO reductase activation proteins and both the small and large NOR subunits (C and B), which show similarity to cytochrome oxidases. There have been few studies of nitric oxide reduction in the Ectothiorhodospiraceae, but these three strains appear to at least have the genetic capability for nitric oxide reduction. In addition to the overall genomic distinction, these genetic differences are another reason to consider these strains as a separate species.
3.3.5. HIPIP
In Chromatiaceae the iron–sulfur protein HiPIP was shown to be the electron donor to the photosynthetic reaction center PufLM and the electron acceptor from the BC1 complex [62,63]. The majority of Ectothiorhodospiraceae contain either three or four HiPIP isozymes, with the exception of Ets. mongolicus and Trs. sibirica, which have only one. In addition, Hlr. halophila has genes for four HiPIP isoenzymes, two of which are abundant in cell extracts [64]. The gene for one of these isozymes is mixed in with the photosynthetic genes, suggesting that it is the electron donor in this species as it is in Chromatiaceae and presumably Ectothiorhodospiraceae. It is likely that it acts as a mediator of electrons between the bc1 complex and photosynthetic reaction center in these bacteria. HiPIP II actually forms a strong complex with the reaction center and rapidly reacts with soluble HiPIP [65]. On the other hand, the green-colored Halorhodospira species do not have HiPIP and are likely to utilize cytochrome c5, which is present in all Halorhodospira species characterized to date, but not in the Ectothiorhodospiraceae. Cytochrome c5 is known as a soluble mediator in the green sulfur bacteria in the family Chlorobiaceae and the membrane bound c5 is a possible component of the bc complex [66].
In the green-colored Halorhodospira species, there are no HiPIP genes at all, which is an important difference to the red-colored Halorhodospira species. In this case, the electron mediator between the bc1 complex and photosynthetic reaction center must be different. Perhaps due to the difficulty in growing Hlr. halochloris and Hlr. abdelmalekii, no experimental studies of electron transfer have been published. However, all of the Halorhodospira species produce a soluble cytochrome c5 similar to those found in Chlorobiaceae species and which is thought to couple the bc complex to the photosynthetic reaction center. It is proposed that this cytochrome c5 is the electron mediator in these two species.
3.3.6. Cytochrome b5
Cytochrome b5, which is a well-known constituent of eukaryotic cells, has for the first time found in Ect. vacuolata, where its function is unknown [67]. It differs from its eukaryotic counterparts in having a cysteine disulfide and the presence of a signal peptide, which suggests that it is located in the periplasm. At least seven examples are now known from the genome sequences of Ectothiorhodospiraceae. These are Ect. vacuolata DSM 2111T, Ect. PHS-1, Ect. shaposhnikovii DSM 243T, Ect. mobilis DSM 237T, Ect. BSL-9, Ect. haloalkaliphila ATCC 51935T and Ect. marina DSM 241T.
The 10-heme cytochromes, MtrA and MtrF (or OmcA), as well as the outer membrane porin MtrB, are involved in the reduction and solubilization of insoluble iron and manganese minerals primarily in Shewanella species, as well as in other bacteria [68]. MtrAB are occasionally found in the purple sulfur bacteria, such as Ect. vacuolata, Ect. shaposhnikovii, Ect. haloakaliphila, Ect. BSL-9 and Ect. marina. In the species of Ectothiorhodospiraceae, these two genes are also associated with OmcA, as in Shewanella. Cytochrome c4 or HiPIP are sometimes found associated with them, suggesting oxidation of FeII rather than reduction of FeIII, since they have high redox potentials. A HiPIP gene is adjacent to the MtrABF cluster in Ect. vacuolata DSM 2111T and Hrs. halophila, although not in other species considered herein.
3.3.7. Arsenic Oxidation
The Ectothiorhodospiraceae and the Halorhodospiraceae are generally capable of oxidizing arsenic (III) to arsenic (V), but only a few species are capable of using As(III) as sole electron donor for growth, including Ect. species strain PHS-1 and Ect. species strain BSL-9 [40]. The enzymes involved are the molybdopterin proteins ArxA and ArrA, which are closely related. Most Ectothiorhodospiraceae have arrABC genes, but only Ect. PHS-1, Ect. BSL-9, Ect. shaposhnikovii DSM 243T, Ect. B14B, Ect. A-7Y, Ect. A-7R, and Hrs. halophila SL1T and BN9630, have five to eight arx genes. The authors of [40] examined four of these species plus Ect. vacuolata for oxidation of As(III) and checked for As(III) dependent growth and found that only PHS-1 and BSL-9 would grow with arsenic. Therefore, there has to be more to the story because Ect. shaposhnikovii and Hrs. halophila have the requisite genes but failed to grow.
3.3.8. Photoactive Yellow Protein—PYP
Another interesting difference between the red- and green-colored Halorhodospira strains is that only the red strains have a pair of photoactive yellow proteins (PYP) [69]. It is completely absent in the green-colored strains but has been discovered in the Thio-rhodospira sibirica genome as well as Halochromatium salexigens (Chromatiaceae). The only proven role of PYP in purple bacteria is to reverse the effects of red light on the bacteriophytochrome in the hybrid protein PPR in Rhodocista centenaria, which is a PYP/bacteriophytochrome/histidine kinase [70]. The Chromatiaceae do not have PPR, but some species do have a hybrid PYP/bacteriophytochrome/diguanylate cyclase/phosphodiesterase, which is called PPD [71]. Interaction among the separate domains has not been demonstrated in PPD and the functional role of the PYP domain is likely to be different than it is in PPR.
3.3.9. Photosynthesis Gene Clusters
As other phototrophic Proteobacteria, Ectothiorhodospiraceae and Halorhodospira species have photosynthesis gene clusters, including genes for carotenoid and bacteriochlorophyll biosynthesis, the photosynthetic reaction center and antenna proteins, as well as regulatory and sensory proteins. The structure of the gene clusters and the arrangement of genes show clear differences between Ectothiorhodospira and Halorhodospira species (Figure 3).
