Overcoming Antibiotic Resistance with Novel Paradigms of Antibiotic Selection
Abstract
1. Background of the Problem
2. Overview of Conventional Antimicrobial Susceptibility Tests
Culture-Based Antimicrobial Susceptibility Tests
3. Overview of Methods That Detect Genes Conferring Antibiotic Resistance
4. Mechanisms Underlying Failure of Conventional Antimicrobial Susceptibility Tests
4.1. General Considerations of Minimum Inhibitory Concentrations and Thresholds for Non-Blood Borne Infections
4.2. Biofilms and Bacterial Multicellularity Are Not Considered by Conventional Antimicrobial Susceptibility Tests
4.3. Antibiotic Resistance within Multispecies Communities Is Not Considered by Conventional Antimicrobial Susceptibility Tests
4.4. Bacterial Regulation through Teazeled Receptors Is Not Considered by Conventional Antimicrobial Susceptibility Tests
4.5. Currently Unknown Pathogens Are Not Analyzed by Conventional Antimicrobial Susceptibility Tests
5. Emerging Methods for Antibiotic Selection
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. Bionanoscience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Dobbin, C.; Maley, M.; Harkness, J.; Benn, R.; Malouf, M.; Glanville, A.; Bye, P. The impact of pan-resistant bacterial pathogens on survival after lung transplantation in cystic fibrosis: Results from a single large referral centre. J. Hosp. Infect. 2004, 56, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Somayaji, R.; Parkins, M.D.; Shah, A.; Martiniano, S.L.; Tunney, M.M.; Kahle, J.S.; Waters, V.J.; Elborn, J.S.; Bell, S.C.; Flume, P.A.; et al. Antimicrobial susceptibility testing (AST) and associated clinical outcomes in individuals with cystic fibrosis: A systematic review. J. Cyst. Fibros. 2019, 18, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Waters, V.J.; Kidd, T.J.; Canton, R.; Ekkelenkamp, M.B.; Johansen, H.K.; LiPuma, J.J.; Bell, S.C.; Elborn, J.S.; Flume, P.A.; VanDevanter, D.R.; et al. Reconciling Antimicrobial Susceptibility Testing and Clinical Response in Antimicrobial Treatment of Chronic Cystic Fibrosis Lung Infections. Clin. Infect. Dis. 2019, 69, 1812–1816. [Google Scholar] [CrossRef]
- Rex, J.H.; Pfaller, M.A. Has Antifungal Susceptibility Testing Come of Age? Clin. Infect. Dis. 2002, 35, 982–989. [Google Scholar] [CrossRef]
- Kubicek-Sutherland, J.Z.; Heithoff, D.M.; Ersoy, S.C.; Shimp, W.R.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Host-dependent Induction of Transient Antibiotic Resistance: A Prelude to Treatment Failure. EBioMedicine 2015, 2, 1169–1178. [Google Scholar] [CrossRef]
- Doern, G.V.; Brecher, S.M. The Clinical Predictive Value (or Lack Thereof) of the Results of In Vitro Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2011, 49, S11–S14. [Google Scholar] [CrossRef]
- Hayes, J.F. Fighting Back against Antimicrobial Resistance with Comprehensive Policy and Education: A Narrative Review. Antibiotics 2022, 11, 644. [Google Scholar] [CrossRef]
- Carey, E.; Chen, H.-Y.P.; Baker, D.; Blankenhorn, R.; Vega, R.J.; Ho, M.; Munro, S. The association between non-ventilator associated hospital acquired pneumonia and patient outcomes among U.S. Veterans. Am. J. Infect. Control 2022. [Google Scholar] [CrossRef]
- Jenkins-Lonidier, L. Pulmonary Infections, Including Ventilator-Associated Pneumonia. Crit. Care Nurs. Clin. N. Am. 2021, 33, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.D.; McCants, D.; Velamuri, S. Using machine learning for process improvement in sepsis management. J. Healthcare Qual. Res. 2022; in press. [Google Scholar] [CrossRef] [PubMed]
- Vasala, A.; Hytönen, V.P.; Laitinen, O.H. Modern Tools for Rapid Diagnostics of Antimicrobial Resistance. Front. Cell. Infect. Microbiol. 2020, 10, 308. [Google Scholar] [CrossRef]
- Thrift, W.J.; Ronaghi, S.; Samad, M.; Wei, H.; Nguyen, D.G.; Cabuslay, A.S.; Groome, C.E.; Santiago, P.J.; Baldi, P.; Hochbaum, A.I.; et al. Deep Learning Analysis of Vibrational Spectra of Bacterial Lysate for Rapid Antimicrobial Susceptibility Testing. ACS Nano 2020, 14, 15336–15348. [Google Scholar] [CrossRef]
