Abstract
The Anaplasmataceae family encompasses obligate intracellular α-proteobacteria of human and veterinary medicine importance. This study performed multi-locus sequencing to characterize Ehrlichia and Anaplasma in coati’s blood samples in Midwestern Brazil. Twenty-five samples (25/165—15.1%) were positive in the screening PCR based on the dsb gene of Ehrlichia spp. and were characterized using 16S rRNA, sodB, groEL, and gltA genes and the 23S-5S intergenic space region (ITS). Phylogenetic analyses based on all six molecular markers positioned the sequences into a new clade, with a common origin of Ehrlichia ruminantium. Haplotype analyses of 16S RNA sequences revealed the presence of two distinct Ehrlichia genotypes. Six samples (6/165, 3.6%) were positive in the screening nPCR for the 16S rRNA gene of Anaplasma spp. and were submitted to an additional PCR targeting the ITS for molecular characterization. Phylogenetic analyses based on both 16S rRNA gene and ITS positioned the Anaplasma sp. detected in the present study in a large clade with other Anaplasma sp. previously detected in ticks and wild animals and in a clade with ‘Candidatus Anaplasma brasiliensis’, respectively. Based on distinct molecular markers, the present work described a putative novel Anaplasmataceae agent, namely ‘Candidatus Ehrlichia dumleri’, and Anaplasma sp. closely related to the previously described ‘Candidatus Anaplasma brasiliensis’.
1. Introduction
The Anaplasmataceae family (Order Rickettsiales) encompasses obligate intracellular α-proteobacteria that can cause important diseases in humans and animals [1,2]. Wild and domestic carnivores are considered an important source of tick-borne agents, especially in anthropized areas, where close contact between domestic/wild animals and humans can occur. Procyonidae mammals, including ring-tailed coatis Nasua nasua from Brazil, can easily adapt to peri-urban areas [3,4], favoring the exchange of ectoparasites and vector-borne pathogens among humans and animals. In the United States, raccoons (Procyon lotor) are incriminated as important reservoirs of Anaplasma phagocytophilum, a zoonotic agent transmitted by ticks from Ixodes persulcatus complex [1,5,6,7]. More recently, an unusual case of human anaplasmosis was reported in the Amazon, French Guiana, caused by ‘Candidatus Anaplasma sparouinense’, a putative novel Anaplasma sp. closely related to Anaplasma genotypes previously detected in Amblyomma coelebs ticks collected from coatis, rats, and sloths (‘Candidatus Anaplasma amazonensis’) [2].
Although coatis from urban forest fragments at Campo Grande city, Mato Grosso do Sul state, Brazil seem to present tick species diversity lower than those from natural areas, Amblyomma dubitatum, Amblyomma sculptum, and Amblyomma ovale were identified parasitizing this gregarious species in [8]. In fact, coatis can be found infected with Trypanosoma cruzi and Leishmania spp., two zoonoses of extreme importance in Public Health [9,10]. These findings demonstrate the importance of constant surveillance on vector-borne agents in wild fauna, especially those with zoonotic potential. The present study aimed to detect the presence of Ehrlichia and Anaplasma spp. in coati’s blood samples molecularly and assess the phylogenetic positioning of the detected agents based on a multi-locus sequencing approach.
2. Materials and Methods
2.1. Ethical Statement
All experimental procedures were approved by the ‘Instituto Chico Mendes de Biodiversidade’ (ICMBio) (SISBIO 49662-8) and by the Ethics Committee on Animal Use of the School of Agricultural and Veterinary Sciences, UNESP (CEUA FCAV/UNESP 06731/19), Ethics Committee on Animal Use of the Universidade Católica Dom Bosco (CEUA UCDB 001/2018) and Air Force Cooperation Agreement (Nº01/GAP-CG/2018).
2.2. Blood Sampling
Between March 2018 and January 2019, coatis (N. nasua) were sampled every three months for 10 consecutive nights (with an interval of one day—Saturday) in two Cerrado areas located in Campo Grande city, Mato Grosso do Sul State, Midwestern Brazil. All captures and recaptures were performed by convenience, and recapture times were performed by chance. The first sampling spot, Parque Estadual do Prosa (PEP) (−20.44987, −54.56529) is a conservation unit of 135 hectares. The second sampling spot was a Brazilian Air Force Private Area (VBA) (−20.47163, −54.65405), a complex of 197 hectares divided into a military operational area and a residential area inhabited by at least 730 humans with domestic animals such as dogs.
All capture procedures were previously described [4,8,11]. After chemical restraint, animals were marked with numbered colored earrings and had a microchip implanted in the subcutaneous tissue between the shoulder blades. They were measured, and the age was estimated according to the reference literature [12]. Blood was sampled from the femoral vein using tubes containing EDTA (Ethylenediamine tetraacetic acid) and then placed into RNAse/DNAse free cryotubes and stored in an −80 °C freezer until molecular analyses.
2.3. DNA Extraction from Coatis’ Blood Samples and Conventional PCR (PCR) for Mammalian Endogenous Control
DNA was extracted from 200 μL of blood using Illustra Blood Mini Kit (GE Healthcare®, Chicago, IL, USA), according to the manufacturer’s instructions. To evaluate the quality of the extracted DNA and avoid false negative results, DNA samples were tested by a PCR targeting the mammalian gapdh [13]. Ultra-pure sterile water (Life Technologies®, Carlsbad, CA, USA) was used as a negative control in PCR assays.
2.4. Screening and Characterization of Anaplasmataceae Agents
Screening for Ehrlichia spp. was performed using a PCR targeting the dsb gene (~409 bp) [14]. Positive samples were submitted to the following PCR or nested PCR (nPCR) protocols for additional molecular characterization: 16S rRNA (~1470 bp) [15], groEL (~680 bp) [16], sodB (~ 600 bp) [17], gltA (~800 bp) [18] and ITS 23S–5S (~300 bp) [19]. Screening for Anaplasma spp. was performed using an nPCR targeting the 16S rRNA gene (~546 bp) [20]. Positive samples were submitted to PCR protocols for additional molecular characterization targeting the ITS 23S–5S (~300 bp) [19] and groEL (~340 bp) [21]. The PCR reactions contained 10X PCR buffer (Life Technologies®, Carlsbad, CA, USA), 1 mM MgCl2 (Life Technologies®, Carlsbad, CA, USA), 0·2 mM deoxynucleotide triphosphate (dNTPs) mixture (Life Technologies®, Carlsbad, CA, USA), 1·5 U Taq DNA Polymerase (Life Technologies®, Carlsbad, CA, USA), and 0·5 µM of each primer (Integrated DNA Technologies®, Coralville, IA, USA) (Table S1). Ehrlichia canis (Jaboticabal strain) and Anaplasma phagocytophilum (Webster strain) DNA were used as positive controls. Ultra-pure sterile water (Life Technologies®, Carlsbad, CA, USA) was used as a negative control in all PCR assays.
