Effects of Helicobacter pylori and Nitrate-Reducing Bacteria Coculture on Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Isolation and Identification of Nitrate-Reducing Bacteria
2.2.1. Isolation
2.2.2. Nitrate Reduction Test
2.2.3. Bacteria Identification
2.3. Bacterial Species
2.4. Cytokine Measurement
2.4.1. Sample Preparation
2.4.2. Cytokine Measurement in Stimulated Culture Supernatants
2.4.3. Cytokine mRNA Transcription in Stimulated Cells
2.5. Measurement of MAPK Activity
2.5.1. Sample Preparation
2.5.2. Western Blot
2.6. Cell Cycle Measurement
2.6.1. Sample Preparation
2.6.2. Measurement and Analysis of Cell Cycle Progression
2.7. Statistical Analysis
3. Results
3.1. Cytokine Measurement in Stimulated Culture Supernatants
3.1.1. Actinomyces Genus
3.1.2. Rothia Genus
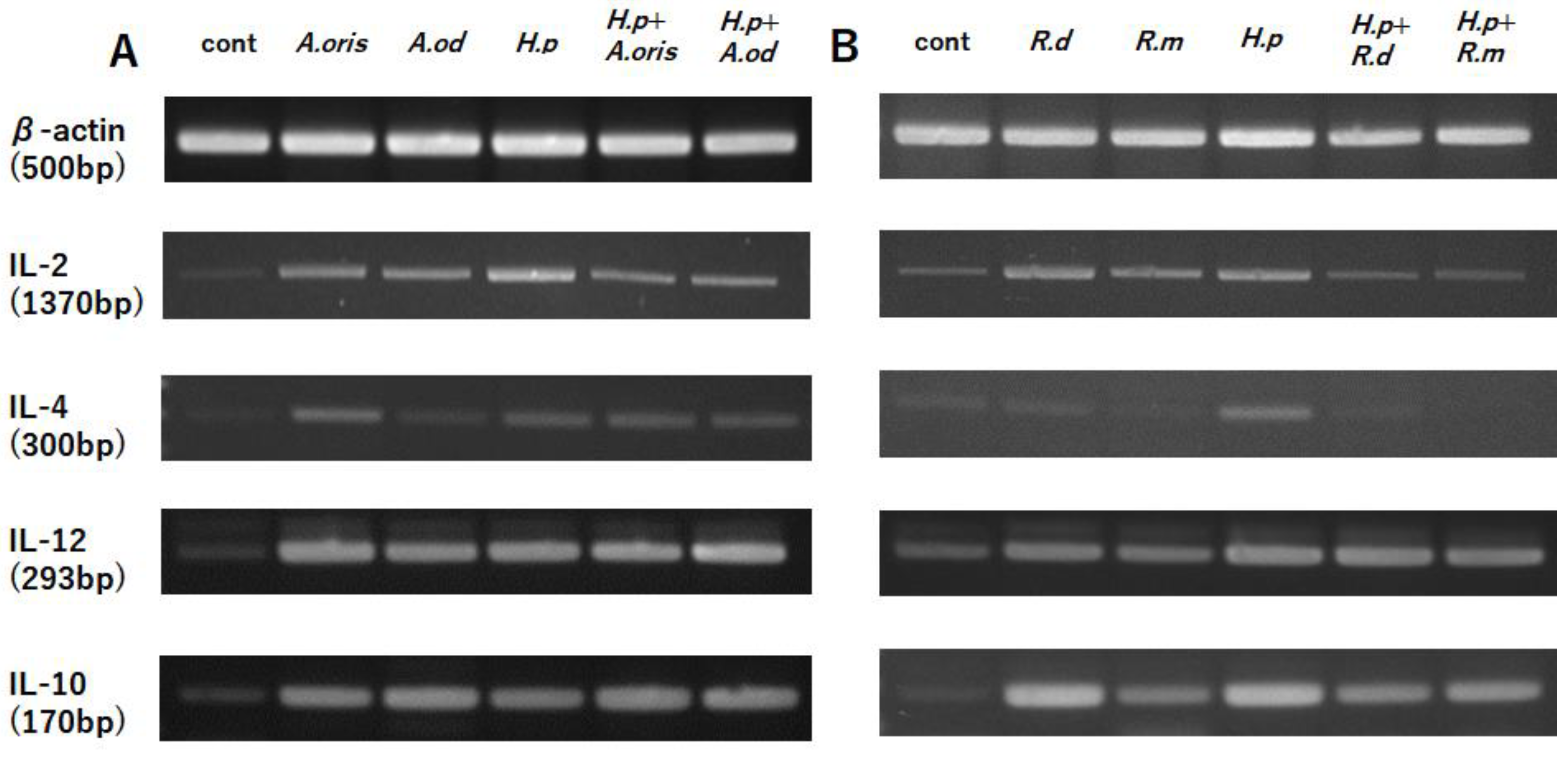
3.2. Cytokine mRNA Transcription
3.2.1. Actinomyces Genus
3.2.2. Rothia Genus
3.3. MAPK Activity
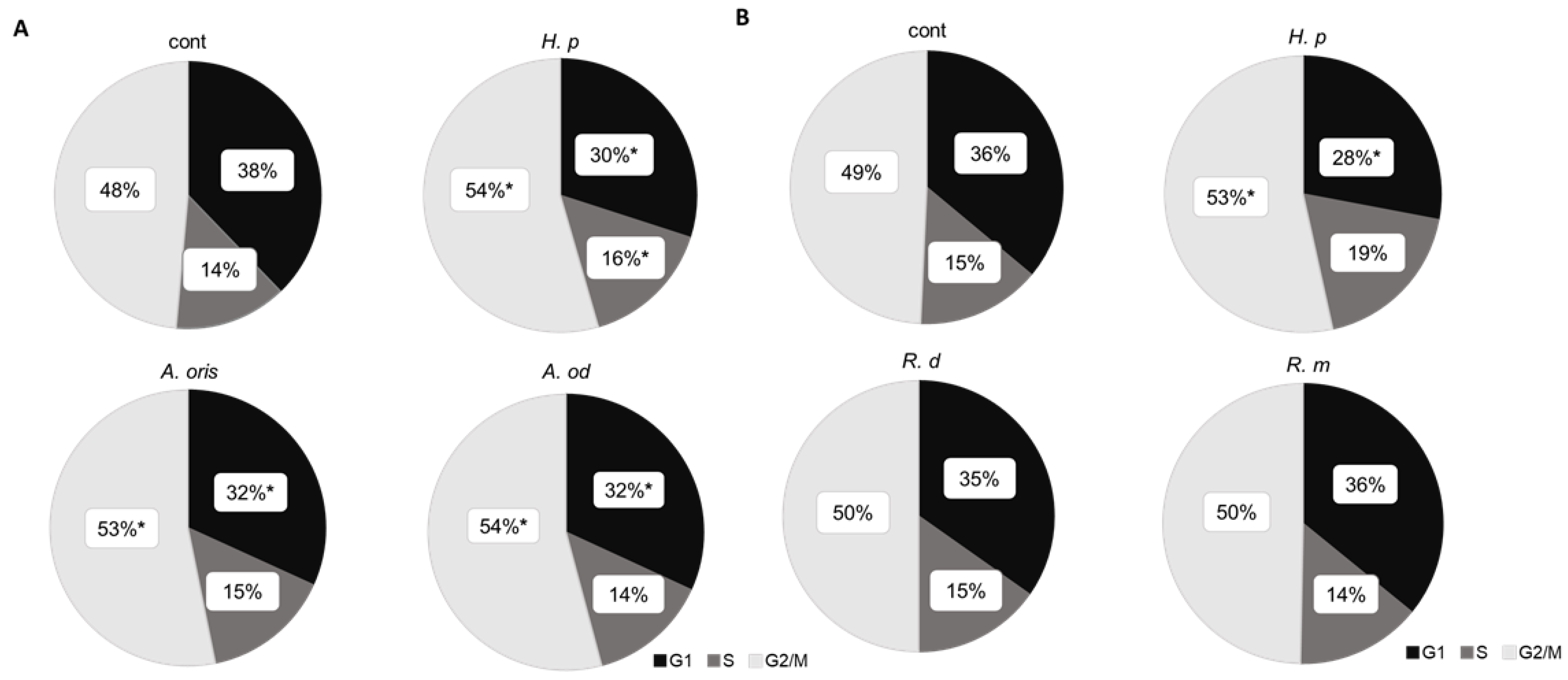
3.4. Cell Cycle
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zamani, M.; Ebrahimtabar, F.; Zamani, V.; Miller, W.H.; Alizadeh-Navaei, R.; Shokri-Shirvani, J.; Derakhshan, M.H. Systematic review with meta-analysis: The worldwide prevalence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2018, 47, 868–876. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Infection with Helicobacter pylori. In Schistosomes, Liver Flukes and Helicobacter pylori; IARC Mono-graphs: Lyon, France, 1994; Volume 61, pp. 177–240. [Google Scholar]