- Citron, D.M.; Ostovari, M.I.; Karlsson, A.; Goldstein, E.J. Evaluation of the E test for susceptibility testing of anaerobic bacteria. J. Clin. Microbiol. 1991, 29, 2197–2203. [Google Scholar] [CrossRef] [PubMed]
- Steinberger-Levy, I.; Shifman, O.; Zvi, A.; Ariel, N.; Beth-Din, A.; Israeli, O.; Gur, D.; Aftalion, M.; Maoz, S.; Ber, R. A Rapid Molecular Test for Determining Yersinia pestis Susceptibility to Ciprofloxacin by the Quantification of Differentially Expressed Marker Genes. Front. Microbiol. 2016, 7, 763. [Google Scholar] [CrossRef]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.-G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.-C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef]
- Choi, J.; Jung, Y.-G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef]
- Khan, Z.A.; Siddiqui, M.F.; Park, S. Current and Emerging Methods of Antibiotic Susceptibility Testing. Diagnostics 2019, 9, 49. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Marvin, L.F.; Roberts, M.A.; Fay, L.B. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin. Chim. Acta 2003, 337, 11–21. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Burnham, C.-A.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef]
- Kasas, S.; Malovichko, A.; Villalba, M.I.; Vela, M.E.; Yantorno, O.; Willaert, R.G. Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics 2021, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Leung, L.M.; Opene, B.; Fondrie, W.E.; Lee, Y.I.; Chandler, C.E.; Yoon, S.H.; Doi, Y.; Ernst, R.K.; Goodlett, D.R. Rapid Microbial Identification and Antibiotic Resistance Detection by Mass Spectrometric Analysis of Membrane Lipids. Anal. Chem. 2019, 91, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Kassim, A.; Omuse, G.; Premji, Z.; Revathi, G. Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: A cross-sectional stud. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 21. [Google Scholar] [CrossRef]
- Martínez, M.L.; Plata-Menchaca, E.P.; Ruiz-Rodríguez, J.C.; Ferrer, R. An approach to antibiotic treatment in patients with sepsis. J. Thorac. Dis. 2020, 12, 1007–1021. [Google Scholar] [CrossRef]
- Luna, C.M. Appropriateness and delay to initiate therapy in ventilator-associated pneumonia. Eur. Respir. J. 2006, 27, 158–164. [Google Scholar] [CrossRef]
- Monk, J.M. Predicting Antimicrobial Resistance and Associated Genomic Features from Whole-Genome Sequencing. J. Clin. Microbiol. 2019, 57, e01610-18. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Obande, G.A.; Banga Singh, K.K. Current and Future Perspectives on Isothermal Nucleic Acid Amplification Technologies for Diagnosing Infections. Infect. Drug Resist. 2020, 13, 455–483. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Bachmann, T.T.; Lüdke, G.; Lisby, J.G.; Kahlmeter, G.; Mohess, A.; Becker, K.; Hays, J.P.; Woodford, N.; Mitsakakis, K.; et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. Microbiol. 2019, 17, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Burnham, C.-A.D.; Leeds, J.; Nordmann, P.; O’Grady, J.; Patel, J. Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 2017, 15, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Satola, S.W.; Read, T.D. Genome-Based Prediction of Bacterial Antibiotic Resistance. J. Clin. Microbiol. 2019, 57, e01405-18. [Google Scholar] [CrossRef] [PubMed]
- Sundsfjord, A.; Simonsen, G.S.; Haldorsen, B.C.; Haaheim, H.; Hjelmevoll, S.-O.; Littauer, P.; Dahl, K.H. Genetic methods for detection of antimicrobial resistance. APMIS 2004, 112, 815–837. [Google Scholar] [CrossRef] [PubMed]
- Maljkovic Berry, I.; Melendrez, M.C.; Bishop-Lilly, K.A.; Rutvisuttinunt, W.; Pollett, S.; Talundzic, E.; Morton, L.; Jarman, R.G. Next Generation Sequencing and Bioinformatics Methodologies for Infectious Disease Research and Public Health: Approaches, Applications, and Considerations for Development of Laboratory Capacity. J. Infect. Dis. 2019, 221, S292–S307. [Google Scholar] [CrossRef]
- Cockerill, F.R. Genetic Methods for Assessing Antimicrobial Resistance. Antimicrob. Agents Chemother. 1999, 43, 199–212. [Google Scholar] [CrossRef]
- Gyssens, I.C.; Kern, W.V.; Livermore, D.M. The role of antibiotic stewardship in limiting antibacterial resistance among hematology patients. Haematologica 2013, 98, 1821–1825. [Google Scholar] [CrossRef]