The amplicons obtained from PCR and nPCR assays were subjected to 1% ethidium bromide-stained agarose gel electrophoresis and results were visualized in UV transilluminator (ChemiDoc MP Imaging System, Bio Rad®, Hercules, CA, USA). Amplicons that presented high intensity on agarose gel (size of the amplicon matching the control, unique and strong bands) were purified using Exosap Life Technologies®, Carlsbad, CA, USA) and subjected to sequencing. The sequencing of amplicons was realized by the Sanger method [22] using ABI PRISM 3730 DNA Analyzer (Applied Biosystems) at Human Genome and Stem Cell Research Center, ‘Instituto de Biociências’, University of São Paulo (USP), São Paulo, SP, Brazil. All obtained sequences were submitted to GenBank under the following accession numbers: Ehrlichia sp. dsb (OP819944–OP819946), Ehrlichia sp. 16S rRNA (OM530509-OM530517), Ehrlichia sp. groEL (OP819939), Ehrlichia sp. sodB (OP903236-OP903238), Ehrlichia sp. 23S-5S (OM717254-OM717255), Ehrlichia sp. gltA (OP819935-OP919938, OP819940-OP819941), Anaplasma sp. 16S rRNA (OP279731-OP279734), and Anaplasma sp. 23S-5S (OP918134).
2.5. Bioinformatic Analyses
Electropherograms were submitted to a quality-screening test using Phred-Phrap software (version 23) [23,24] to obtain consensus sequences from the alignment of sense and antisense sequences. BLASTn tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 4 October 2022) [25] was used to compare the obtained sequences with those previously deposited in the GenBank database to obtain Query-coverage%, E-value, and % identity. For phylogenetic inferences, sequences from the present study were aligned with those retrieved from GenBank using MAFFT software version 7 [26]. The best evolutionary model was chosen using the iqTREE software (available at: http://iqtree.cibiv.univie.ac.at/ accessed on 4 October 2022), under the Akaike Information Criterion (AIC) [27,28]. The phylogenetic analyses were based on Bayesian inference (BI) and performed using MrBayes version 3.1.2 [29] via the CIPRES Science Gateway. Markov chain Monte Carlo simulations were run for 106 generations with a sampling frequency of every 100 generations and a 25% burn-in. The phylogenetic tree edition and rooting (outgroup) were performed using TreeGraph 2.0 beta software [30].
Additionally, an analysis of nucleotide polymorphisms of the sequences obtained in the present study was performed. The number of haplotypes, haplotype diversity (Hd), and nucleotide diversity (Pi) were determined using the program DnaSP 5, version 5.10.01 [31]. Also, the pairwise distance matrix among sequences detected at the present study and other Anaplasmataceae species was estimated using the Mega-X software version 10.1.8 [32].
3. Results
In total, 165 blood samples were collected (63 in PEP and 102 in VBA). In PEP, blood samples were obtained from 47 different individuals (27 females and 20 males; seven infants, 2 subadults and 38 adults). In VBA, samples were obtained from 57 different individuals (32 females and 25 males; 11 infants, 8 sub-adults, and 38 adults) (Table 1). All recapture procedures are previously described [11].

Table 1.
Identification of ring-tailed coatis (Nasua nasua) sampled and recaptured in two areas, namely Parque Estadual do Prosa and Vila da Base Aérea, city of Campo Grande, State of Mato Grosso do Sul, Midwestern Brazil, with the identification number, sex, age and positivity in the screening PCR to Ehrlichia sp. (dsb gene) during subsequent recaptures.
3.1. PCR Assays for Ehrlichia spp. Screening and Characterization
All DNA samples obtained from coati blood (n = 165) were positive in the PCR targeting the gapdh endogenous gene. Out of 165 coatis’ blood samples, 25 (15.1%) were positive in the screening PCR for the dsb gene of Ehrlichia spp. [4/63 samples—6.3% from PEP area (from 4 different coatis) and 21/102 samples—20.6% from VBA (obtained from 16 different coatis). No recaptured animal showed positivity in more than one collection time at PEP, while coatis from VBA showed positivity in more than one recapture (Table 1).
Transiently positive PCR results were detected in three animals from VBA: VBA03 was positive in four out of six samplings; VBA21 was positive in 2/6 samplings, and VBA44 in 3/3 samplings. No animal from PEP showed PCR positive results for more than one sampling (Table 2). Animal VBA03 remained PCR positive after 276 days after the first positive PCR (18 June 2018–20 March 2019); animal VBA21 after 28 days (21 March 2019–23 April 2019); and VBA44 after 174 days (7 November 2018–30 April 2019).

Table 2.
Identification of sampling date (day, month, and year) during subsequent recaptures of ring-tailed coatis (Nasua nasua) positive to Ehrlichia sp. sampled and recaptured in two areas, namely Parque Estadual do Prosa and Vila da Base Aérea, city of Campo Grande, State of Mato Grosso do Sul, Midwestern Brazil. All dates when positive PCR results were found are bolded.
Out of 25 positive samples in the dsb-based PCR screening for Ehrlichia spp., three were properly sequenced, nine were positive in the PCR targeting the near-complete 16S rRNA gene, one for groEL gene, three for sodB gene, two for 23S–5S ITS, and six for gltA gene. Results from Blast analyses, including sequence size, query coverage, E-value, and % of identity, are shown in Table S1—Supplementary Materials.
Phylogenetic analysis inferred by the Bayesian method (BI) and TIM2+I evolutionary model and based on the dsb gene (alignment of 395 bp) grouped the three sequenced samples in a new clade separated (100% posterior probability) from other Ehrlichia genotypes previously detected in wild animals from Brazil and Amblyomma ticks from Brazil and Argentina (Figure 1). The genetic divergence between sequences obtained in the present study and those closely related and retrieved from GenBank ranged from 0.05 to 0.18 (Supplementary Materials—Table S2).

Figure 1.
Phylogenetic tree inferred by the Bayesian method and based on the dsb gene from Ehrlichia spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua) sampled in Campo Grande city, Mato Grosso do Sul state, Brazil. Sequences detected in the present study are highlighted in bold pink (VBA).