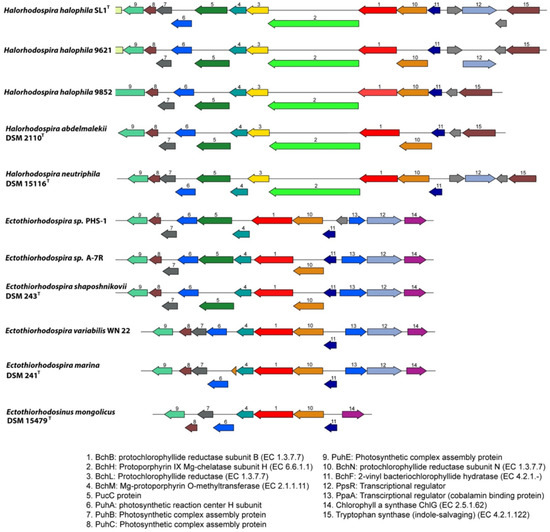
Figure 3.
Comparison of the bchB genomic region between representatives of Halorhodospira and Ectothiorhodospira species. Genes are colored based on their family membership.
While in Halorhodospira species a cluster with ppsR-bchFNBHLM genes (the regulator gene ppsR is absent from Hlr. abdelmalekii and Hlr. halochloris) is present, in Ectothio-rhodospira species an additional regulatory ppaA gene is combined with the ppsR gene (genes 12 and 13 in Figure 3).
The presence of the two regulatory genes ppsR and ppaA in the photosynthetic gene cluster is common to many phototrophic Alpha- and Betaproteobacteria as well as Gemmatimonas. The ppaA gene is absent from Chromatiaceae and Halorhodospiraceae and is found among phototrophic Gammaproteobacteria only in the genus Ectothiorhodospira (Figure 3). Both regulatory genes are absent from Ets. Mongolicus. Quite characteristic for Ectothio-rhodospiraceae, and different to most other purple bacteria, including the Halorhodospiraceae, is a gene cluster ChlG-ppsR-ppaA-bchFNB, with the exclusion of bchL and bchH genes from the common bchFNBHLM gene cluster, as shown for representatives of the genus Ectothiorhodospira (Figure 3). Both bchH and bchL genes are at separate locations in these bacteria. All Halorhodospira species have an additional regulator gene pufQ which is located between bchZ and pufB (bchCXYZ-pufQBALMC). This regulator is found in many phototrophic Alphaproteobacteria but is absent from all Ectothiorhodospiraceae and from Chromatiaceae as well. All Halorhodospira species and Ectothiorhodospiraceae lack the aerobic Mg-protoporphyrin IX monomethylester oxidative cyclase (acsF) and therefore depend on anoxic conditions for bacteriochlorophyll biosynthesis using the anaerobic form of the enzyme (encoded by bchlE).
3.4. Habitats and Environmental Distribution
Quite remarkably, species of phototrophic Ectothiorhodospiraceae and Halorhodospiraceae (Ectothiorhodospira, Halorhodospira, Thiorhodospira and Ectothiorhodosinus species), including their chemotrophic relatives, are characteristic inhabitants of marine and saline waters worldwide and preferably develop in alkaline and saline soda lakes. They are phylogenetically related to alkaliphilic chemotrophic sulfur-oxidizing bacteria of the genera Thioalkalivibrio, Alkalilimnicola, Alkalispirillum and are important sulfur-oxidizing chemotrophic bacteria in many alkaline soda lakes [72,73]. A recent review summarizes their occurrence in various types of salt and soda lakes in different geographic regions [74].
Although Halorhodospira halophila was first isolated from Summer Lake, Oregon [45,46] and Hlr. neutriphila originates from a marine saltern [49], the great majority of the studied strains originate from African soda lakes, most prominently those of the Wadi el-Natrun. This contains all strains of the green-colored species and a second strain of Hlr. halophila (9630). Several other strains assigned to this species (including, among others, BN9620, BN9622 and BN9624) are likely distinct on the species level from Hlr. halophila. In addition to strains from the Wadi el-Natrun, two isolates from Mongolian soda lakes (M38 and M39old) belong to this presumably new species.
The first intensively studied soda lakes with mass developments of red-colored and green-colored Halorhodospira species were those in the Wadi el-Natrun in Egypt [35,50,75]. While pH-optima of the type strain of Hlr. halophila SL1T (isolated from the highly alkaline and saline Summer Lake in Oregon) were found to be at 7.4–7.9 [45,46], isolates from the Wadi el-Natrun had pH optima at 8.5–9.0. Two more haloalkaliphilic species, the green-colored, bacteriochlorophyll b-producing Hlr. halochloris [47] and Hlr. abdelmalekii [48], originate from these soda lakes.
In addition, isolates of the moderate halophilic and alkaliphilic Ectothiorhodospira species Ect. haloalkaliphila [14,35] and Ect. variabilis, which is most closely related to Ect. halo-alkaliphila [36], were found in soda lakes of the Wadi el-Natrun and also in soda lakes from Siberia and Mongolia [36].
Halorhodospira halophila and two morphological distinct bacteria assigned to the genus Ectothiorhodospira were present in Mongolian soda lakes with high salinities (>15% salts), while other species of Ectothiorhodospira and Thiorhodospira were isolated from lakes with lower salinities of up to 5.5–6.0% salts. For the first time the moderately halophilic Ectothiorhodosinus mongolicus (salt optimum 1–7%) was isolated from one of these lakes [42]. Additionally, Thiorhodospira sibirica [43] and Ect. magna [44] were isolated from soda lakes of remote areas in Asia in Siberia, Mongolia and the Transbaikal region, but have so far not been found in other locations. Possibly the remote origin of these bacteria indicates their separate evolution and is a reason for their distant relationship to other strains of the Ectothio-rhodospiraceae/Halorhodospiraceae families.
Other Ectothiorhodospira species, especially Ect. marina and Ect. mobilis, prefer marine habitats, where they have been regularly observed and repeatedly been isolated from.
In recent years, proof of the presence of Ectothiorhodospiraceae in alkaline and saline lakes worldwide has been obtained by analysis of clone libraries. The analysis of 16S rRNA gene clone libraries from several lakes of the Wadi el-Natrun (Lake Fazda, Lake Hamra and Lake UmRisha) revealed a great diversity of sequences related to Ectothio-rhodospiraceae species, including Hlr. halochloris, and Ect. haloalkaliphila [76]. In addition, communities of phototrophic purple bacteria in Chilean salt lakes of the Atacama Desert, Laguna Chaxa and Laguna Tebenquiche that were studied by clone libraries of pufLM gene sequences were found to contain bacteria related to Ect. mobilis, Ect. variabilis and Hlr. halophila as closest relatives [77,78].