- Uemura, N.; Okamoto, S.; Yamamoto; Matsumura, N.; Yamaguchi, S.; Yamakido, M.; Taniyama, K.; Sasaki, N.; Schlemper, R.J. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001, 345, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.Y. Helicobacter pylori infection is the primary cause of gastric cancer. J. Gastroenterol. 2000, 35, 90–97. [Google Scholar] [PubMed]
- de Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Wroblewski, L.E.; Peek, R.M., Jr.; Wilson, K.T. Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef]
- Blaser, M.J.; Atherton, J.C. Helicobacter pylori persistence: Biology and disease. J. Clin. Investig. 2004, 113, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Weng, M.T.; Chiu, Y.T.; Wei, P.Y.; Chiang, C.W.; Fang, H.L.; Wei, S.C. Microbiota and gastrointestinal cancer. J. Formos Med. Assoc. 2019, 118, S32–S41. [Google Scholar] [CrossRef]
- Eun, C.S.; Kim, B.K.; Han, D.S.; Kim, S.Y.; Kim, K.M.; Choi, B.Y.; Song, K.S.; Kim, Y.S.; Kim, J.F. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014, 19, 407–416. [Google Scholar] [CrossRef]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Ferreira, R.M.; Pereira-Marques, J.; Pinto-Ribeiro, I.; Costa, J.L.; Carneiro, F.; Machado, J.C.; Figueiredo, C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018, 67, 226–236. [Google Scholar] [CrossRef]
- Sheh, A.; Fox, J.G. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes 2013, 4, 505–531. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Vaghari-Tabari, M.; Ferns, G.A.; Qujeq, D.; Andevari, A.N.; Sabahi, Z.; Moein, S. Signaling, metabolism, and cancer: An important relationship for therapeutic intervention. J. Cell Physiol. 2021, 236, 5512–5532. [Google Scholar] [CrossRef] [PubMed]
- Alipour, M. Molecular Mechanism of Helicobacter pylori-Induced Gastric Cancer. J. Gastrointest. Cancer 2021, 52, 23–30. [Google Scholar] [CrossRef]