Phylogenetic analysis inferred by the Bayesian method (BI) and HKY+I+G evolutionary model and based on the 16S rRNA gene (alignment of 1201 bp) grouped the sequences detected in the present study in a new large clade (92% posterior probability) (Figure 2). The genetic divergence between the Ehrlichia genotypes detected in coatis from PEP and ‘Candidatus Ehrlichia occidentalis’ and Ehrlichia sp. Eira barbara ranged from 1.2 to 1.7% (Supplementary File—Table S3).
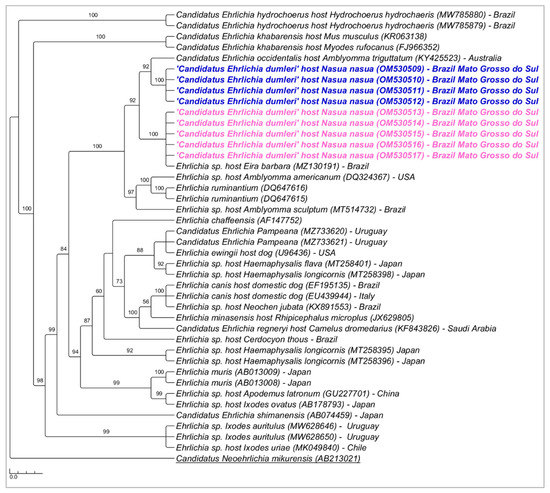
Figure 2.
Phylogenetic tree inferred by the Bayesian method and based on the 16S rRNA gene from Ehrlichia spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua) sampled in Campo Grande city, Mato Grosso do Sul state, Brazil. Sequences detected in the present study are highlighted in bold blue (PEP—genotype 1) and pink (VBA—genotype 2).
Phylogenetic analyses inferred by the Bayesian method (BI) on the groEL gene (alignment of 530 bp and HKY+G4 as an evolutionary model) (Figure 3), sodB gene (alignment of 592 bp and TPM2+G4 as an evolutionary model) (Figure 4), and 23S–5S intergenic region (alignment of 401 bp and HKY+G4 as an evolutionary model) (Figure 5) grouped the sequences detected in the present study in a new clade sharing the same origin with E. ruminantium, with posterior probabilities ranging from 78 to 98%. The genetic divergence between Ehrlichia genotypes detected in coatis from the present study and E. ruminantium ranged from 0.02 to 0.16 for groEL, sodB, and 23S-5S molecular markers (Supplementary Materials).

Figure 3.
Phylogenetic tree inferred by the Bayesian method and based on the groEL gene from Ehrlichia spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua) sampled in Campo Grande city, Mato Grosso do Sul state, Brazil. Sequences detected in the present study are highlighted in bold pink (VBA).

Figure 4.
Phylogenetic tree inferred by the Bayesian method and based on the sodB gene from Ehrlichia spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua) sampled in Campo Grande city, Mato Grosso do Sul state, Brazil. Sequences detected in the present study are highlighted in bold pink (VBA).

Figure 5.
Phylogenetic tree inferred by the Bayesian method and based on the 23S–5S intergenic region from Ehrlichia spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua) sampled in Campo Grande city, Mato Grosso do Sul state, Brazil. Sequences detected in the present study are highlighted in bold pink (VBA) and blue (PEP).
Phylogenetic analysis inferred by the Bayesian method (BI) and TN+G4 evolutionary model and based on the gltA gene (alignment of 650 bp) showed similar topology to 16S rRNA-based phylogenetic inference, positioning sequences from the present study in a new large clade (92% posterior probability) divided into two sub-clades (Figure 6). The genetic divergence between sequences from these two sub-clades ranged from 0.01 to 0.16 (Supplementary Materials).

Figure 6.
Phylogenetic tree inferred by the Bayesian method and based on the gltA gene from Ehrlichia spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua) sampled in Campo Grande city, Mato Grosso do Sul state, Brazil. Sequences detected in the present study are highlighted in bold pink (VBA) and blue (PEP).
In order to evaluate the genetic distance between the two Ehrlichia sp. sub-clades detected in the present study (PEP x VBA), median joining network (MJN) and haplotype analyses with fourteen 16S rRNA sequences that clustered together or in a clade near those sequences detected in coatis (Table 3, Figure 7) was performed. As a result, seven genotypes were found with 34 variable sites. In the MJN analysis, sequences detected in coatis from VBA and the one detected in E. barbara, Mato Grosso, Brazil (MZ130191) were represented by the same haplotype (haplotype #4). On the other hand, sequences detected in PEP clustered in two different genotypes (haplotype #1 and #2), also apart from haplotype 3, which was represented by ‘Ca. Ehrlichia occidentalis’ (KY425523). Sequences positioned in a clade near sequences detected in coatis (E. ruminantium DQ647615, DQ647616, and Ehrlichia sp. DQ324367) in the BI were represented by different genotypes (#5, #6, and #7) (Figure 7). The genetic distance (p-distance) between the two detected haplotypes detected from coatis (VBA xPEP) ranged from 0.010 to 0.012 (Supplementary Materials).

Table 3.
Polymorphisms and diversity of 16Sr RNA sequences of Ehrlichia spp. obtained from coatis (Nasua nasua) sampled in the city of Campo Grande, Mato Grosso do Sul state, Brazil.
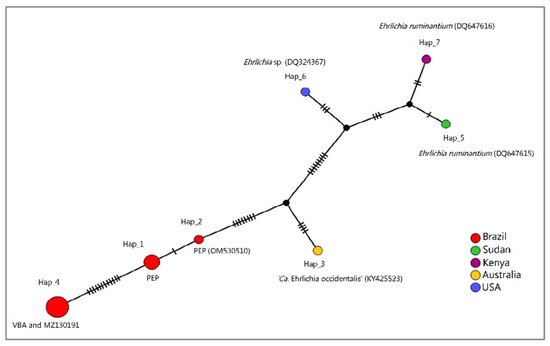
Figure 7.
Median joining network containing Ehrlichia 16S rRNA sequences representatives of the two clades where sequences detected in coatis were positioned in the Bayesian Inference. While the lines between haplotypes represent mutational steps, the black circles indicate median vectors.