4. Systematic Conclusions
The comparison of a large number of genome sequences of phototrophic purple sulfur bacteria in the present study clearly demonstrated the need to separate Halorhodospira species and relatives from the Ectothiorhodospiraceae and to place them in the new family Halorhodospiraceae. The comparison of the genomic phylogeny, including considerations of ANI, demonstrated that the phototrophic Ectothiorhodospiraceae as currently known represent two separate families, being almost equally distant from the Chromatiaceae family. Based on the significant differences of the extremely halophilic species from all other Ecto-thiorhodospiraceae, we propose to recognize these species as members of a new family, the Halorhodospiraceae fam. nov.
The herein proposed reclassification of the families of phototrophic purple sulfur bacteria with the separation of Halorhodospira species from the Ectothiorhodospiraceae family and the existence of three families of purple sulfur bacteria within the Gammaproteobacteria requires a careful reconsideration of species assignment to these families and sheds new light on the assignment of a number of chemotrophic species and genera to the Ectothiorhodospiraceae which have been made in recent years but do not warrant to be included within this family.
Among those bacteria that have been assigned to the Ectothiorhodospiraceae which without doubt cannot be considered as members of this family are Acidiferrobacter thio-oxydans [79], now classified with Acidiferribacteraceae and Acidiferribacterales [80,81] and Oceanococcus atlanticus [82]. They do not fit into any of the three phototrophic Chromatiales families (Figure 1). In addition, Halofilum ochraceum [83], with ANI values to all considered Ectothiorhodospiraceae of only 65–68%, should not be included in Ectothiorhodospiraceae or Halorhodospiraceae. In addition, a few bacteria for which genome sequences at present are not available, should not be included in Ectothiorhodospiraceae or Halorhodospiraceae based on information available for 16S rRNA gene sequences and other properties. One of these species is Natronocella acetinitrilica [84]. Two other species are Methylonatrum kenyense, which is a gammaproteobacterium considered to be of unknown affiliation (genus incertae sedis), and Methylohalomonas lacus [85], which may be related to Thioalkali-spira species of the Thioalkalispiraceae [86].
Other species/genera that have been assigned to the Ectothiorhodospiraceae are distantly related to the phototrophic Chromatiaceae, Ectothiorhodospiraceae or Halorhodospiraceae by deep-branching lineages both in genome-based and 16S rRNA-based phylogenetic trees (Figure 1 and Figure 2). These are Thiohalospira halophila, Thiohalomonas denitrificans, Thiogranum longum, Thioalbus denitrificans, Inmirania thermothiophila and the acidiphilic Acidihalobacter prosperous and related species, which therefore should not be considered as belonging to either Ectothiorhodospiraceae or Halorhodospiraceae. Their placement in one or more new separate families needs to be evaluated and these bacteria will not be further considered in the present discussion.
Halophilic and alkaliphilic chemotrophic bacteria that are most closely related to phototrophic Ectothiorhodospiraceae are species of the genus Thioalkalivibrio, particularly those belonging to the Thioalkalivibrio sulfidophilus cluster (Figure 1 and Figure 2). Species of both clusters of Thioalkalivibrio are the only known chemotrophic bacteria that can be con-sidered to be included in the Ectothiorhodospiraceae.
The new family Halorhodospiraceae is represented by two major groups of phototrophic bacteria, the red-colored Hlr. halophila and relatives and the green-colored Hlr. halochloris and relatives. Phenotypic and phylogenetic differences suggest a separation of the green-colored species in a separate genus, with ANI values of the type species of 70.4–71.3 to strains of Hlr. halophila. Although Hlr. neutriphila appears phylogenetically and phenotypically (characteristically by the preference for neutral pH and high G + C content of 72 mol%) distinct from Hlr. halophila, an ANI >77% to Hlr. halophila (Table 3) and 16S rRNA identity of 96.5% to the type strain of Hlr. halophila SL1 support recognizing this bacterium as a distinct species of the genus Halorhodospira. This is in line with proposals made by others suggesting genus delineations at ANI values close to 74% [51,87].
According to both the phylogenomic and the 16S rRNA tree, a phylogenetic cluster distinct to these phototrophic bacteria is represented by a group of chemotrophic bacteria, including chemotrophic alkaliphilic and halotolerant species, the nitrifying Nitrococcus mobilis, Aquisalimonas asiatica, Spiribacter salinus, Spiribacter (Halopeptonella) vilamensis and Arhodomonas aquaeolei (Figure 1 and Figure 2). According to 16S rRNA phylogeny, this cluster includes Alkalispirillum mobilis and Alkalilimnicola ehrlichii and relatives as well, which, according to the genomic tree, form a separate lineage closer to Halorhodospira. The phylogenetic distance of the whole group of chemotrophic relatives suggests the exclusion of these bacteria from the Halorhodospiraceae and placing them within one or more separate new families. Accordingly, the Halorhodospiraceae are represented exclusively by phototrophic bacteria.
Based on the present data, Ectothiorhodosinus mongolicus [42] and Thiorhodospira sibirica [43] represent genera distinct from Ectothiorhodospira. Ect. magna [44], though distantly related to other species of the genus, is placed inside a group of species together with Ect. shaposhnikovii and Ect. vacuolata. ANI values among all studied strains/species of the genus Ectothiorhodospira are >74.9% which is in line with a proposed genus demarcation of approximately 74% [88]. Other strains (strains A-7Y, A-7R and B14B) with ANI values to related Ectothiorhodospira species below 90% (87.8–89.4) presumably represent a new species of this genus. Although little information is known, and genome sequences are not available for Ect. salini and “Ect. imhoffii”, both species appear most closely related to Ect. mobilis and Ect. marismotui according to 16S rRNA gene sequences (Figure 2; [88,89]).
Characteristics of the three families.
There are three families of the Chromatiales that are characterized by the presence of phototrophic purple sulfur bacteria forming coherent clusters that are, with very few exceptions, clearly distinct from their chemotrophic relatives. These families can be distinguished by a number of phenotypic properties as well as on the basis of genomic properties.
The Chromatiaceae primarily live in fresh water or marine habitats; they are phototrophic and primarily autotrophic. When oxidizing sulfide and thiosulfate, intermediate sulfur is stored in the periplasmic space, enclosed within a proteinaceous membrane containing one to five small, related sulfur globule proteins. The thiosulfate dehydrogenase, SoxA, and its electron-acceptor, SoxX, exist as separate subunits. The small protective thiol, glutathione, is present as the amide.