- Rodloff, A.; Bauer, T.; Ewig, S.; Kujath, P.; Müller, E. Susceptible, Intermediate, and Resistant—The Intensity of Antibiotic Action. Dtsch. Arztebl. Int. 2008, 105, 657–662. [Google Scholar] [CrossRef]
- Kiem, S.; Schentag, J.J. Interpretation of Antibiotic Concentration Ratios Measured in Epithelial Lining Fluid. Antimicrob. Agents Chemother. 2008, 52, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Müller, M.; Derendorf, H. Rational dosing of antibiotics: The use of plasma concentrations versus tissue concentrations. Int. J. Antimicrob. Agents 2002, 19, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Vouloumanou, E.K.; Samonis, G.; Vardakasa, K.Z. Fosfomycin. Clin. Microbiol. Rev. 2016, 29, 321–347. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Walkty, A.J.; Karlowsky, J.A. Fosfomycin: A First-Line Oral Therapy for Acute Uncomplicated Cystitis. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 2082693. [Google Scholar] [CrossRef]
- Levison, M.E.; Levison, J.H. Pharmacokinetics and Pharmacodynamics of Antibacterial Agents. Infect. Dis. Clin. N. Am. 2009, 23, 791–815. [Google Scholar] [CrossRef] [PubMed]
- Rybak, M.J. Pharmacodynamics: Relation to antimicrobial resistance. Am. J. Infect. Control 2006, 34, S38–S45. [Google Scholar] [CrossRef] [PubMed]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Kuti, J.L. OPTIMIZING ANTIMICROBIAL PHARMACODYNAMICS: A GUIDE FOR YOUR STEWARDSHIP PROGRAM. Rev. Méd. Clín. Las Condes 2016, 27, 615–624. [Google Scholar] [CrossRef]
- Craig, W.A. State-of-the-Art Clinical Article: Pharmacokinetic/Pharmacodynamic Parameters: Rationale for Antibacterial Dosing of Mice and Men. Clin. Infect. Dis. 1998, 26, 1–12. [Google Scholar] [CrossRef]
- Landersdorfer, C.B.; Nation, R.L. Limitations of Antibiotic MIC-Based PK-PD Metrics: Looking Back to Move Forward. Front. Pharmacol. 2021, 12, 770518. [Google Scholar] [CrossRef]
- Coenye, T.; Goeres, D.; Van Bambeke, F.; Bjarnsholt, T. Should standardized susceptibility testing for microbial biofilms be introduced in clinical practice? Clin. Microbiol. Infect. 2018, 24, 570–572. [Google Scholar] [CrossRef] [PubMed]
- Piliouras, P.; Ulett, G.C.; Ashhurst-Smith, C.; Hirst, R.G.; Norton, R.E. A comparison of antibiotic susceptibility testing methods for cotrimoxazole with Burkholderia pseudomallei. Int. J. Antimicrob. Agents 2002, 19, 427–429. [Google Scholar] [CrossRef] [PubMed]
- Wanger, A.; Chávez, V. Antibiotic Susceptibility Testing in Practical Handbook of Microbiology; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Kumar, A.; Alam, A.; Rani, M.; Ehtesham, N.Z.; Hasnain, S.E. Biofilms: Survival and defense strategy for pathogens. Int. J. Med. Microbiol. 2017, 307, 481–489. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Post, J.C.; Hiller, N.L.; Nistico, L.; Stoodley, P.; Ehrlich, G.D. The role of biofilms in otolaryngologic infections: Update 2007. Curr. Opin. Otolaryngol. Head Neck Surg. 2007, 15, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.H.; Kim, S.T.; Kang, I.G.; Lee, J.H.; Cha, H.E.; Kim, D.Y. Comparison of tonsillar biofilms between patients with recurrent tonsillitis and a control group. Acta Otolaryngol. 2012, 132, 1115–1120. [Google Scholar] [CrossRef]
- Weeks, J.R.; Staples, K.J.; Spalluto, C.M.; Watson, A.; Wilkinson, T.M.A. The Role of Non-Typeable Haemophilus influenzae Biofilms in Chronic Obstructive Pulmonary Disease. Front. Cell. Infect. Microbiol. 2021, 11, 720742. [Google Scholar] [CrossRef]
- Sillanpää, J.; Chang, C.; Singh, K.V.; Montealegre, M.C.; Nallapareddy, S.R.; Harvey, B.R.; Ton-That, H.; Murray, B.E. Contribution of Individual Ebp Pilus Subunits of Enterococcus faecalis OG1RF to Pilus Biogenesis, Biofilm Formation and Urinary Tract Infection. PLoS ONE 2013, 8, e68813. [Google Scholar] [CrossRef]
- Tetz, G.V.; Artemenko, N.K.; Tetz, V.V. Effect of DNase and Antibiotics on Biofilm Characteristics. Antimicrob. Agents Chemother. 2009, 53, 1204–1209. [Google Scholar] [CrossRef]