3.2. PCR Assays for Anaplasma sp. Screening and Characterization
Out of 165 coati blood samples, six (3.6%) were positive in the screening nPCR for the 16S rRNA gene of Anaplasma spp. [1/63 samples—1.6% from PEP area and 5/102 samples—4.9% from VBA (obtained from five different coatis). No recaptured animal showed positivity in more than one sampling time. Co-positivity for Ehrlichia spp. and Anaplasma spp. was not found in the sampled animals. Four samples were properly sequenced, and Blast analyses showed 100% of query coverage, 0.0 E-value, and 100% of identity with Anaplasma spp. sequences previously detected in ticks and wild animals from Brazil (KY499191; KY391804; KY499201; KY499183). Only one sample amplified for the 23S–5S ITS and Blast results showed 100% of query coverage, 0.0 E-value, and 100% of identity with ‘Candidatus Anaplasma brasiliensis’. All six samples failed to amplify the groEL gene fragment.
Phylogenetic analysis inferred by the Bayesian method (BI) and TPM2+I+G evolutionary model and based on the 16S rRNA gene (alignment of 698 bp) grouped the sequences detected in the present study in a large clade (100% posterior probability) with other sequences of Anaplasma sp. detected in ticks and wild animals from Brazil (KY499191; KY391804; KY499201; KY499183) (Figure 8). Phylogenetic analysis inferred by the Bayesian method (BI) and evolutionary model K3P+G4 and based on the 23S–5S ITS region (alignment of 384 bp) grouped the sequences detected in the present study in a large clade (70% posterior probability) with ‘Candidatus Anaplasma brasiliensis’ (Figure 9).
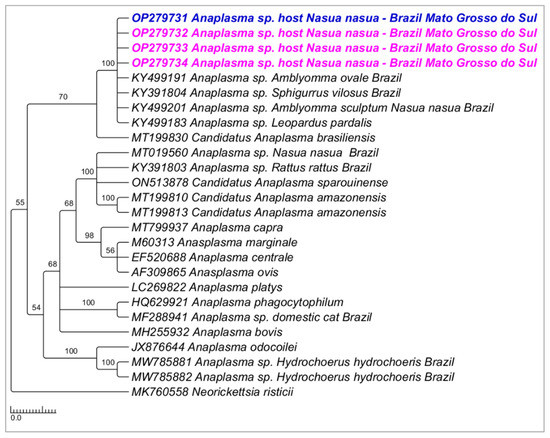
Figure 8.
Phylogenetic tree inferred by the Bayesian method and based on the 16S rRNA gene from Anaplasma spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua). Sequences detected in the present study are highlighted in bold pink (VBA) and blue (PEP).

Figure 9.
Phylogenetic tree inferred by the Bayesian method and based on the 23S–5S ITS region from Anaplasma spp. sequences obtained from blood from ring-tailed coatis (Nasua nasua). Sequences detected in the present study are highlighted in bold pink (VBA) and blue (PEP).
4. Discussion
Previous studies conducted in Brazil indicated the occurrence of putative new genotypes of Ehrlichia spp. and Anaplasma spp. in different orders of wild animals [1,33,34,35,36,37,38,39,40,41,42]. These epidemiological studies indicate an abundant diversity of Anaplasmataceae agents circulating in wild mammals, pointing out the occurrence of putative new species. Regarding this aspect, constant surveillance of Anaplasmataceae agents in biological samples from wild animals shows great importance.
Herein, we were able to detect Ehrlichia DNA in 25 (15.1%) blood samples from coatis using a dsb-based PCR screening. Such findings represent a higher percentage of positivity when compared with two previous studies performed in Brazil. In the Brazilian Pantanal, a previous study reported the occurrence of antibodies to E. canis in one coati (1/31, 3.2%) (titer of 640) as well as the molecular detection of a Ehrlichia 16S rDNA genotype phylogenetically related to multiple Ehrlichia spp. previously detected in ticks and wild rodents from Brazil [39]. Moreover, Ehrlichia/Anaplasma 16S rDNA was detected in 1/18 coati blood sampled in Iguazu National Park, southern Brazil, albeit without sequencing [43]. In the present study, we were able to sample a higher number of animals when compared with the two above-mentioned studies from Brazil (165 blood samples analyzed herein, in contrast with 31 blood samples performed by Sousa et al. [39] and 18 by Collere et al. [43]). This higher number of sampling procedures might explain the higher percentage of positive animals found in the present study. The two previous studies and the present one were performed in different biomes: while the study conducted by Sousa et al. [39] took place in Pantanal, the latter performed by Collere et al. [43] was carried out in the Atlantic forest. The present study was conducted in the Cerrado biome, an area characterized by grassland to a nearly closed canopy of medium-height trees overlying grass with a tropical savanna climate [4]. Although these three studies were performed with the same animal species, it is known that tick diversity changes according to the studied biome [3,8,39,44,45]. Animals from the present study were found parasitized by non-identified Amblyomma sp. larvae, Amblyomma dubitatum, and Amblyomma sculptum nymphs, and A. sculptum and Amblyomma ovale adults [8]. Since the vector of the detected Ehrlichia sp. is not known yet, the studied biome and, consequently, tick diversity, together with the higher number of sampled animals, might have influenced the higher percentage of positive animals in the Cerrado biome. It is also worth mentioning that the applied technique might have influenced the percentage of positive animals. For instance, nested PCR is highly sensitive, and in the case of E. canis detection, it is capable of detecting 0.2 pg of purified DNA, while a conventional PCR can only detect values above 20 pg [46]. Quantitative PCR (qPCR) can also be used to increase the sensibility of the diagnostic tests and screening, and in some cases, showed a higher sensitivity compared to previously described gel-based PCR, RNA, PCR-ELISA, and hybridization assays [47]. However, the ability of previously standardized qPCR protocols to catch novel Anaplasmataceae agents should be further investigated. In a previous study, a multiplex qPCR based on the groEL gene and validated for the main Ehrlichia and Anaplasma species of medical and veterinary importance [37] was unable to catch putative novel Ehrlichia and Anaplasma genotypes described in mammals from the Superorder Xenarthra in Brazil [41].