- Wang, F.; Meng, W.; Wang, B.; Qiao, L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014, 45, 196–202. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- Figueiredo, C.A.; Marques, C.R.; dos Santos Costa, R.; da Silva, H.B.; Alcantara-Neves, N.M. Cytokines, cytokine gene polymorphisms and Helicobacter pylori infection: Friend or foe? World J. Gastroenterol. 2014, 20, 5235–5243. [Google Scholar] [CrossRef]
- Bagheri, V.; Memar, B.; Momtazi, A.A.; Sahebkar, A.; Gholamin, M.; Abbaszadegan, M.R. Cytokine networks and their association with Helicobacter pylori infection in gastric carcinoma. J. Cell Physiol. 2018, 233, 2791–2803. [Google Scholar] [CrossRef]
- Wang, Z. Regulation of Cell Cycle Progression by Growth Factor-Induced Cell Signaling. Cells. 2021, 10, 3327. [Google Scholar] [CrossRef]
- Thornton, T.M.; Rincon, M. Non-classical p38 map kinase functions: Cell cycle checkpoints and survival. Int. J. Biol. Sci. 2009, 5, 44–51. [Google Scholar] [CrossRef]
- Chiou, C.C.; Chan, C.C.; Kuo, Y.P.; Chan, E.C. Helicobacter pylori inhibits activity of cdc2 kinase and delays G2/M to G1 progression in gastric adenocarcinoma cell. Scand. J. Gastroenterol. 2011, 38, 147–152. [Google Scholar] [CrossRef] [PubMed]


| Species | Number of Strains |
|---|---|
| Actinomyces oris | 2 |
| Actinomyces odontolyticus | 3 |
| Rothia mucilaginosa | 5 |
| Rothia dentocariosa | 3 |
| Streptococcus salivarius | 2 |
| Streptococcus vestibularis | 2 |
| Streptococcus oralis | 1 |
| Streptococcus sinensis | 1 |
| Staphylococcus aureus | 3 |
| Bacillus subtilis | 1 |
| Neisseria subflava | 1 |
| Klebsiella pneumoniae | 1 |