Ectothiorhodospiraceae primarily live in shallow alkaline desert soda lakes and also marine shallow waters and tolerate up to about 5–7% salt. They are primarily phototrophic and autotrophic. When oxidizing sulfide and thiosulfate, the intermediate sulfur is excreted into the growth medium without a membrane. SoxA and SoxX exist as separate subunits. Glutathione is present as the amide.
The Halorhodospiraceae are found in soda lakes, require elevated salt concentrations for growth and some species can even live in saturated brines. Intermediate elemental sulfur is excreted into the growth medium. SoxX is fused to the N-terminus of SoxA. Glutathione is not amidated. They are phototrophic bacteria forming two distinct groups of species, red-colored species producing bacteriochlorophyll a and green-colored species producing bacteriochlorophyll b. The red-colored species likely use the small iron–sulfur protein HiPIP as mediator between the bc1 complex and the photosynthetic reaction center PufLM. On the other hand, the green species are likely to use the small soluble cytochrome c5 as mediator.
5. Emended Description of the Family Ectothiorhodospiraceae Imhoff 1984a, 33VP
Ec.to.thi’o.rho.do.spi.ra’ce.ae. M.L. fem. n. Ectothiorhodospira type genus of the family; -aceae ending to denote a family; M. L. fem. pl. n. Ectothiorhodospiraceae the Ectothio-rhodospira family.
The family constitutes slightly or moderately halophilic phototrophic bacteria growing under alkaline conditions and their chemotrophic relatives. They are Gram-negative and belong to the Gammaproteobacteria. Cells are spiral-, vibrioid- or rod-shaped, motile by means of polar flagella and divide by binary fission. They are either phototrophic purple sulfur bacteria that perform anoxygenic photosynthesis with bacteriochlorophylls and carotenoids as photosynthetic pigments or aerobic chemolithotrophic sulfur-oxidizing bacteria, some of which may use nitrate as alternative electron acceptors. Growth of phototrophic representatives is preferably anaerobic in the light, with reduced sulfur compounds as electron donors. Sulfide is oxidized to elemental sulfur and is deposited outside the cells, eventually also in the peripheral periplasmic space of the cell body. The final oxidation product is sulfate. Ectothiorhodospiraceae are found in marine and moderately saline environments containing sulfide and having an alkaline to extremely alkaline pH. Glycine betaine, but not ectoine, is the major compatible solute in these bacteria.
The mol% G + C of the DNA is from 53.8–68.4 (genome sequence).
Type genus: Ectothiorhodospira Pelsh 1936, 120.
6. Description of the Family Halorhodospiraceae. fam. nov.
Ha.lo.rho.do.spi.ra’ce.ae. M.L. fem. n. Halorhodospira type genus of the family; -aceae ending to denote a family; M. L. fem. pl. n. Halorhodospiraceae the family of Halorhodospira.
The family constitutes moderately or extremely halophilic and extremely halotolerant phototrophic bacteria growing under alkaline conditions. They are Gram-negative and belong to the Gammaproteobacteria. Cells are spiral-, vibrioid- or rod-shaped, motile by means of polar flagella and divide by binary fission. They are phototrophic purple sulfur bacteria that perform anoxygenic photosynthesis with bacteriochlorophylls and carotenoids as photosynthetic pigments and have internal photosynthetic membranes as lamellar stacks continuous with the cytoplasmic membrane. Photosynthetic pigments are bacteriochlorophyll a or b and carotenoids. Growth of phototrophic representatives is preferably anaerobic in the light, with reduced sulfur compounds as electron donors. Sulfide is oxidized to elemental sulfur and is deposited outside the cells. The final oxidation product is sulfate. Halorhodospiraceae are found in saline, preferably extremely saline environments containing sulfide and having an alkaline to extremely alkaline pH. They are regular inhabitants and represent major groups of the bacterial populations of soda lakes of various salinities. Species of this family are the most halophilic eubacteria. Glycine betaine, ectoine and trehalose accumulate as compatible solutes in response to salt and osmotic stress.
The mol% G + C of the DNA is 55.8–72.0 (genome sequence).
Type genus: Halorhodospira Imhoff and Süling 1996, 112; Imhoff and Süling 1997, 915.VP.
6.1. Emended Description of the Genus Halorhodospira
Ha’lo.rho’do. spi’ra. Gr.gen. n. halos of the salt; Gr. n. rhodon the rose; Gr. n. spira the spiral; M.L fem. n. Halorhodospira, the spiral rose from salt lakes.
Cells are spiral- or rod-shaped, 0.6–1.2 µm in diameter, motile by bipolar flagella and multiply by binary fission. They are Gram-negative and belong to the Gammaproteobacteria and grow photoautotrophically under anoxic conditions with reduced sulfur compounds as electron donors, or photoheterotrophically with a limited number of simple organic compounds. Sulfide is oxidized to elemental sulfur, which is deposited outside the cells and may be further oxidized to sulfate. Internal photosynthetic membranes appear as lamellar stacks continuous with the cytoplasmic membrane. Photosynthetic pigments are bacteriochlorophyll a and carotenoids. Growth is dependent on highly saline and alkaline conditions. Greater than 10% (w/v) total salt concentration is required for optimal growth in all known species, some of which even grow in saturated salt solutions. Glycine betaine, ectoine and trehalose accumulate as compatible solutes in response to salt and osmotic stress. Growth factors are not required. Storage products are polysaccharides, poly-β-hydroxybutyrate and polyphosphate. Halorhodospira species are found in hypersaline and extremely saline environments, preferably with moderately to extremely alkaline pH (up to pH 11–12), that contain sulfide and are exposed to light, such as salt flats, salt lakes and soda lakes, but some species may also inhabit salterns and coastal lagoons.
The mol% G + C of the DNA is from 67.9–72.0 (genome sequence).
Type species: Halorhodospira (Hlr.) halophila (Raymond and Sistrom) Imhoff and Süling 1996, 110.
6.2. Description of the Genus Halochlorospira gen. nov.
Ha’lo.chlo’ro. spi’ra. Gr.gen. n. halos of the salt; Gr. n. chloros green; Gr. n. spira the spiral; M.L fem. n. Halochlorospira, the green spiral from salt lakes.