- Tetz, V.V.; Korobov, V.P.; Artemenko, N.K.; Lemkina, L.M.; Panjkova, N.V.; Tetz, G.V. Extracellular phospholipids of isolated bacterial communities. Biofilms 2004, 1, 149–155. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W. Microbial biofilms in intertidal systems: An overview. Cont. Shelf Res. 2000, 20, 1257–1273. [Google Scholar] [CrossRef]
- Stewart, P.S.; William Costerton, J. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Reffuveille, F.; Fernández, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P. Multicellular resistance: Biofilms. Trends Microbiol. 2001, 9, 204. [Google Scholar] [CrossRef] [PubMed]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Søgaard-Andersen, L.; van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115–124. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents—How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef]
- Stewart, P.S.; Zhang, T.; Xu, R.; Pitts, B.; Walters, M.C.; Roe, F.; Kikhney, J.; Moter, A. Reaction–diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. npj Biofilms Microbiomes 2016, 2, 16012. [Google Scholar] [CrossRef] [PubMed]
- Williamson, K.S.; Richards, L.A.; Perez-Osorio, A.C.; Pitts, B.; McInnerney, K.; Stewart, P.S.; Franklin, M.J. Heterogeneity in Pseudomonas aeruginosa Biofilms Includes Expression of Ribosome Hibernation Factors in the Antibiotic-Tolerant Subpopulation and Hypoxia-Induced Stress Response in the Metabolically Active Population. J. Bacteriol. 2012, 194, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- Sønderholm, M.; Koren, K.; Wangpraseurt, D.; Jensen, P.Ø.; Kolpen, M.; Kragh, K.N.; Bjarnsholt, T.; Kühl, M. Tools for studying growth patterns and chemical dynamics of aggregated Pseudomonas aeruginosa exposed to different electron acceptors in an alginate bead model. npj Biofilms Microbiomes 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Yang, S.; Hay, I.D.; Cameron, D.R.; Speir, M.; Cui, B.; Su, F.; Peleg, A.Y.; Lithgow, T.; Deighton, M.A.; Qu, Y. Antibiotic regimen based on population analysis of residing persister cells eradicates Staphylococcus epidermidis biofilms. Sci. Rep. 2016, 5, 18578. [Google Scholar] [CrossRef]
- Lewis, K. Persister Cells: Molecular Mechanisms Related to Antibiotic Tolerance. In Antibiotic Resistance; Springer: Berlin/Heidelberg, Germany, 2012; pp. 121–133. [Google Scholar]
- Wood, T.K. Combatting bacterial persister cells. Biotechnol. Bioeng. 2016, 113, 476–483. [Google Scholar] [CrossRef]
- Oliver, A.; Sánchez, J.M.; Blázquez, J. Characterization of the GO system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2002, 217, 31–35. [Google Scholar] [CrossRef]
- Mandsberg, L.F.; Ciofu, O.; Kirkby, N.; Christiansen, L.E.; Poulsen, H.E.; Høiby, N. Antibiotic Resistance in Pseudomonas aeruginosa Strains with Increased Mutation Frequency Due to Inactivation of the DNA Oxidative Repair System. Antimicrob. Agents Chemother. 2009, 53, 2483–2491. [Google Scholar] [CrossRef]
- Penesyan, A.; Paulsen, I.T.; Kjelleberg, S.; Gillings, M.R. Three faces of biofilms: A microbial lifestyle, a nascent multicellular organism, and an incubator for diversity. npj Biofilms Microbiomes 2021, 7, 80. [Google Scholar] [CrossRef]
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van de Wiele, C.; De Spiegeleer, B. Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In vitro and In vivo. Front. Neurosci. 2017, 11, 183. [Google Scholar] [CrossRef]
- Solano, C.; Echeverz, M.; Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014, 18, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Sionov, R.V.; Steinberg, D. Targeting the Holy Triangle of Quorum Sensing, Biofilm Formation, and Antibiotic Resistance in Pathogenic Bacteria. Microorganisms 2022, 10, 1239. [Google Scholar] [CrossRef] [PubMed]
- Pumbwe, L.; Skilbeck, C.A.; Wexler, H.M. Presence of Quorum-sensing Systems Associated with Multidrug Resistance and Biofilm Formation in Bacteroides fragilis. Microb. Ecol. 2008, 56, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Subhadra, B.; Oh, M.H.; Choi, C.H. RND efflux pump systems in Acinetobacter, with special emphasis on their role in quorum sensing. J. Bacteriol. Virol. 2019, 49, 1–11. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, B.; Grenier, D.; Yi, L. Regulatory Mechanisms of the LuxS/AI-2 System and Bacterial Resistance. Antimicrob. Agents Chemother. 2019, 63, e01186-19. [Google Scholar] [CrossRef]
