Aggregatibacter actinomycetemcomitans and Filifactor alocis as Associated with Periodontal Attachment Loss in a Cohort of Ghanaian Adolescents
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
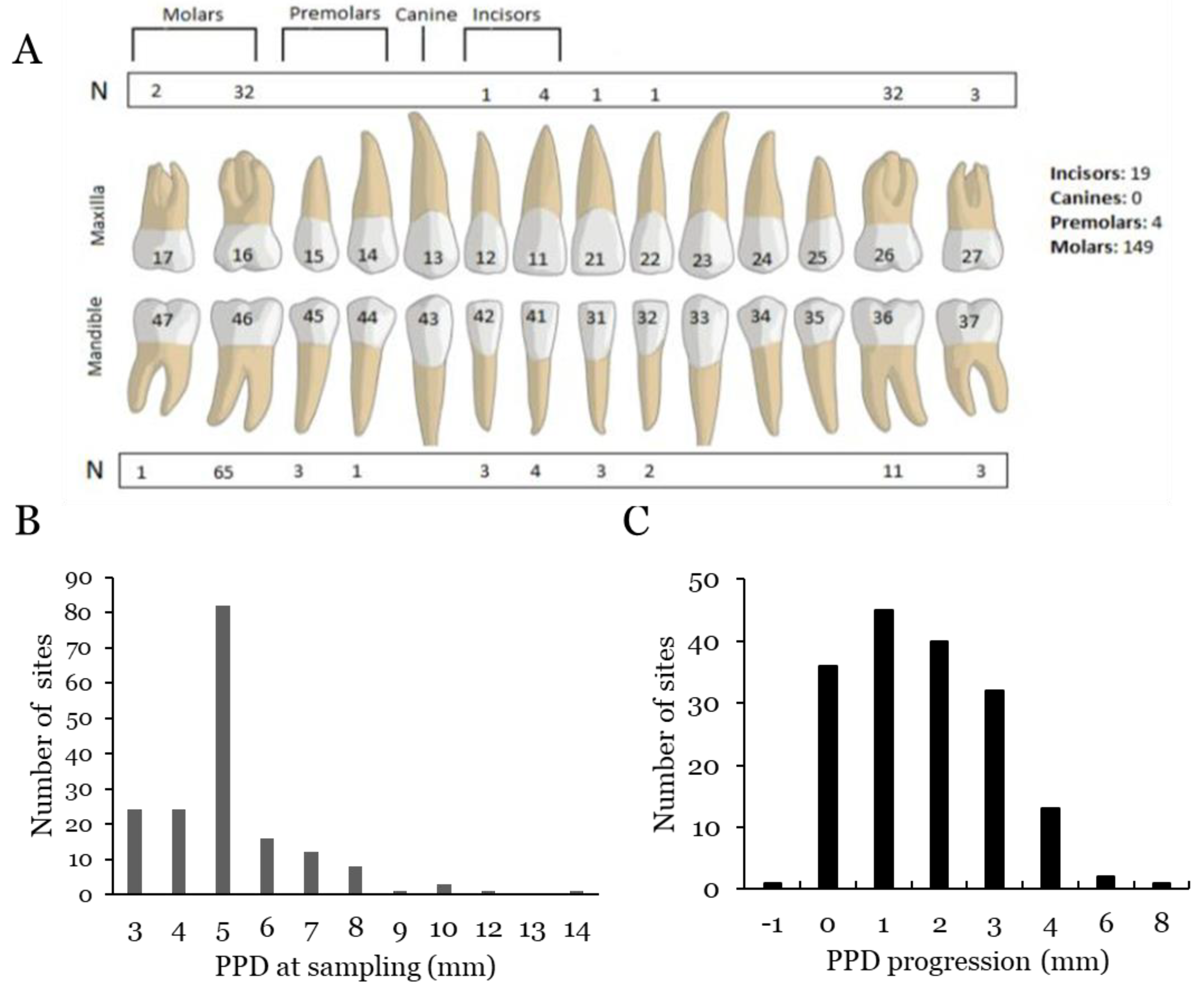
2.2. Clinical Assessments
2.3. Sampling
2.4. DNA Isolation
2.5. Microbiological Analyses
2.5.1. Quantitative PCR
2.5.2. Conventional PCR
2.6. Statistical Analyses
2.7. Ethics Statement
3. Results
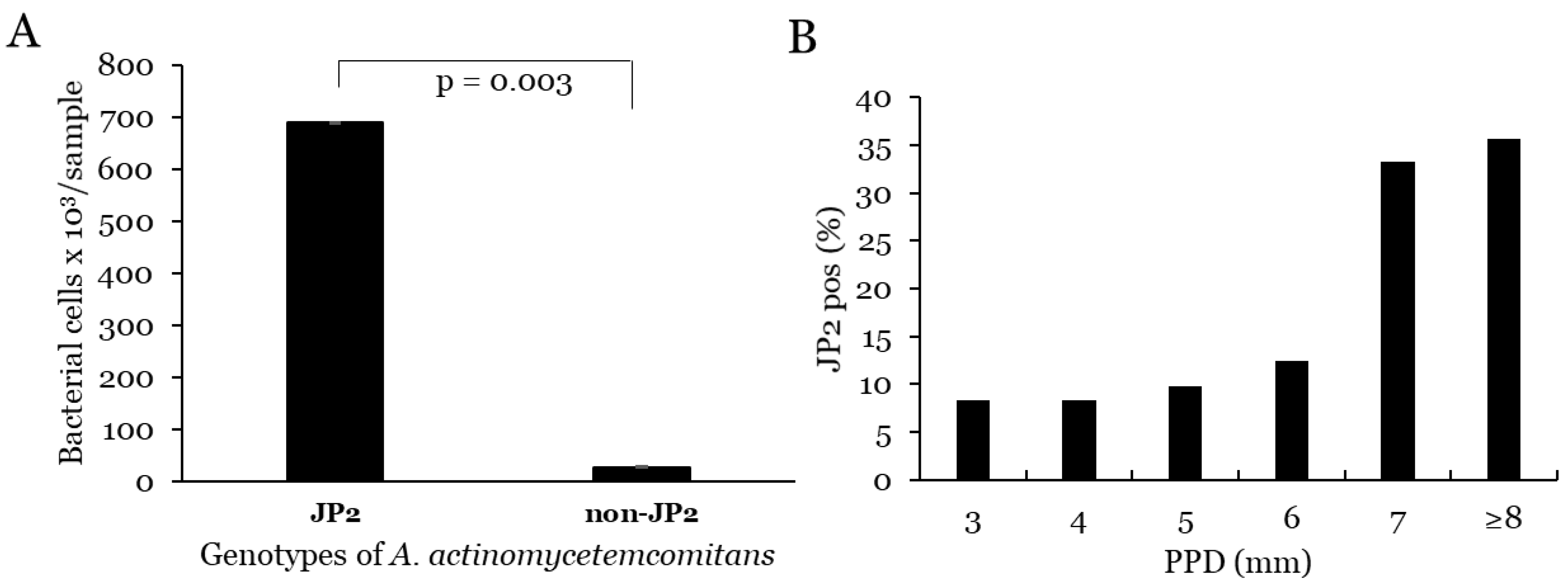
3.1. A. actinomycetemcomitans
3.1.1. Prevalence and Levels of A. ctinomycetemcomitans
3.1.2. Correlation with PPD and Progression for A. actinomycetemcomitans
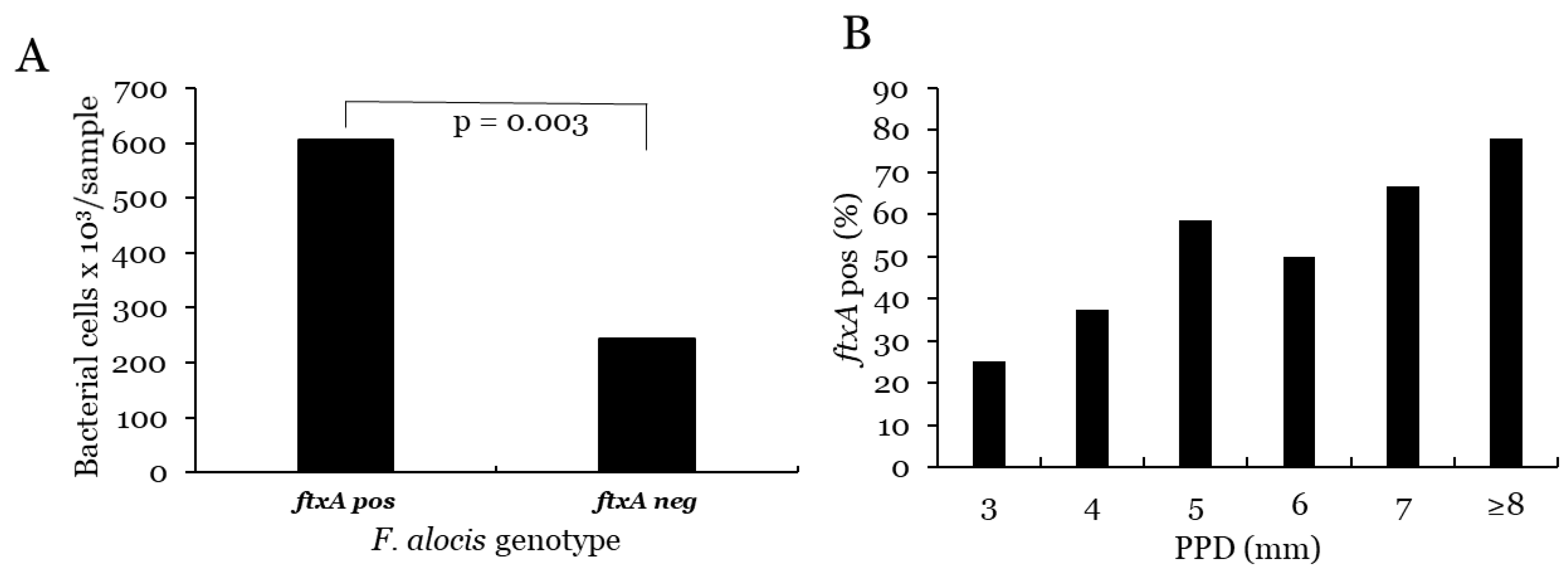
3.2. F. alocis
3.2.1. Prevalence and Levels of F. alocis
3.2.2. Correlation with PPD and Progression for F. alocis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Könönen, E.; Gursoy, M.; Gursoy, U. Periodontitis: A Multifaceted Disease of Tooth-Supporting Tissues. J. Clin. Med. 2019, 8, 1135. [Google Scholar] [CrossRef] [PubMed]
- Yucel-Lindberg, T.; Båge, T. Inflammatory mediators in the pathogenesis of periodontitis. Expert Rev. Mol. Med. 2013, 15, e7. [Google Scholar] [CrossRef] [PubMed]
- Susin, C.; Haas, A.N.; Albandar, J.M. Epidemiology and demographics of aggressive periodontitis. Periodontology 2000 2014, 65, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Norderyd, O.; Kochi, G.; Papias, A.; Köhler, A.A.; Helkimo, A.N.; Brahm, C.O.; Lindmark, U.; Lindfors, N.; Mattsson, A.; Rolander, B. Oral health of individuals aged 3-80 years in Jönköping, Sweden during 40 years (1973–2013). II. Review of clinical and radiographic findings. Swed. Dent. J. 2015, 39, 69–86. [Google Scholar] [PubMed]
- Zhang, X.; Wang, X.; Wu, J.; Wang, M.; Hu, B.; Qu, H.; Zhang, J.; Li, Q. The global burden of periodontal diseases in 204 countries and territories from 1990 to 2019. Oral Dis. 2022. [Google Scholar] [CrossRef]