Cells are spiral- or rod-shaped, 0.5–1.2 µm in diameter, motile by bipolar flagella and multiply by binary fission. They are Gram-negative and belong to the Gammaproteobacteria and grow photoautotrophically under anoxic conditions with reduced sulfur compounds as electron donors, or photoheterotrophically with a limited number of simple organic compounds. Internal photosynthetic membranes appear as lamellar stacks continuous with the cytoplasmic membrane. Photosynthetic pigments are bacteriochlorophyll b and carotenoids. Sulfide is oxidized to elemental sulfur, which is deposited outside the cells and may be further oxidized to sulfate. Growth is dependent on highly saline and alkaline conditions. Greater than 10% (w/v) total salt concentration is required for optimal growth by all known species, some of which grow in saturated salt solutions. Glycine betaine, ectoine and trehalose accumulate as compatible solutes in response to salt and osmotic stress. Growth factors not required. Storage products are polysaccharides, poly-β-hydroxybutyrate and polyphosphate. Halochlorospira species are found in hypersaline and extremely saline environments with slightly to extremely alkaline pH (up to pH 11–12) that contain sulfide and are exposed to light, such as salt flats, salt lakes and soda lakes.
The mol% G + C of the DNA is from 55.8–62.9 (genome sequence).
Type species: Halochlorospira (Hcs.) halochloris (Imhoff and Trüper, 1977).
6.3. Description of Halochlorospira halochloris comb. nov.
The description is entirely the same as for Halorhodospira halochloris.
6.4. Descrition of Halochlorospira abdelmalekii comb. nov.
The description is entirely the same as for Halorhodospira abdelmalekii.
Author Contributions
Conceptualization, T.E.M., J.F.I. and J.A.K.; methodology, J.F.I. and J.A.K.; validation, J.F.I. and J.A.K.; investigation, J.F.I. and J.A.K.; data curation, J.A.K.; writing T.E.M., J.F.I. and J.A.K.; supervision, J.F.I. and J.A.K.; funding acquisition, J.A.K. All authors have read and agreed to the published version of the manuscript +. + Terrance E. Meyer passed away on 1 August 2020, while the manuscript was in the final stages of preparation.
Funding
This work was partially sponsored by the Wilson Enhancement Fund for Applied Research in Science at Bellevue University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under the accession numbers provided in Table 1.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Pfennig, N.; Trüper, H.G. Higher Taxa of the Phototrophic Bacteria. Int. J. Syst. Bacteriol. 1971, 21, 17–18. [Google Scholar] [CrossRef][Green Version]
- Fowler, V.J.; Pfennig, N.; Schubert, W.; Stackebrandt, E. Towards a phylogeny of phototrophic purple sulfur bacteria? 16S rRNA oligonucleotide cataloguing of 11 species of Chromatiaceae. Arch. Microbiol. 1984, 139, 382–387. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Fowler, V.J.; Schubert, W.; Imhoff, J.F. Towards a phylogeny of phototrophic purple sulfur bacteria—The genus Ectothiorhodospira. Arch. Microbiol. 1984, 137, 366–370. [Google Scholar] [CrossRef]
- Imhoff, J.F. Reassignment of the Genus Ectothiorhodospira Pelsh 1936 to a New Family, Ectothiorhodospiraceae fam. nov., and Emended Description of the Chromatiaceae Bavendamm 1924. Int. J. Syst. Bacteriol. 1984, 34, 338–339. [Google Scholar] [CrossRef]
- Imhoff, J.F. Family Ectothiorhodospiraceae. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: New York, NY, USA, 2005; Volume 2, Part B; pp. 41–43. [Google Scholar]
- Imhoff, J.F. Genus Ectothiorhodospira. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: New York, NY, USA, 2005; Volume 2, Part B, pp. 43–48. [Google Scholar]
- Imhoff, J.F. Genus Halorhodospira. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: New York, NY, USA, 2005; Volume 2, Part B, pp. 49–52. [Google Scholar]
- Imhoff, J.F. The family Ectothiorhodospiraceae. In The Prokaryotes. An Evolving Electronic Resource for the Microbiological Community, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer Verlag: New York, NY, USA, 1999. [Google Scholar]
- Thiemann, B.; Imhoff, J.F. Differentiation of Ectothiorhodospiraceae Based on Their Fatty Acid Composition. Syst. Appl. Microbiol. 1996, 19, 223–230. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Bias-Imhoff, U. Lipids, Quinones and Fatty Acids of Anoxygenic PHOTOTROPHIC bacteria. In Anoxygenic Photosynthetic Bacteria; Blankenship, R.E., Madigan, M.T., Bauer, C.E., Eds.; Kluwer Academic Publ.: Dordrecht, The Netherlands, 1995; pp. 179–205. [Google Scholar]
- Zahr, M.; Fobel, B.; Mayer, H.; Imhoff, J.F.; Campos, V.P.; Weckesser, J. Chemical composition of the lipopolysaccharides of Ectothiorhodospira shaposhnikovii, Ectothiorhodospira mobilis and Ectothiorhodospira halophila. Arch. Microbiol. 1992, 157, 499–504. [Google Scholar]
- Weckesser, J.; Drews, G.; Mayer, H. Lipopolysaccharides of photosynthetic bacteria. Ann. Rev. Microbiol. 1979, 33, 215–239. [Google Scholar] [CrossRef]
- Weckesser, J.; Mayer, H.; Schulz, G. Anoxygenic Phototrophic Bacteria: Model Organisms for Studies on Cell Wall Macromolecules. In Anoxygenic Photosynthetic Bacteria; Blankenship, R.E., Madigan, M.T., Bauer, C.E., Eds.; Kluwer Academic Publ.: Dordrecht, The Netherlands, 1995; pp. 207–230. [Google Scholar]