- Maseda, H.; Sawada, I.; Saito, K.; Uchiyama, H.; Nakae, T.; Nomura, N. Enhancement of the mexAB—oprM Efflux Pump Expression by a Quorum-Sensing Autoinducer and Its Cancellation by a Regulator, MexT, of the mexEF—oprN Efflux Pump Operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; El Jaziri, M. Quorum-Sensing Mechanisms and Bacterial Response to Antibiotics in P. aeruginosa. Curr. Microbiol. 2016, 73, 747–753. [Google Scholar] [CrossRef]
- Kanamaru, K.; Kanamaru, K.; Tatsuno, I.; Tobe, T.; Sasakawa, C. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 2000, 38, 805–816. [Google Scholar] [CrossRef]
- Rahmati, S.; Yang, S.; Davidson, A.L.; Zechiedrich, E.L. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol. 2002, 43, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Sitnikov, D.M.; Schineller, J.B.; Baldwin, T.O. Control of cell division in Escherichia coli: Regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc. Natl. Acad. Sci. USA 1996, 93, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Thapa, T.; Leuzzi, R.; Ng, Y.K.; Baban, S.T.; Adamo, R.; Kuehne, S.A.; Scarselli, M.; Minton, N.P.; Serruto, D.; Unnikrishnan, M. Multiple Factors Modulate Biofilm Formation by the Anaerobic Pathogen Clostridium difficile. J. Bacteriol. 2013, 195, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Tetz, V. Introducing the sporobiota and sporobiome. Gut Pathog. 2017, 9, 38. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus subtilis: Their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef]
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus Endospores to Extreme Terrestrial and Extraterrestrial Environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef]
- Paredes-Sabja, D.; Shen, A.; Sorg, J.A. Clostridium difficile spore biology: Sporulation, germination, and spore structural proteins. Trends Microbiol. 2014, 22, 406–416. [Google Scholar] [CrossRef]
- Lawley, T.D.; Clare, S.; Walker, A.W.; Goulding, D.; Stabler, R.A.; Croucher, N.; Mastroeni, P.; Scott, P.; Raisen, C.; Mottram, L.; et al. Antibiotic Treatment of Clostridium difficile Carrier Mice Triggers a Supershedder State, Spore-Mediated Transmission, and Severe Disease in Immunocompromised Hosts. Infect. Immun. 2009, 77, 3661–3669. [Google Scholar] [CrossRef]
- Yoon, S.S.; Brandt, L.J. Treatment of Refractory/Recurrent C. difficile-associated Disease by Donated Stool Transplanted Via Colonoscopy. J. Clin. Gastroenterol. 2010, 44, 562–566. [Google Scholar] [CrossRef]
- Khanolkar, R.A.; Clark, S.T.; Wang, P.W.; Hwang, D.M.; Yau, Y.C.W.; Waters, V.J.; Guttman, D.S. Ecological Succession of Polymicrobial Communities in the Cystic Fibrosis Airways. mSystems 2020, 5, e00809–e00822. [Google Scholar] [CrossRef]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef] [PubMed]
- Little, W.; Black, C.; Smith, A.C. Clinical Implications of Polymicrobial Synergism Effects on Antimicrobial Susceptibility. Pathogens 2021, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Combes, A.; Figliolini, C.; Trouillet, J.-L.; Kassis, N.; Wolff, M.; Gibert, C.; Chastre, J. Incidence and Outcome of Polymicrobial Ventilator-Associated Pneumonia. Chest 2002, 121, 1618–1623. [Google Scholar] [CrossRef]
- Adamowicz, E.M.; Flynn, J.; Hunter, R.C.; Harcombe, W.R. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J. 2018, 12, 2723–2735. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- O’Brien, T.J.; Figueroa, W.; Welch, M. Decreased efficacy of antimicrobial agents in a polymicrobial environment. ISME J. 2022, 16, 1694–1704. [Google Scholar] [CrossRef]
- Bottery, M.J.; Pitchford, J.W.; Friman, V.-P. Ecology and evolution of antimicrobial resistance in bacterial communities. ISME J. 2021, 15, 939–948. [Google Scholar] [CrossRef]
- Yurtsev, E.A.; Chao, H.X.; Datta, M.S.; Artemova, T.; Gore, J. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol. Syst. Biol. 2013, 9, 683. [Google Scholar] [CrossRef]
- Bottery, M.J.; Wood, A.J.; Brockhurst, M.A. Selective Conditions for a Multidrug Resistance Plasmid Depend on the Sociality of Antibiotic Resistance. Antimicrob. Agents Chemother. 2016, 60, 2524–2527. [Google Scholar] [CrossRef]