We observed a different prevalence of positive animals according to the sampling spot [4/63 samples—6.3% from the PEP area (from four different coatis) and 21/102 samples—20.6% from VBA (obtained from 16 different coatis). Although sampling areas are in Campo Grande city, Cerrado biome, they present very different characteristics. PEP is a preserved forest area, being an important refuge for wildlife, where many species of mammals can be found, such as ring-tailed coatis, anteaters, capybaras, opossums, bats, and also many species of birds and reptiles [4,48,49,50,51]. Contrary, VBA is a more anthropized area (residential area), surrounded by three forest fragments and one area used for military training. There is a residential complex inhabited by many families and domestic animals. The houses are not fenced, and they all have a trash can at the front. Coatis have access to the outside of the houses, and they were observed during field activities walking around the houses and also searching for food in the dumpsters [8]. This difference in the prevalence between the two areas may be explained by a phenomenon called ‘dilution effect’ [52,53,54]. Preserved areas (e.g., PEP) are characterized by high species richness of different hosts for ectoparasites and pathogens or are likely to contain a high proportion of hosts that are inefficient in transmitting the disease agent to a feeding vector. With a higher abundance of weakly competent reservoir species, the stronger the dilution effect is, which may decrease the probability of disease transmission [54].
Interestingly, the present study shows, for the first time, transient positivity for Ehrlichia sp. in coati blood samples. Three animals sampled at VBA were PCR-positive at different times of sampling. All three animals were positive in March 2019 and two animals were in April 2019. Due to the small number of recaptured/positive animals, and since the captures were performed by chance on different days, it is not possible to infer any type of correlation according to the sampling period and PCR positivity. Besides that, the results found herein are influenced by the limit of the detection of the PCR protocols used. Nevertheless, these data indicate that N. nasua may play a role in the maintenance of this new Anaplasmataceae agent in the wild environment.
Phylogenetic analyses positioned Ehrlichia sp. detected from coatis in the present study in a new clade, originated as a sister clade to E. ruminantium. It is noteworthy that all phylogenetic inferences based on different molecular markers showed the same topology, with the establishment of a new distinct clade. Interestingly, two branches were observed on the phylogenetic inferences based on 16S rRNA and gltA genes and 23S-5S intergenic region, according to the sampling spot (PEP and VBA). These findings may indicate the occurrence of distinct genotypes of a unique putative novel Anaplasmataceae agent, which is proposed here as ‘Candidatus Ehrlichia dumleri’, in honor of Dr. John Stephen Dumler and his great contribution to the knowledge of this group of proteobacteria. Indeed, while the genetic divergence between these two genotypes ranged from 0.010 to 0.013, values ranging from 1.2 to 1.3% were found when comparing the newly described genotypes and ‘Candidatus Ehrlichia occidentalis’. Moreover, the haplotype analyses based on the 16S rRNA sequences corroborate the presence of two distinct genotypes, also according to the sampling spot.
Recently, several ‘Candidatus Ehrlichia’ have been proposed and detected in biological samples from wild animals by using different molecular target genes. For instance, ‘Candidatus Ehrlichia regneryi’ was detected in spleen samples of dromedary camels (Camelus dromedarius) in Saudi Arabia using two genetic markers (16SrRNA and groEL) [55]. ‘Candidatus Ehrlichia pampeana’ was detected in Haemaphysalis juxtakochi and in gray Brocket Deer (Mazama gouazoubira) from Uruguay using three loci (16S rRNA, dsb, and groEL) [56]. ‘Candidatus Ehrlichia hydrochoerus’ was detected in blood samples from capybaras (Hydrochoerus hydrochaeris) from Brazil, using four loci (16S rRNA, dsb, groEL and sodB) [57]. In the present study, we were able to perform the molecular characterization using six different molecular targets, including a large fragment of 16S rRNA, dsb, sodB, groEL, gltA genes and the 23S-5S intergenic space region (ITS). Phylogenetic analyses based on all six molecular markers showed the same topology, supporting the description of the new ‘Candidatus Ehrlichia dumleri’ presented herein.
The present work describes a low percentage of positive animals for Anaplasma spp. (6/165—3.6%) by using a screening nPCR based on the 16S rRNA gene. Sousa et al. [39] detected Anaplasma spp. in 22% (7/31) coatis sampled in the Brazilian Pantanal, using an nPCR also based on the 16S rRNA gene. Phylogenetic inferences grouped the sequences detected in coatis from Pantanal in two different clades: one clade related to A. phagocytophilum and another one clustering with Anaplasma sp. previously detected in different wild animal species, including rodents, marsupials, and carnivores [39]. The present work showed the detection of an Anaplasma sp. closely related to ‘Candidatus Anaplasma brasiliensis’, which was previously detected in anteaters in the state of São Paulo [41], and Anaplasma sp. detected in other wild animals from Brazil. Recently, an unusual case of human anaplasmosis caused by ‘Candidatus Anaplasma sparouinense’, which was genetically related to ‘Candidatus Anaplasma amazonensis’ was recently described in sloths from the Brazilian Amazon [41], was reported in the Amazon of French Guiana [2]. Future studies should be performed in order to check if wild animals may represent a source of infection by these novel neotropical Anaplasma to humans.
5. Conclusions
Anaplasmataceae agents were detected in blood samples from free-ranging coatis sampled in Midwestern Brazil. A higher positivity for Ehrlichia spp. was found in the sampled coatis when compared to that one found for Anaplasma spp. Phylogenetic analyses based on six molecular markers (16S rRNA, dsb, sodB, groEL, gltA, and the 23S-5S ITS) positioned the novel Ehrlichia sp. detected in the sampled coatis into a new clade and supported the description of ‘Candidatus Ehrlichia dumleri’, a putative novel Anaplasmataceae species that appeared as a sister clade to E. ruminantium. Phylogenetic analyses based on both 16S rRNA gene and 23S–5S intergenic region grouped the Anaplasma sp. detected in coatis with the previously described ‘Candidatus Anaplasma brasiliensis’.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10122379/s1, Table S1: Blast results from positive animals for Ehrlichia sp., with GenBank accession number from the present study, localization, gene, fragment size, organism, % of query coverage, E-value, % of identity and GenBank accession number of the pathogen; Table S2: Pairwise genetic distances matrix among Ehrlichia sp. that clustered close to sequences detected at the present study at based on dsb gene. Pairwise genetic distances were obtained using the p-distance method in MEGA X. Sequences detected at the present study are highlighted in bold pink (Air Force Private Area); Table S3: Pairwise genetic distances matrix among Ehrlichia sp. that clustered close to sequences detected at the present study at based on large fragment of 16SrRNA gene. Pairwise genetic distances were obtained using the p-distance method in MEGA X. Sequences detected at the present study are highlighted in bold blue (Parque Estadual do Prosa) and pink (Air Force Private Area); Table S4: Pairwise genetic distances matrix among Ehrlichia sp. sequence that clustered close to sequence detected at the present study at based on groEL gene. Pairwise genetic distances were obtained using the p-distance method in MEGA X. Sequence detected at the present study is highlighted in bold pink (Air Force Private Area); Table S5: Pairwise genetic distances matrix among Ehrlichia sp. sequences that clustered close to sequences detected at the present study at based on sodB gene. Pairwise genetic distances were obtained using the p-distance method in MEGA X. Sequences detected at the present study are highlighted in bold pink (Air Force Private Area); Table S6: Pairwise genetic distances matrix among Ehrlichia sp. sequences that clustered close to sequences detected at the present study at based on 23S-5S intergenic region. Pairwise genetic distances were obtained using the p-distance method in MEGA X. Sequences detected at the present study are highlighted in bold pink (Air Force Private Area); Table S7: Pairwise genetic distances matrix among Ehrlichia sp. sequences that clustered close to sequences detected at the present study at based on gltA gene. Pairwise genetic distances were obtained using the p-distance method in MEGA X. Sequences detected at the present study are highlighted in bold blue (Parque Estadual do Prosa) and pink (Air Force Private Area).