- Imhoff, J.F.; Süling, J. The phylogenetic relationship among Ectothiorhodospiraceae: A reevaluation of their taxonomy on the basis of 16S rDNA analyses. Arch. Microbiol. 1996, 165, 106–113. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Rahn, T.; Künzel, S.; Neulinger, S.C. Phylogeny of Anoxygenic Photosynthesis Based on Sequences of Photosynthetic Reaction Center Proteins and a Key Enzyme in Bacteriochlorophyll Biosynthesis, the Chlorophyllide Reductase. Microorganisms 2019, 7, 576. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Rahn, T.; Künzel, S.; Neulinger, S.C. Photosynthesis Is Widely Distributed among Proteobacteria as Demonstrated by the Phylogeny of PufLM Reaction Center Proteins. Front. Microbiol. 2018, 8, 2679. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Rahn, T.; Künzel, S.; Keller, A.; Neulinger, S.C. Osmotic Adaptation and Compatible Solute Biosynthesis of Phototrophic Bacteria as Revealed from Genome Analyses. Microorganisms 2019, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.; Han, J.; Lapidus, A.; Cheng, J.F.; Goodwin, L.; Pitluck, S.; Peters, L.; Land, M.L.; Hauser, L.; Vogl, K.; et al. The draft genome of Thiorhodospira sibirica ATCC 700588. 2011; unpublished. [Google Scholar]
- Saltikov, C.W.; Zargar, K.; Conrad, A.; Bernick, D.; Lowe, T.M.; Stolc, V.; Hoeft, S.; Oremland, R.S.; Stolz, J. Ectothiorhodospira sp. PHS-1, whole genome shotgun sequence. 2012; unpublished. [Google Scholar]
- Bryant, D.A.; Huntemann, M.; Han, J.; Chen, A.; Kyrpides, N.; Mavromatis, K.; Markowitz, V.; Palaniappan, K.; Ivanova, N.; Schaumberg, A.; et al. Ectothiorhodospira haloalkaliphila ATCC 51935, whole genome shotgun sequence. DOE Joint Genome Institute. 2013; unpublished. [Google Scholar]
- Hernandez-Maldonado, J.; Stoneburner, B.; Boren, A.; Miller, L.; Rosen, M.; Oremland, R.S.; Saltikov, C.W. Genome sequence of the photoarsenotrophic bacterium Ectothiorhodospira sp. strain BSL-9, isolated from a hypersaline alkaline arsenic-rich extreme environment. Genome Announc. 2016, 4, e01139-16. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Majid, S.; Deole, R.; Brettin, T.S.; Bruce, D.; Delano, S.F.; Detter, J.C.; Gleasner, C.D.; Han, C.S.; Misra, M.; et al. Complete genome sequence of Halorhodospira halophila SL1. Stand. Genom. Sci. 2013, 8, 206–214. [Google Scholar] [CrossRef]
- Tsukatani, Y.; Hirose, Y.; Harada, J.; Yonekawa, C.; Tamiaki, H. Unusual features in the photosynthetic machinery of Halorhodospira halochloris DSM 1059 revealed by complete genome sequencing. Photosynth. Res. 2019, 140, 311–319. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Muntyan, M.S.; Pantaleeva, A.N.; Muyzer, G. Thioalkalivibrio sulfidophilus sp. nov., a haloalkaliphilic, sulfur-oxidizing gammaproteobacterium from alkaline habitats. Int. J. Syst. Evol. Microbiol. 2012, 62, 1884–1889. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Hashwa, F.; Trüper, H.G. Isolation of extremely halophilic phototrophic bacteria from the alkaline Wadi Natrun, Egypt. Arch. Hydrobiol. 1978, 84, 381–388. [Google Scholar]
- Gorlenko, V.M.; Bryantseva, I.A.; Rabold, S.; Tourova, T.P.; Rubtsova, D.; Smirnova, E.; Thiel, V.; Imhoff, J.F. A novel alkaliphilic and halophilic purple sulfur bacterium Ectothiorhodospira variabilis from soda lakes. Int. J. Syst. Evol. Microbiol. 2009, 59, 658–664. [Google Scholar] [CrossRef]
- Ventura, S.; Viti, C.; Pastorelli, R.; Giovannetti, L. Revision of species delineation in the genus Ectothiorhodospira. Int. J. Syst. Evol. Microbiol. 2000, 50, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S.; Giovanetti, L.; Gori, A.; Viti, C.; Materassi, R. Total DNA restriction pattern and quinone composition of members of the family Ectothiorhodospiraceae. System. Appl. Microbiol. 1993, 16, 405–410. [Google Scholar] [CrossRef]
- Tourova, T.P.; Spiridonova, E.M.; Berg, I.; Slobodova, N.V.; Boulygina, E.S.; Sorokin, D.Y. Phylogeny and evolution of the family Ectothiorhodospiraceae based on comparison of 16S rRNA, cbbL and nifH gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2387–2398. [Google Scholar] [CrossRef] [PubMed]
- McCann, S.H.; Boren, A.; Hernandez-Maldonado, J.; Stoneburner, B.; Saltikov, C.W.; Stolz, J.F.; Oremland, R.S. Arsenite as an Electron Donor for Anoxygenic Photosynthesis: Description of Three Strains of Ectothiorhodospira from Mono Lake, California and Big Soda Lake, Nevada. Life 2016, 7, E1. [Google Scholar] [CrossRef] [PubMed]
- Gorlenko, V.M.; Burganskaya, E.I.; Bryantseva, I.A. Phototrophic communities of the Berikei highly mineralized mesothermal sulfide springs (Dagestan, Russia). Microbiology 2019, 88, 146–155. [Google Scholar] [CrossRef]
- Gorlenko, V.M.; Bryantseva, I.A.; Panteleeva, E.E.; Turova, T.P.; Kolganova, T.V.; Makhneva, Z.K.; Moskalenko, A.A. Ectothio-rhodosinus mongolicum gen. nov., sp. nov., a new purple sulfur bacterium from a soda lake in Mongolia. Microbiology (Moscow) 2003, 73, 66–73. [Google Scholar] [CrossRef]
- Bryantseva, I.A.; Gorlenko, V.M.; Kompantseva, E.I.; Imhoff, J.F.; Süling, J.; Mityushina, L. Thiorhodospira sibirica gen. nov., sp. nov., a new alkaliphilic purple sulfur bacterium from a Siberian soda lake. Int. J. Syst. Evol. Microbiol. 1999, 49, 697–703. [Google Scholar] [CrossRef]
- Bryantseva, I.A.; Tourova, T.P.; Kovaleva, O.L.; Kostrikina, N.A.; Gorlenko, V.M. Ectothiorhodospira magna sp. nov., a new large alkaliphilic purple sulfur bacterium. Microbiology 2010, 79, 780–790. [Google Scholar] [CrossRef]