- Perlin, M.H.; Clark, D.R.; McKenzie, C.; Patel, H.; Jackson, N.; Kormanik, C.; Powell, C.; Bajorek, A.; Myers, D.A.; Dugatkin, L.A.; et al. Protection of Salmonella by ampicillin-resistant Escherichia coli in the presence of otherwise lethal drug concentrations. Proc. R. Soc. B Biol. Sci. 2009, 276, 3759–3768. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS Matrix: The “House of Biofilm Cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [PubMed]
- Schaar, V.; Uddback, I.; Nordstrom, T.; Riesbeck, K. Group A streptococci are protected from amoxicillin-mediated killing by vesicles containing -lactamase derived from Haemophilus influenzae. J. Antimicrob. Chemother. 2014, 69, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Vitse, J.; Devreese, B. The Contribution of Membrane Vesicles to Bacterial Pathogenicity in Cystic Fibrosis Infections and Healthcare Associated Pneumonia. Front. Microbiol. 2020, 11, 630. [Google Scholar] [CrossRef]
- Devos, S.; Stremersch, S.; Raemdonck, K.; Braeckmans, K.; Devreese, B. Intra- and Interspecies Effects of Outer Membrane Vesicles from Stenotrophomonas maltophilia on β-Lactam Resistance. Antimicrob. Agents Chemother. 2016, 60, 2516–2518. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Fontaine, C.; Xavier, J.B. Altruistic cell death and collective drug resistance. Mol. Syst. Biol. 2012, 8, 627. [Google Scholar] [CrossRef]
- Tanouchi, Y.; Pai, A.; Buchler, N.E.; You, L. Programming stress-induced altruistic death in engineered bacteria. Mol. Syst. Biol. 2012, 8, 626. [Google Scholar] [CrossRef]
- O’Brien, T.J.; Welch, M. A Continuous-Flow Model for in vitro Cultivation of Mixed Microbial Populations Associated With Cystic Fibrosis Airway Infections. Front. Microbiol. 2019, 10, 2713. [Google Scholar] [CrossRef]
- Vega, N.M.; Gore, J. Collective antibiotic resistance: Mechanisms and implications. Curr. Opin. Microbiol. 2014, 21, 28–34. [Google Scholar] [CrossRef]
- Meredith, H.R.; Srimani, J.K.; Lee, A.J.; Lopatkin, A.J.; You, L. Collective antibiotic tolerance: Mechanisms, dynamics and intervention. Nat. Chem. Biol. 2015, 11, 182–188. [Google Scholar] [CrossRef]
- Lopes, S.P.; Ceri, H.; Azevedo, N.F.; Pereira, M.O. Antibiotic resistance of mixed biofilms in cystic fibrosis: Impact of emerging microorganisms on treatment of infection. Int. J. Antimicrob. Agents 2012, 40, 260–263. [Google Scholar] [CrossRef]
- Briaud, P.; Camus, L.; Bastien, S.; Doléans-Jordheim, A.; Vandenesch, F.; Moreau, K. Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci. Rep. 2019, 9, 16564. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, A.P.; Lopes, S.P.; Pereira, M.O. Insights into Cystic Fibrosis Polymicrobial Consortia: The Role of Species Interactions in Biofilm Development, Phenotype, and Response to In-Use Antibiotics. Front. Microbiol. 2017, 7, 2146. [Google Scholar] [CrossRef] [PubMed]
- Orazi, G.; Jean-Pierre, F.; O’Toole, G.A. Pseudomonas aeruginosa PA14 Enhances the Efficacy of Norfloxacin against Staphylococcus aureus Newman Biofilms. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef] [PubMed]
- Galera-Laporta, L.; Garcia-Ojalvo, J. Antithetic population response to antibiotics in a polybacterial community. Sci. Adv. 2020, 6, eaaz5108. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Lv, X.; Niu, X.; Yu, F.; Zuo, J.; Bao, Y.; Yin, H.; Huang, C.; Nawaz, S.; Zhou, W.; et al. Effect of nutritional and environmental conditions on biofilm formation of avian pathogenic Escherichia coli. J. Appl. Microbiol. 2022, 132, 4236–4251. [Google Scholar] [CrossRef]
- Tetz, V.; Tetz, G. Novel Cell Receptor System of Eukaryotes Formed by Previously Unknown Nucleic Acid-Based Receptors. Receptors 2022, 1, 13–53. [Google Scholar] [CrossRef]
- Tetz, V.; Tetz, G. Novel prokaryotic system employing previously unknown nucleic acids-based receptors. Microb. Cell Fact. 2022, 21, 202. [Google Scholar] [CrossRef]
- Tetz, V.V.; Tetz, G.V. Effect of Extracellular DNA Destruction by DNase I on Characteristics of Forming Biofilms. DNA Cell Biol. 2010, 29, 399–405. [Google Scholar] [CrossRef]