| Echerichia fergusonii | 1 |
| Primer Name | Sequence | Tm (°C) |
|---|---|---|
| β-actinF | 5′-GAGGCATCCTCACCCTGAAG-3′ | 58 |
| β-actinR | 5′-TCTTGGGATGGGGAGTCTGT-3′ | |
| IL-2F | 5′-CGTAATAGTTCTGGAACTAAAGGG-3′ | 56 |
| IL-2R | 5′-TGGGAAGCACTTAATTATCAAGTC-3′ | |
| IL-12F | 5′-ATTTAACGTTTGTCTGCCAGGATGT-3′ | 58 |
| IL-12R | 5′-ACATGGGAACTAGCATCTTGTTCTC-3′ | |
| IL-4F | 5′-AGCCTCACAGAGCAGAAGAC-3′ | 60 |
| IL-4R | 5′-CCGTTTCAGGAGTCGGATCA-3′ | |
| IL-10F | 5′-CGCTTCCCGAAGTAACAAGG-3′ | 58 |
| IL-10R | 5′-CACTGGGTAGCTTCTTTCGG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojima, H.; Kuraoka, S.; Okanoue, S.; Okada, H.; Gotoh, K.; Matsushita, O.; Watanabe, A.; Yokota, K. Effects of Helicobacter pylori and Nitrate-Reducing Bacteria Coculture on Cells. Microorganisms 2022, 10, 2495. https://doi.org/10.3390/microorganisms10122495
Ojima H, Kuraoka S, Okanoue S, Okada H, Gotoh K, Matsushita O, Watanabe A, Yokota K. Effects of Helicobacter pylori and Nitrate-Reducing Bacteria Coculture on Cells. Microorganisms. 2022; 10(12):2495. https://doi.org/10.3390/microorganisms10122495
Chicago/Turabian StyleOjima, Hinako, Sakiko Kuraoka, Shyoutarou Okanoue, Hiroyuki Okada, Kazuyoshi Gotoh, Osamu Matsushita, Akari Watanabe, and Kenji Yokota. 2022. "Effects of Helicobacter pylori and Nitrate-Reducing Bacteria Coculture on Cells" Microorganisms 10, no. 12: 2495. https://doi.org/10.3390/microorganisms10122495
APA StyleOjima, H., Kuraoka, S., Okanoue, S., Okada, H., Gotoh, K., Matsushita, O., Watanabe, A., & Yokota, K. (2022). Effects of Helicobacter pylori and Nitrate-Reducing Bacteria Coculture on Cells. Microorganisms, 10(12), 2495. https://doi.org/10.3390/microorganisms10122495