Author Contributions
Data curation: M.R.A., R.Z.M., H.M.H., L.P., A.C.C., W.T.G.B. and G.C.d.M.; funding acquisition: M.R.A., H.M.H. and R.Z.M.; investigation: M.R.A., R.Z.M., H.M.H., L.P., A.C.C., W.T.G.B. and G.C.d.M.; methodology: M.R.A., R.Z.M., H.M.H., L.P., A.C.C., W.T.G.B. and G.C.d.M.; project administration: M.R.A., H.M.H. and R.Z.M.; supervision: M.R.A., H.M.H. and R.Z.M.; writing—original draft: M.R.A. and L.P.; writing—review and editing: M.R.A., R.Z.M., H.M.H., L.P., A.C.C., W.T.G.B. and G.C.d.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by FAPESP (Foundation for Research Support of the State of São Paulo—Process #2018/02753-0; #2020/12037-0) and CNPq (National Council for Scientific and Technological Development; Productivity Grant to MRA [CNPq Process # 303701/2021-8)]. LP received a scholarship from FAPESP (2019/15150-4).
Data Availability Statement
All obtained sequences were submitted to GenBank with following accession numbers: Ehrlichia sp. dsb (OP819944–OP819946), Ehrlichia sp. 16S rRNA (OM530509-OM530517), Ehrlichia sp. groEL (OP819939), Ehrlichia sp. sodB (OP903236-OP903238), Ehrlichia sp. 23S-5S (OM717254-OM717255), Ehrlichia sp. gltA (OP819935-OP919938, OP819940-OP819941), Anaplasma sp. 16S rRNA (OP279731-OP279734), and Anaplasma sp. 23S-5S (OP918134) and could be accessed through https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 4 October 2022).
Acknowledgments
The authors are especially thankful to ‘Programa de Pós-Graduação em Ciências Veterinárias’ from FCAV-Unesp and InsanaHuna Research Group (www.insanahuna.com (accessed on 4 October 2022)) for the fieldwork support and to the reviewers whose suggestions significantly improved the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- André, M.R. Diversity of Anaplasma and Ehrlichia/Neoehrlichia agents in terrestrial wild carnivorous worldwide: Implications for human and domestic animal health and wildlife conservation. Frontiers 2018, 5, 293. [Google Scholar]
- Duron, O.; Koual, R.; Musset, L.; Buysse, M.; Lambert, Y.; Jaulhac, B.; Blanchet, D.; Alsibai, K.D.; Lazrek, Y.; Epelboin, L.; et al. Novel Chronic Anaplasmosis in Splenectomized Patient, Amazon Rainforest. Emerg. Infect. Dis. 2022, 28, 1673–1676. [Google Scholar] [CrossRef] [PubMed]
- Estevam, L.G.T.M.; Junior, A.A.F.; Silvestre, B.T.; Hemetrio, N.S.; Almeida, L.R.; Oliveira, M.M.; Silva, S.M.; Ribeiro, M.F.B.; Silveira, J.A.G. Seven years of evaluation of ectoparasites and vector-borne pathogens among ring-tailed coatis in an urban park in southeastern Brazil. Vet. Parasitol. Reg. Stud. Rep. 2020, 21, 100442. [Google Scholar] [CrossRef]
- Barreto, W.T.G.; Miraglia, H.H.; Macedo, G.C.; Rucco, A.C.; Assis, W.O.; Oliveira-Santos, L.G.; Porfírio, G.E.O. Density and survivorship of the South American coati (Nasua nasua) in urban areas in Central–Western Brazil. HYSTRIX 2021, 32, 82–88. [Google Scholar] [CrossRef]
- Levin, M.L.; Nicholson, W.L.; Massung, R.F.; Sumner, J.W.; Fish, D. Comparison of the reservoir competence of medium-sized mammals and Peromyscus leucopus for Anaplasma phagocytophilum in Connecticut. Vector Borne Zoonotic Dis. 2002, 2, 125–136. [Google Scholar] [CrossRef]
- Yabsley, M.J.; Murphy, S.M.; Luttrell, M.P.; Little, S.E.; Massung, R.F.; Stallknecht, D.E.; Conti, L.A.; Blackmore, C.G.M.; Durden, L.A. Experimental and field studies on the suitability of raccoons (Procyon lotor) as hosts for tick-borne pathogens. Vector Borne Zoonotic Dis. 2008, 8, 491–503. [Google Scholar] [CrossRef]
- Woldehiwet, Z. The natural history of Anaplasma phagocytophilum. Vet. Parasitol. 2010, 8, 491–503. [Google Scholar] [CrossRef]
- Perles, L.; Martins, T.F.; Barreto, W.T.G.; Carvalho de Macedo, G.; Herrera, H.M.; Mathias, L.A.; Labruna, M.B.; Barros-Battesti, D.M.; Machado, R.Z.; André, M.R. Diversity and Seasonal Dynamics of Ticks on Ring-Tailed Coatis Nasua nasua (Carnivora: Procyonidae) in Two Urban Areas from Midwestern Brazil. Animals 2022, 12, 293. [Google Scholar] [CrossRef]
- Porfirio, G.E.O.; Santos, F.M.; Macedo, G.C.; Barreto, W.T.G.; Campos, J.B.V.; Meyers, A.C.; André, M.R.; Perles, L.; Oliveira, C.E.; Xavier, S.C.C.; et al. Maintenance of Trypanosoma cruzi, T. evansi and Leishmania spp. by domestic dogs and wild mammals in a rural settlement in Brazil-Bolivian border. Int. J. Parasitol. Parasites Wildl. 2018, 7, 398–404. [Google Scholar] [CrossRef]
- Santos, F.M.; Macedo, G.C.; Barreto, W.T.G.; Oliveira-Santos, L.G.R.; Garcia, C.M.; Mourão, G.M.; Porfírio, G.E.O.; Marino, E.D.; André, M.R.; Perles, L.; et al. Outcomes of Trypanosoma cruzi and Trypanosoma evansi infections on health of Southern coati (Nasua nasua), crab-eating fox (Cerdocyon thous), and ocelot (Leopardus pardalis) in the Brazilian Pantanal. PLoS ONE 2018, 13, e0201357. [Google Scholar] [CrossRef]