- Kassebaum, N.J.; Smith, A.G.C.; Bernabé, E.; Fleming, T.D.; Reynolds, A.E.; Vos, T.; Murray, C.J.L.; Marcenes, W.; GBD 2015 Oral Health Collaborators. Global, Regional, and National Prevalence, Incidence, and Disability-Adjusted Life Years for Oral Conditions for 195 Countries, 1990–2015: A Systematic Analysis for the Global Burden of Diseases, Injuries, and Risk Factors. J. Dent. Res. 2017, 96, 380–387. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89 (Suppl. S1), S159–S172. [Google Scholar] [CrossRef]
- Graetz, C.; Mann, L.; Krois, J.; Sälzer, S.; Kahl, M.; Springer, C.; Schwendicke, F. Comparison of periodontitis patients’ classification in the 2018 versus 1999 classification. J. Clin. Periodontol. 2019, 46, 908–917. [Google Scholar] [CrossRef]
- Bouziane, A.; Hamdoun, R.; Abouqal, R.; Ennibi, O. Global prevalence of aggressive periodontitis: A systematic review and meta-analysis. J. Clin. Periodontol. 2020, 47, 406–428. [Google Scholar] [CrossRef]
- Yoshida, A.; Bouziane, A.; Erraji, S.; Lakhdar, L.; Rhissassi, M.; Miyazaki, H.; Ansai, T.; Iwasaki, M.; Ennibi, O. Etiology of aggressive periodontitis in individuals of African descent. Jpn. Dent. Sci. Rev. 2021, 57, 20–26. [Google Scholar] [CrossRef]
- Kissa, J.; El Houari, B.; Amine, K.; Chemlali, S.; Khlil, N.; Mikou, S.; Gharibi, A.; El Ouadnassi, I.; Rifki, C.; Albandar, J.M. Prevalence of periodontal disease in young Moroccans: A national survey. J. Periodontol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Haubek, D.; Ennibi, O.-K.; Poulsen, K.; Væth, M.; Poulsen, S.; Kilian, M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet 2008, 371, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Åberg, C.H.; Kwamin, F.; Claesson, R.; Dahlén, G.; Johansson, A.; Haubek, D. Progression of attachment loss is strongly associated with presence of the JP2 genotype of Aggregatibacter actinomycetemcomitans: A prospective cohort study of a young adolescent population. J. Clin. Periodontol. 2014, 41, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Rylev, M.; Kilian, M. Prevalence and distribution of principal periodontal pathogens worldwide. J. Clin. Periodontol. 2008, 35, 346–361. [Google Scholar] [CrossRef]
- Dahlén, G.; Claesson, R.; Åberg, C.H.; Haubek, D.; Johansson, A.; Kwamin, F. Subgingival bacteria in Ghanaian adolescents with or without progression of attachment loss. J. Oral Microbiol. 2014, 6, 23977. [Google Scholar] [CrossRef] [PubMed]