- Raymond, J.C.; Sistrom, W.R. The isolation and preliminary characterization of a halophilic photosynthetic bacterium. Arch. Microbiol. 1967, 59, 255–268. [Google Scholar] [CrossRef]
- Raymond, J.C.; Sistrom, W.R. Ectothiorhodospira halophila: A new species of the genus Ectothiorhodospira. Archiv. Mikrobiol. 1969, 69, 121–126. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Trüper, H.G. Ectothiorhodospira halochloris sp. nov., a new extremely halophilic phototrophic bacterium containing bacteriochlorophyll b. Arch. Microbiol. 1977, 114, 115–121. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Truper, H.G. Ectothiorhodospira abdelmalekii sp. nov., a new halophilic and alkaliphilic phototrophic bacterium. Zentbl. Bakteriol. 1981, 2, 228–234. [Google Scholar] [CrossRef]
- Hirschler-Réa, A.; Matheron, R.; Riffaud, C.; Mouné, S.; Eatock, C.; Herbert, R.A.; Willison, J.C.; Caumette, P. Isolation and characterization of spirilloid purple phototrophic bacteria forming red layers in microbial mats of Mediterranean salterns: Description of Halorhodospira neutriphila sp. nov. and emendation of the genus Halorhodospira. Int. J. Syst. Evol. Microbiol. 2003, 53, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F.; Sahl, H.G.; Soliman, G.S.; Trüper, H.G. The Wadi Natrun: Chemical composition and microbial mass developments in alkaline brines of Eutrophic Desert Lakes. Geomicrobiol. J. 1979, 1, 219–234. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Singh, K.S.; Kirksey, J.; Hoff, W.D.; Deole, R. Draft Genome Sequence of the Extremely Halophilic Phototrophic Purple Sulfur Bacterium Halorhodospira halochloris. J. Genom. 2014, 2, 118–120. [Google Scholar] [CrossRef][Green Version]
- Brune, D.C. Isolation and characterization of sulfur globule proteins from Chromatium vinosum and Thiocapsa roseopersicina. Arch. Microbiol. 1995, 163, 391–399. [Google Scholar] [CrossRef]
- Pattaragulwanit, K.; Brune, D.C.; Trüper, H.G.; Dahl, C. Molecular genetic evidence for extracytoplasmic localization of sulfur globules in Chromatium vinosum. Arch. Microbiol. 1998, 169, 434–444. [Google Scholar] [CrossRef]
- Dahl, C.; Franz, B.; Hensen, D.; Kesselheim, A.; Zigann, R. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: Identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 2013, 159, 2626–2638. [Google Scholar] [CrossRef]
- Dahl, C. Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life 2015, 67, 268–274. [Google Scholar] [CrossRef]
- Fahey, R.C. Novel Thiols of Prokaryotes. Annu. Rev. Microbiol. 2001, 55, 333–356. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, R.G.; Newton, G.L.; Sherrill, C.; Fahey, R.C. Glutathione amide and its perthiol in anaerobic sulfur bacteria. J. Bacteriol. 1996, 78, 4742–4746. [Google Scholar] [CrossRef] [PubMed]
- van Petegem, F.; De Vos, D.; Savvides, S.; Vergauwen, B.; van Beeumen, J. Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: Crystallographic analysis of a glutathione amide reductase. J. Mol. Biol. 2007, 374, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Kerfeld, C.A.; Melnicki, M.R. Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 2016, 31, 66–75. [Google Scholar] [CrossRef]
- MacCready, J.S.; Tran, L.; Basalla, J.L.; Hakim, P.; Vecchiarelli, A.G. The McdAB system positions α-carboxysomes in proteobacteria. Mol. Microbiol. 2021, 116, 277–297. [Google Scholar] [CrossRef]
- Venturoli, G.; Mamedov, M.D.; Mansy, S.S.; Musiani, F.; Strocchi, M.; Francia, F.; Semenov, A.Y.; Cowan, J.A.; Ciurli, S. Electron transfer from HiPIP to the photooxidized tetraheme cytochrome subunit of Allochromatium vinosum reaction center: New insights from site-directed mutagenesis and computational studies. Biochemistry 2004, 43, 437–445. [Google Scholar] [CrossRef]
- Nagashima, K.V.P.; Matsuura, K.; Shimada, K.; Verméglio, A. High-Potential Iron−Sulfur Protein (HiPIP) Is the Major Electron Donor to the Reaction Center Complex in Photosynthetically Growing Cells of the Purple Bacterium Rubrivivax gelatinosus. Biochemistry 2002, 41, 14028–14032. [Google Scholar] [CrossRef]
- Przysiecki, C.T.; Meyer, T.E.; Cusanovich, M.A. Circular dichroism and redox properties of high redox potential ferredoxins. Biochemistry 1985, 24, 2542–2549. [Google Scholar] [CrossRef]
- Lieutaud, C.; Alric, J.; Bauzan, M.; Nitschke, W.; Schoepp-Cothenet, B. Study of the high-potential iron sulfur protein in Halo-rhodospira halophila confirms that it is distinct from cytochrome c as electron carrier. Proc. Natl. Acad. Sci. USA 2005, 102, 3260–3265. [Google Scholar] [CrossRef]
- Azai, C.; Tsukatani, Y.; Itoh, S.; Oh-Oka, H. C-type cytochromes in the photosynthetic electron transfer pathways in green sulfur bacteria and heliobacteria. Photosynth. Res. 2010, 104, 189–199. [Google Scholar] [CrossRef]
- Kostanjevecki, V.; Leys, D.; van Driessche, G.; Meyer, T.E.; Cusanovich, M.A.; Fischer, U.; Guisez, Y.; van Beeumen, J. Structure and characterization of Ectothiorhodospira vacuolata cytochrome b(558), a prokaryotic homologue of cytochrome b(5). J. Biol. Chem. 1999, 274, 35614–35620. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Schicklberger, M.; Gescher, J. Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl. Environ. Microbiol. 2012, 78, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.E.; Kyndt, J.A.; Memmi, S.; Moser, T.; Colón-Acevedo, B.; Devreese, B.; Van Beeumen, J.J. The growing family of photoactive yellow proteins and their presumed functional roles. Photochem. Photobiol. Sci. 2012, 11, 1495–1514. [Google Scholar] [CrossRef] [PubMed]