- Perles, L.; Macedo, G.G.; Barreto, W.T.G.; Francisco, G.V.; Herrera, H.M.; Barros-Battesti, D.M.; Machado, R.Z.; André, M.R. Longitudinal dynamics and health impact of Hepatozoon procyonis (Apicomplexa: Hepatozoidae) on naturally infected ring-tailed coatis Nasua nasua (Carnivora: Procyonidae) from Midwestern Brazil. Ticks Tick-Borne Dis. 2022, 13, 101982. [Google Scholar] [CrossRef] [PubMed]
- Olifiers, N.; Bianchi, R.C.; D’Andrea, P.S.; Mourão, G.; Gompper, M.E. Estimating age of carnivores from the Pantanal region of Brazil. Wildl. Biol. 2010, 16, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Birkenheuer, A.J.; Levy, M.G.; Breitschwerdt, E.B. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B. canis DNA in canine blood samples. J. Clin. Microbiol. 2003, 41, 4172–4177. [Google Scholar] [CrossRef] [PubMed]
- Doyle, C.; Labruna, M.B.; Breitschwerdt, E.B.; Tang, Y.; Corstvet, R.E.; Hegarty, B.C.; Bloch, K.C.; Li, P.; Walker, D.H.; McBride, J.W. Detection of medically important Ehrlichia by quantitative multicolor TaqMan real-time polymerase chain reaction of the dsb gene. J. Mol. Diagn. 2005, 7, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Greiman, S.E.; Tkach, V.V.; Pulis, E.; Fayton, T.J.; Curran, S.S. Large Scale Screening of Digeneans for Neorickettsia Endosymbionts Using Real-Time PCR Reveals New Neorickettsia Genotypes, Host Associations and Geographic Records. PLoS ONE 2014, 9, e98453. [Google Scholar] [CrossRef]
- Müller, A.; Monti, G.; Otth, C.; Sepúlveda, P.; Bittencourt, P.; Nachum-Biala, Y.; Gutierrez, R.; Harrus, S. “Candidatus Neoehrlichia chilensis” sp. nov.: Molecular detection and characterization of a novel Anaplasmataceae in wild rodents from Valdivia, southern Chile. Transb. Emerg. Dis. 2018, 65, 357–362. [Google Scholar] [CrossRef]
- Victoria, L.O.; Montilla, H.J.; Qurollo, B.A.; Maggi, R.G.; Hegarty, B.C.; Tornquist, S.J.; Breitschwerdt, E.B. Potentially novel Ehrlichia species in horses, Nicaragua. Emerg. Infect. Dis. 2015, 21, 335–338. [Google Scholar] [CrossRef]
- Li, H.; Zheng, Y.C.; Ma, L.; Jia, N.; Jiang, B.G.; Jiang, R.R. Human infection with a novel tick-borne Anaplasma species in China: A surveillance study. Lancet Infect. Dis. 2015, 15, 663–670. [Google Scholar] [CrossRef]
- Rejmanek, D.; Bradburd, G.; Foley, J. Molecular characterization reveals distinct genospecies of Anaplasma phagocytophilum from diverse North American hosts. J. Med. Microbiol. 2012, 61, 204–212. [Google Scholar] [CrossRef]
- Massung, R.F.; Slater, K.; Owens, J.H.; Nicholson, W.L.; Mather, T.N.; Solberg, V.B.; Olson, J.G. Nested PCR assay for detection of granulocytic ehrlichiae. J. Clin. Microbiol. 1998, 36, 1090–1095. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Y.; Zhang, F.; Zhang, W.; Wang, J.; Cui, Y.; Wang, R.; Jian, F.; Zhang, L.; Ning, C. Molecular and phylogenetic analysis of Anaplasma spp. in sheep and goats from six provinces of China. J. Vet. Sci. 2016, 17, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1997, 74, 5463–5467. [Google Scholar] [CrossRef]
- Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. Base-calling of automated sequencer traces usingPhred. I. Accuracy assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive, sequence choice and visualization. Briefings in Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Buckley, T.R.; Thorne, J. Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Trifinopoulus, J.; Nguyen, L.T.; Haeseler, V.A.; Minh, B.Q. W-IQ-TREE: A fast-online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 12, 1572–1574. [Google Scholar] [CrossRef]
- Stover, B.C.; Muller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analyses across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- André, M.R.; Adania, C.H.; Machado, R.Z.; Allegretti, S.M.; Felippe, P.A.N.; Silva, K.F.; Kagaghi, A.C. Molecular and serologic detection of Ehrlichia spp. in endangered Brazilian wild capitve felids. J. Wildl. Dis. 2010, 3, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- André, M.R.; Dumler, J.S.; Scorpio, D.G.; Teixeira, R.H.F.; Allegretti, S.M.; Machado, R.Z. Molecular detection of tick-borne bacterial agents in Brazilian and exotic captive carnivores. Ticks Tick-Borne Dis. 2012, 3, 247–253. [Google Scholar] [CrossRef]
- André, M.R.; Calchi, A.C.; Perles, L.; Gonçalves, L.R.; Uccella, L.; Lemes, J.R.B.; Nantes, W.A.G.; Santos, F.M.; Porfírio, G.E.O.; Barros-Battesti, D.M.; et al. Novel Ehrlichia and Hepatozoon genotypes in white-eared opossums (Didelphis albiventris) and associated ticks from Brazil. Ticks Tick-Borne Dis. 2022, 13, 102022. [Google Scholar] [CrossRef]
- Sacchi, A.B.V.; Duarte, J.M.B.; André, M.R.; Machado, R.Z. Prevalence and molecular characterization of Anaplasmataceae agents in free-ranging Brazilian marsh deer (Blastocerus dichotomus). Comp. Immunol. Microbiol. Infect. Dis. 2012, 4, 325–334. [Google Scholar] [CrossRef]