- Åberg, C.H.; Kwamin, F.; Claesson, R.; Johansson, A.; Haubek, D. Presence of JP2 and Non-JP2 Genotypes of Aggregatibacter actinomycetemcomitans and attachment loss in adolescents in Ghana. J. Periodontol. 2012, 83, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Brogan, J.M.; Lally, E.T.; Poulsen, K.; Kilian, M.; Demuth, D.R. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: Analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 1994, 62, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Markowitz, K.; Furgang, D.; Fairlie, K.; Ferrandiz, J.; Nasri, C.; McKiernan, M.; Gunsolley, J. Aggregatibacter actinomycetemcomitans and Its Relationship to Initiation of Localized Aggressive Periodontitis: Longitudinal Cohort Study of Initially Healthy Adolescents. J. Clin. Microbiol. 2007, 45, 3859–3869. [Google Scholar] [CrossRef]
- Fine, D.H.; Markowitz, K.; Fairlie, K.; Tischio-Bereski, D.; Ferrendiz, J.; Furgang, D.; Paster, B.J.; Dewhirst, F.E. A Consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis Is Present in Sites Prior to Bone Loss in a Longitudinal Study of Localized Aggressive Periodontitis. J. Clin. Microbiol. 2013, 51, 2850–2861. [Google Scholar] [CrossRef]
- Arunraj, R.; Neelakandan, A.; Potluri, R.; Yadalam, P.K.; Chakraborty, P.; Saravanan, A. The varied proportion of Filifactor alocis in periodontal health and disease in the South Indian subpopulation. Contemp. Clin. Dent. 2021, 12, 433. [Google Scholar] [CrossRef]
- Oscarsson, J.; Claesson, R.; Bao, K.; Brundin, M.; Belibasakis, G. Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA. Toxins 2020, 12, 687. [Google Scholar] [CrossRef] [PubMed]
- Bao, K.; Claesson, R.; Gehrig, P.; Grossmann, J.; Oscarsson, J.; Belibasakis, G.N. Proteomic Characterization of the Oral Pathogen Filifactor alocis Reveals Key Inter-Protein Interactions of Its RTX Toxin: FtxA. Pathogens 2022, 11, 590. [Google Scholar] [CrossRef] [PubMed]
- Ozuna, H.; Snider, I.; Belibasakis, G.N.; Oscarsson, J.; Johansson, A.; Uriarte, S.M. Aggregatibacter actinomycetemcomitans and Filifactor alocis: Two exotoxin-producing oral pathogens. Front. Oral Health 2022, 3, 981343. [Google Scholar] [CrossRef] [PubMed]