- Nijland, R.; Hall, M.J.; Burgess, J.G. Dispersal of Biofilms by Secreted, Matrix Degrading, Bacterial DNase. PLoS ONE 2010, 5, e15668. [Google Scholar] [CrossRef]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar]
- Ibáñez de Aldecoa, A.L.; Zafra, O.; González-Pastor, J.E. Mechanisms and Regulation of Extracellular DNA Release and Its Biological Roles in Microbial Communities. Front. Microbiol. 2017, 8, 1390. [Google Scholar] [CrossRef] [PubMed]
- Sumby, P.; Barbian, K.D.; Gardner, D.J.; Whitney, A.R.; Welty, D.M.; Long, R.D.; Bailey, J.R.; Parnell, M.J.; Hoe, N.P.; Adams, G.G.; et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. USA 2005, 102, 1679–1684. [Google Scholar] [CrossRef] [PubMed]
- Terekhov, S.S.; Mokrushina, Y.A.; Nazarov, A.S.; Zlobin, A.; Zalevsky, A.; Bourenkov, G.; Golovin, A.; Belogurov, A.; Osterman, I.A.; Kulikova, A.A.; et al. A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding. Sci. Adv. 2020, 6, eaaz9861. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Johnson, N.; Cizmas, L.; McDonald, T.J.; Kim, H. A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 2016, 150, 702–714. [Google Scholar] [CrossRef]
- Quirós, P.; Colomer-Lluch, M.; Martínez-Castillo, A.; Miró, E.; Argente, M.; Jofre, J.; Navarro, F.; Muniesa, M. Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Human Fecal Samples. Antimicrob. Agents Chemother. 2014, 58, 606–609. [Google Scholar] [CrossRef]
- Rajwa, B.; Dundar, M.M.; Akova, F.; Bettasso, A.; Patsekin, V.; Dan Hirleman, E.; Bhunia, A.K.; Robinson, J.P. Discovering the unknown: Detection of emerging pathogens using a label-free light-scattering system. Cytom. Part A 2010, 77, 1103–1112. [Google Scholar] [CrossRef]
- Locey, K.J.; Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 5970–5975. [Google Scholar] [CrossRef]
- Tetz, G.; Vecherkovskaya, M.; Zappile, P.; Dolgalev, I.; Tsirigos, A.; Heguy, A.; Tetz, V. Complete Genome Sequence of Kluyvera intestini sp. nov., Isolated from the Stomach of a Patient with Gastric Cancer. Genome Announc. 2017, 5, e01184-17. [Google Scholar] [CrossRef]
- Vecherkovskaya, M.F.; Tetz, G.V.; Tetz, V.V. Complete Genome Sequence of the Streptococcus sp. Strain VT 162, Isolated from the Saliva of Pediatric Oncohematology Patients. Genome Announc. 2014, 2, e00647-14. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Tetz, V.; Tetz, G. Draft Genome Sequence of Bacillus obstructivus VT-16-70 Isolated from the Bronchoalveolar Lavage Fluid of a Patient with Chronic Obstructive Pulmonary Disease. Genome Announc. 2017, 5, e01754-16. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Vikina, D.; Brown, S.; Zappile, P.; Dolgalev, I.; Tsirigos, A.; Heguy, A.; Tetz, V. Draft Genome Sequence of Streptococcus halitosis sp. nov., Isolated from the Dorsal Surface of the Tongue of a Patient with Halitosis. Microbiol. Resour. Announc. 2019, 8, e01704-18. [Google Scholar] [CrossRef] [PubMed]
- Tetz, V.; Tetz, G. Draft Genome Sequence of the Uropathogenic Herbaspirillum frisingense Strain ureolyticus VT-16-41. Genome Announc. 2017, 5, e00279-17. [Google Scholar] [CrossRef] [PubMed]
- Webb, K.; Zain, N.M.M.; Stewart, I.; Fogarty, A.; Nash, E.F.; Whitehouse, J.L.; Smyth, A.R.; Lilley, A.K.; Knox, A.; Williams, P.; et al. Porphyromonas pasteri and Prevotella nanceiensis in the sputum microbiota are associated with increased decline in lung function in individuals with cystic fibrosis. J. Med. Microbiol. 2022, 71, 001481. [Google Scholar] [CrossRef]
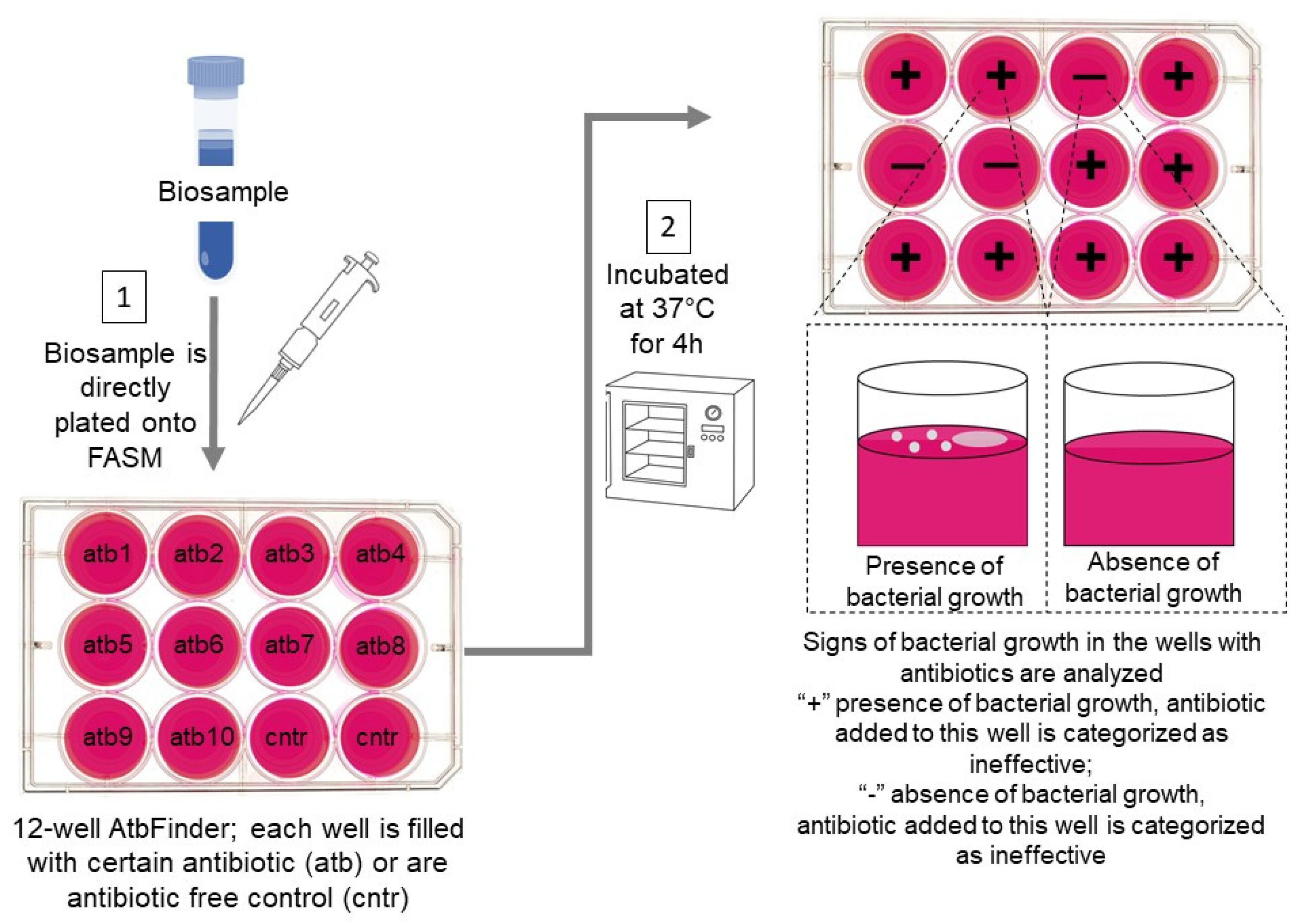
- Tetz, G.; Tetz, V. Evaluation of a New Culture-Based AtbFinder Test-System Employing a Novel Nutrient Medium for the Selection of Optimal Antibiotics for Critically Ill Patients with Polymicrobial Infections within 4 h. Microorganisms 2021, 9, 990. [Google Scholar] [CrossRef]
- Tetz, G.; Tetz, V. No TitleDraft Genome Sequence of Chryseobacterium mucoviscidosis sp. nov. Strain VT16-26, Isolated from the Bronchoalveolar Lavage Fluid of a Patient with Cystic Fibrosis. Genome Announc. 2018, 6, e01473-17. [Google Scholar] [CrossRef]