- Kyndt, J.A.; Fitch, J.C.; Meyer, T.E.; Cusanovich, M.A. The Photoactivated PYP Domain of Rhodospirillum centenum Ppr Accelerates the Recovery of the Bacteriophytochrome Domain after White Light Illumination. Biochemistry 2007, 46, 8256–8262. [Google Scholar] [CrossRef] [PubMed]
- Kyndt, J.A.; Fitch, J.C.; Meyer, T.E.; Cusanovich, M.A. Thermochromatium tepidum Photoactive Yellow Protein/Bacteriophytochrome/Diguanylate Cyclase: Characterization of the PYP Domain. Biochemistry 2005, 44, 4755–4764. [Google Scholar] [CrossRef]
- Tourova, T.P.; Slobodova, N.V.; Bumazhkin, B.K.; Kolganova, T.V.; Muyzer, G.; Sorokin, D.Y. Analysis of community composition of sulfur-oxidizing bacteria in hypersaline and soda lakes using soxB as a functional marker. FEMS Microbiol. Ecol. 2013, 84, 280–289. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Gorlenko, V.M.; Namsaraev, B.B.; Namsaraev, Z.B.; Lysenko, A.M.; Eshinimaev, B.T.; Khmelenina, V.N.; Trotsenko, Y.A.; Kuenen, J.G. Prokaryotic communities of the north-eastern Mongolian soda lakes. Hydrobiologia 2004, 522, 235–248. [Google Scholar] [CrossRef]
- Imhoff, J.F. Anoxygenic Phototrophic Bacteria from Extreme Environments. In Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects; Hallenbeck, P.C., Ed.; Springer: Cham, Switzerland, 2017; pp. 427–480. [Google Scholar]
- Jannasch, H.W. Die bakterielle Rotfärbung der Salzseen des Wadi Natrun. Arch. Hydrobiol. 1957, 53, 425–433. [Google Scholar] [CrossRef]
- Mesbah, N.M.; Abou-El-Ela, S.H.; Wiegel, J. Novel and unexpected prokaryotic diversity in water and sediments of the alkaline, hypersaline lakes of the Wadi an Natrun, Egypt. Microbiol. Ecol. 2007, 54, 598–617. [Google Scholar] [CrossRef]
- Tank, M.; Thiel, V.; Imhoff, J.F. Phylogenetic relationship of phototrophic purple sulfur bacteria according to pufL and pufM genes. Int. Microbiol. 2009, 12, 175–185. [Google Scholar]
- Thiel, V.; Tank, M.; Neulinger, S.C.; Gehrmann, L.; Dorador, C.; Imhoff, J.F. Unique communities of anoxygenic phototrophic bacteria in saline lakes of Salar de Atacama (Chile). Evidence for a new phylogenetic lineage of phototrophic Gammaproteobacteria from pufLM gene analyses. FEMS Microbiol. Ecol. 2010, 74, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, K.B.; Hedrich, S.; Johnson, D.B. Acidiferrobacter thiooxydans, gen. nov. sp. nov.; an acidophilic, thermo-tolerant, facultatively anaerobic iron- and sulfur-oxidizer of the family Ectothiorhodospiraceae. Extremophiles 2011, 15, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Shinohara, A.; Fukui, M. Sulfurifustis variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 3709–3713. [Google Scholar] [CrossRef]
- Issotta, F.; Moya-Beltran, A.; Mena, C.; Covarrubias, P.C.; Thyssen, C.; Bellenberg, S.; Sand, W.; Quatrinia, R.; Vera, M. Insights into the biology of acidophilic members of the Acidiferrobacteraceae family derived from comparative genomic analyses. Res. Microbiol. 2018, 169, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Lai, Q.; Liu, X.; Sun, F.; Du, Y.; Li, G.; Shao, Z. Maricoccus atlantica gen. nov. sp. nov., isolated from deep sea sediment of the Atlantic Ocean. Antonie van Leeuwenhoek 2013, 104, 1073–1081, Erratum in Antonie van Leeuwenhoek 2014, 105, 439. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Zhao, J.-X.; Sang, J.; Chen, G.-J.; Du, Z.-J. Halofilum ochraceum gen. nov., sp. nov., a gammaproteobacterium isolated from a marine solar saltern. Int. J. Syst. Evol. Microbiol. 2017, 67, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; van Pelt, S.; Tourova, T.P.; Takaichi, S.; Muyzer, G. Acetonitrile degradation under haloalkaline conditions by Natronocella acetinitrilica gen. nov., sp.no. Microbiology 2007, 153, 1157–1164. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Trotsenko, Y.A.; Doronina, N.V.; Tourova, T.P.; Galinski, E.A.; Kolganova, T.V.; Muyzer, G. Methylohalomonas lacus gen. nov., sp. nov. and Methylonatrum kenyenese gen. nov., sp. nov., methylotrophic Gammaproteobacteria from hypersaline lakes. Int. J. Syst. Evol. Microbiol. 2007, 57, 2762–2769. [Google Scholar] [CrossRef]
- Mori, K.; Suzuki, K.-I.; Urabe, T.; Sugihara, M.; Tanaka, K.; Hamada, M.; Hanada, S. Thioprofunndum hispidum sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing gammaproteobacterium isolated from the hydrothermal field on Suiyo Seamount, and proposal of Thioalkalispiraceae fam. nov. in the order Chromatiales. Int. J. Syst. Evol. Microbiol. 2011, 61, 2412–2418. [Google Scholar] [CrossRef]
- Barco, R.A.; Garrity, G.M.; Scott, J.J.; Amend, J.P.; Nealson, K.H.; Emerson, D. A genus definition for Bacteria and Archaea based on a standard genome relatedness index. mBio 2020, 11, e02475-19. [Google Scholar] [CrossRef]
- Cai, J.; Guan, Y.; Li, F.; Zhao, Y.; Feng, C.; Tang, N. Biomass and pigments production of a newly isolated photosynthetic bacterium Ectothiorhodospira shaposhnikovii P2 from saline wastewater. Int. J. Environ. Sci. Technol. 2018, 16, 7487–7496. [Google Scholar] [CrossRef]
- Ramana, V.V.; Sasikala, C.; Ramaprasad, E.V.V.; Ramana, C.V. Description of Ectothiorhodospira salini sp. nov. J. Gen. Appl. Microbiol. 2010, 56, 313–319. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).