- Möller, A.J. Microbiological examination of root canals and periapical tissues of human teeth. Methodological studies. Odontol. Tidskr. 1966, 74, 1–380. [Google Scholar]
- Rylev, M.; Abduljabar, A.B.; Reinholdt, J.; Ennibi, O.-K.; Haubek, D.; Birkelund, S.; Kilian, M. Proteomic and immunoproteomic analysis of Aggregatibacter actinomycetemcomitans JP2 clone strain HK1651. J. Proteom. 2011, 74, 2972–2985. [Google Scholar] [CrossRef]
- Aruni, A.W.; Roy, F.; Sandberg, L.; Fletcher, H.M. Proteome variation among Filifactor alocis strains. Proteomics 2012, 12, 3343–3364. [Google Scholar] [CrossRef]
- Jalava, J.; Eerola, E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: Proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Evol. Microbiol. 1999, 49, 1375–1379. [Google Scholar] [CrossRef][Green Version]
- Kirakodu, S.S.; Govindaswami, M.; Novak, M.J.; Ebersole, J.L.; Novak, K.F. Optimizing qPCR for the Quantification of Periodontal Pathogens in a Complex Plaque Biofilm. Open Dent. J. 2008, 2, 49–55. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Rôças, I.N. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol. Immunol. 2003, 18, 263–265. [Google Scholar] [CrossRef]
- Poulsen, K.; Ennibi, O.-K.; Haubek, D. Improved PCR for Detection of the Highly Leukotoxic JP2 Clone of Actinobacillus actinomycetemcomitans in Subgingival Plaque Samples. J. Clin. Microbiol. 2003, 41, 4829–4832. [Google Scholar] [CrossRef]
- Van Der Velden, U.; Abbas, F.; Armand, S.; Loos, B.G.; Timmerman, M.F.; Van Der Weijden, G.A.; Van Winkelhoff, A.J.; Winkel, E.G. Java project on periodontal diseases. The natural development of periodontitis: Risk factors, risk predictors and risk determinants. J. Clin. Periodontol. 2006, 33, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Ennibi, O.K.; Claesson, R.; Akkaoui, S.; Reddahi, S.; Kwamin, F.; Haubek, D.; Johansson, A. High salivary levels of JP2 genotype of Aggregatibacter actinomycetemcomitans is associated with clinical attachment loss in Moroccan adolescents. Clin. Exp. Dent. Res. 2019, 5, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A. Aggregatibacter actinomycetemcomitans Leukotoxin: A Powerful Tool with Capacity to Cause Imbalance in the Host Inflammatory Response. Toxins 2011, 3, 242–259. [Google Scholar] [CrossRef]