- Tetz, G.; Tetz, V.; Vecherkovskaya, M. Complete Genome Sequence of Paenibacillus sp. Strain VT 400, Isolated from the Saliva of a Child with Acute Lymphoblastic Leukemia. Genome Announc. 2015, 3, e00894-15. [Google Scholar] [CrossRef]
- Tetz, G.; Vecherkovskaya, M.; Hahn, A.; Tsifansky, M.; Koumbourlis, A.; Tetz, V. AtbFinder Diagnostic Test System Improves Optimal Selection of Antibiotic Therapy in Persons with Cystic Fibrosis. Preprint 2022, 2022100141. [Google Scholar] [CrossRef]
- Tetz, G.; Kardava, K.; Vecherkovskaya, M.; Tsifansky, M.; Tetz, V. Treatment of chronic relapsing urinary tract infection with antibiotics selected by AtbFinder. Authorea 2022. [Google Scholar] [CrossRef]
- Ersoy, S.C.; Heithoff, D.M.; Barnes, L.; Tripp, G.K.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing. EBioMedicine 2017, 20, 173–181. [Google Scholar] [CrossRef]
| Important Considerations Overseen by Conventional ASTs | Results of the Lack of Consideration by Conventional ASTs |
|---|---|
| Real-life antibiotic contraptions at the site of infection | Use of MIC and susceptibility breakpoints based on pharmacokinetic/pharmacodynamic (PK/PD) of antibiotics in blood by conventional AST is misleading, since concentrations of antibiotics in different tissues differ from those in blood |
| Modulation of time-dependent, concentration dependent, and dependent on total drug exposure with the area under the curve (AUC) mechanism of antibiotic action | |
| Biofilm type of growth | Misses the effects of surface membrane like layer and extracellular polymeric substances on antibiotic tolerance |
| Persister tolerance | |
| Altered antibiotic response of cells with reduced metabolic activity | |
| Effects of quorum sensing on antibiotic response | |
| Effects of TezRs receptors on antibiotic response | |
| Polymicrobial type of infection | Unique transcriptomic activity of antibiotic resistance genes (ARG) within polymicrobial community |
| Collective response to antibiotics | |
| Mutual food addiction | |
| Role of “supporting bacteria” or “accessory pathogen” in antibiotic response of the lead pathogen | |
| Accounting for unknown pathogens | The role of currently unknown pathogens in infection |
| Characteristics | Ideal AST | Culture-Based AST | Rapid AST for Pure Cultures | Nucleic Acid-Based Amplification |
|---|---|---|---|---|
| Recaptures environment at the site of infection | Yes | No | No | Yes |
| Confirmation of antibiotic resistance | Yes | No | No | No |
| Works without need for pure bacterial culture | Yes | No | No | Yes/No |
| Time from biosample to results (h) | <8 | 48–120 | 30–40 | 2–8 |
| Places patient on guided antibiotic therapy at day 1 | Yes | No | No | Yes |
| Appropriate for fastidious/unknown bacteria | Yes | No | No | Yes/No |
| Accounts for biofilm-type of growth | Yes | No | No | No |
| Accounts for modulation of antibiotic response of lead pathogen by other bacteria at the site of infection | Yes | No | No | No |
| Accounts for not-yet culturable bacteria | Yes | No | No | Yes/No |
| Considers antibiotics’ PK/PD in different organs | Yes | No | No | No |
| Accounts for collective response to antibiotics | Yes | No | No | No |
| Accounts for activity of genes of antibiotic resistance | Yes | No | No | No |
| Slow growth and persisters | Yes | No | No | No |
| Antibiotic stewardship | Should reduce the number of used antibiotics | Neutral | Neutral | Increased antibiotic use |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tetz, G.; Tetz, V. Overcoming Antibiotic Resistance with Novel Paradigms of Antibiotic Selection. Microorganisms 2022, 10, 2383. https://doi.org/10.3390/microorganisms10122383
Tetz G, Tetz V. Overcoming Antibiotic Resistance with Novel Paradigms of Antibiotic Selection. Microorganisms. 2022; 10(12):2383. https://doi.org/10.3390/microorganisms10122383
Chicago/Turabian StyleTetz, George, and Victor Tetz. 2022. "Overcoming Antibiotic Resistance with Novel Paradigms of Antibiotic Selection" Microorganisms 10, no. 12: 2383. https://doi.org/10.3390/microorganisms10122383
APA StyleTetz, G., & Tetz, V. (2022). Overcoming Antibiotic Resistance with Novel Paradigms of Antibiotic Selection. Microorganisms, 10(12), 2383. https://doi.org/10.3390/microorganisms10122383