- Kelk, P.; Claesson, R.; Chen, C.; Sjöstedt, A.; Johansson, A. IL-1beta secretion induced by Aggregatibacter (Actinobacillus) actinomycetemcomitans is mainly caused by the leukotoxin. Int. J. Med. Microbiol. 2008, 298, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Åberg, C.H.; Kelk, P.; Johansson, A. Aggregatibacter actinomycetemcomitans: Virulence of its leukotoxin and association with aggressive periodontitis. Virulence 2014, 6, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Miralda, I.; Vashishta, A.; Rogers, M.N.; Lamont, R.J.; Uriarte, S.M. The emerging oral pathogen, Filifactor alocis, extends the functional lifespan of human neutrophils. Mol. Microbiol. 2022, 117, 1340–1351. [Google Scholar] [CrossRef]
- Armstrong, C.L.; Klaes, C.K.; Vashishta, A.; Lamont, R.J.; Uriarte, S.M. Filifactor alocis manipulates human neutrophils affecting their ability to release neutrophil extracellular traps induced by PMA. Innate Immun. 2018, 24, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Kelk, P.; Claesson, R.; Hanstrom, L.; Lerner, U.H.; Kalfas, S.; Johansson, A. Abundant secretion of bioactive interleukin-1beta by human macrophages induced by Actinobacillus actinomycetemcomitans leukotoxin. Infect Immun. 2005, 73, 453–458. [Google Scholar] [CrossRef]
- Kelk, P.; Moghbel, N.S.; Hirschfeld, J.; Johansson, A. Aggregatibacter actinomycetemcomitans Leukotoxin Activates the NLRP3 Inflammasome and Cell-to-Cell Communication. Pathogens 2022, 11, 159. [Google Scholar] [CrossRef]
- Miralda, I.; Uriarte, S.M. Periodontal Pathogens’ strategies disarm neutrophils to promote dysregulated inflammation. Mol. Oral Microbiol. 2020, 36, 103–120. [Google Scholar] [CrossRef]

| PPD | Progresssion | |||||||
|---|---|---|---|---|---|---|---|---|
| N | Mean (mm) | SD | p-Value | N | Mean (mm) | SD | p-Value | |
| Aa pos | 95 | 5.24 | ±1.43 | 0.027 | 95 | 1.83 | ±1.41 | 0.163 |
| Aa neg | 77 | 5.05 | ±1.91 | 75 | 1.57 | ±1.41 | ||
| Aa high | 33 | 5.67 | ±1.80 | 0.027 | 33 | 2.03 | ±1.55 | 0.225 |
| Aa low | 139 | 5.04 | ±1.61 | 137 | 1.64 | ±1.37 | ||
| JP2 | 24 | 5.79 | ±1.58 | 0.013 | 23 | 2.48 | ±1.54 | 0.009 |
| Non-JP2 | 148 | 5.05 | ±1.66 | 147 | 1.60 | ±1.36 | ||
| Fa pos | 157 | 5.24 | ±1.68 | 0.016 | 155 | 1.74 | ±1.41 | 0.384 |
| Fa neg | 14 | 4.21 | ±1.12 | 14 | 1.43 | ±1.45 | ||
| Fa high | 123 | 5.49 | ±1.69 | 0.001 | 121 | 1.93 | ±1.37 | 0.001 |
| Fa low | 48 | 4.31 | ±1.27 | 48 | 1.17 | ±1.39 | ||
| ftxA pos | 90 | 5.52 | ±1.65 | 0.001 | 89 | 1.85 | ±1.60 | 0.398 |
| ftxA neg | 82 | 4.76 | ±1.59 | 81 | 1.57 | ±1.16 | ||
| ftxA-JP2 | 15 | 6.20 | ±1.32 | 0.001 | 15 | 2.93 | ±1.53 | 0.002 |
| Other | 157 | 5.06 | ±1.66 | 155 | 1.60 | ±1.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razooqi, Z.; Höglund Åberg, C.; Kwamin, F.; Claesson, R.; Haubek, D.; Oscarsson, J.; Johansson, A. Aggregatibacter actinomycetemcomitans and Filifactor alocis as Associated with Periodontal Attachment Loss in a Cohort of Ghanaian Adolescents. Microorganisms 2022, 10, 2511. https://doi.org/10.3390/microorganisms10122511
Razooqi Z, Höglund Åberg C, Kwamin F, Claesson R, Haubek D, Oscarsson J, Johansson A. Aggregatibacter actinomycetemcomitans and Filifactor alocis as Associated with Periodontal Attachment Loss in a Cohort of Ghanaian Adolescents. Microorganisms. 2022; 10(12):2511. https://doi.org/10.3390/microorganisms10122511
Chicago/Turabian StyleRazooqi, Zeinab, Carola Höglund Åberg, Francis Kwamin, Rolf Claesson, Dorte Haubek, Jan Oscarsson, and Anders Johansson. 2022. "Aggregatibacter actinomycetemcomitans and Filifactor alocis as Associated with Periodontal Attachment Loss in a Cohort of Ghanaian Adolescents" Microorganisms 10, no. 12: 2511. https://doi.org/10.3390/microorganisms10122511
APA StyleRazooqi, Z., Höglund Åberg, C., Kwamin, F., Claesson, R., Haubek, D., Oscarsson, J., & Johansson, A. (2022). Aggregatibacter actinomycetemcomitans and Filifactor alocis as Associated with Periodontal Attachment Loss in a Cohort of Ghanaian Adolescents. Microorganisms, 10(12), 2511. https://doi.org/10.3390/microorganisms10122511