- Benevenute, J.L.; Dumler, J.S.; Ogrzewalska, M.; Roque, A.L.R.; Mello, V.V.; Sousa, K.C.M.; Gonçalves, L.R.; D’Andrea, P.S.; Sampaio Lemos, E.R.; Machado, R.Z.; et al. Assessment of a quantitative 5’ nuclease real-time polymerase chain reaction using groEl gene for Ehrlichia and Anaplasma species in rodents in Brazil. Ticks Tick-Born Dis. 2017, 8, 646–656. [Google Scholar] [CrossRef]
- Soares, H.S.; Marcili, A.; Barbieri, A.R.M.; Minervino, A.H.H.; Malheiros, A.F.; Gennari, S.M.; Labruna, M.B. Novel Anaplasma and Ehrlichia organisms infecting the wildlife of two regions of the Brazilian Amazon. Acta Trop. 2017, 174, 82–87. [Google Scholar] [CrossRef]
- Sousa, K.C.M.; Calchi, A.C.; Herrera, H.M.; Dumler, M.; Barros-Battesti, D.M.; Machado, R.Z.; André, M.R. Anaplasmataceae agents among wild mammals and ectoparasites in Brazil. Epidemiol. Infect. 2017, 16, 3424–3437. [Google Scholar] [CrossRef]
- Guimarães, A.; Raimundo, J.M.; Silva, A.T.D.; Carpintero, F.M.; Pires, J.R.; Benevenute, J.L.; Machado, R.Z.; André, M.R.; Baldani, C.D. Detection of a puntative novel of Ehrlichia sp. from opossums (Didelphis aurita) from Brazil. Rev. Bras. Parasitol. Vet. 2019, 28, 140–144. [Google Scholar] [CrossRef]
- Calchi, A.C.; Vultão, J.G.; Alves, M.H.; Yogui, D.R.; Desbiez, A.L.J.; Santi, M.D.; de Souza Santana, M.; da Silva, T.M.V.; Werther, K.; Teixeira, M.M.G.; et al. Ehrlichia spp. and Anaplasma spp. in Xenarthra mammals from Brazil, with evidence of novel ‘Candidatus Anaplasma spp.’. Sci. Rep. 2020, 10, 12615. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, P.; Torres, J.M.; Placa, A.J.V.; Mello, V.V.C.D.; Lourenço, E.C.; Herrera, H.M.; Oliveira, C.E.; Hemsley, C.; Titball, R.W.; Machado, R.Z.; et al. Molecular Survey of Anaplasmataceae Agents and Coxiellaceae in Non-Hematophagous Bats and Associated Ectoparasites from Brazil. Parasitologia 2021, 1, 197–209. [Google Scholar] [CrossRef]
- Collere, F.C.M.; Delai, R.M.; Ferrari, L.D.R.; da Silva, L.H.; Fogaça, P.L.; Rodrigues, A.N.; Gonçalves, D.D.; Baggio, R.A.; Moraes, M.F.D.; Hoppe, E.G.L.; et al. ‘Candidatus Mycoplasma haematonasua’ and tick-borne pathogens in ring-tailed coatis (Nasua nasua, Linnaeus, 1976) from the Iguaçu National Park, Paraná State, southern Brazil. Transboud. Emerg. Dis. 2021, 68, 3222–3229. [Google Scholar] [CrossRef] [PubMed]
- Barros-Battesti, D.M.; Baggio, D. Ectoparasites Ixodida Leach, 1817 on wild mammals in the State of Paraná, Brazil. Mem. Inst. Oswaldo Cruz 1992, 87, 291–296. [Google Scholar] [CrossRef][Green Version]
- Labruna, M.B.; Jorge, R.S.P.; Sana, D.A.; Ja’como, A.T.A.; Kashivakura, C.K.; Furtado, M.M.; Ferro, C.; Perez, S.A.; Silveira, L.; Santos, T.S.; et al. Ticks (Acari: Ixodida) on wild carnivores in Brazil. Exp. Appl. Acarol. 2005, 36, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Rikihisa, Y.; Mott, J.M.; Greene, R.; Kim, H.; Zhi, N.; Couto, G.C.; Unver, A.; Bartsch, R. Comparison of nested PCR with immunofluorescentantibody assay for detection of Ehrlichia canis infection in dogs treated with doxycicline. J. Clin. Microbiol. 1997, 35, 1852–1855. [Google Scholar] [CrossRef]
- Carelli, G.; Decaro, N.; Lorusso, A.; Elia, G.; Lorusso, E.; Mari, V.; Buonavoglia, C. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet. Microbiol. 2007, 124, 107–114. [Google Scholar] [CrossRef]
- Costa, E.M.J.; Mauro, R.A. Secondary dispersion in coatis’ feces Nasua nasua e (Linnaeus, 1766) (Mammalia: Procyonidae) in a fragment of Cerrado, Mato Grosso do Sul, Brazil. Neotrop. Biol. Conserv. 2008, 3, 66–72. [Google Scholar]
- Gonçalves, L.R.; Paludo, G.; Bisol, T.B.; Perles, L.; de Oliveira, L.B.; de Oliveira, C.M.; da Silva, T.M.V.; Nantes, W.A.G.; Duarte, M.A.; Santos, F.M.; et al. Molecular detection of piroplasmids in synanthropic rodents, marsupials, and associated ticks from Brazil, with phylogenetic inference of a putative novel Babesia sp. from white-eared opossum (Didelphis albiventris). Parasitol. Res. 2021, 120, 3537–3546. [Google Scholar] [CrossRef]
- Nantes, W.A.G.; Santos, F.M.; de Macedo, G.C.; Barreto, W.T.G.; Gonçalves, L.R.; Rodrigues, M.S.; Chulli, J.V.M.; Rucco, A.C.; Assis, W.O.; Porfírio, G.E.O.; et al. Trypanosomatid species in Didelphis albiventris from urban forest fragments. Parasitol Res. 2021, 120, 223–231. [Google Scholar] [CrossRef]
- Nantes, W.A.G.; Barreto, W.T.G.; Santos, F.M.; Macedo, G.C.; Rucco, A.C.; Assis, W.O.; Porfírio, G.E.O.; Andrade, G.B.; Jansen, A.M.; Herrera, H.M. The influence of parasitism by Trypanosoma cruzi in the hematological parameters of the white ear opossum (Didelphis albiventris) from Campo Grande, Mato Grosso do Sul, Brazil. Int. J. Parasitol. Parasites Wildl. 2019, 9, 16–20. [Google Scholar] [CrossRef]
- Matuschka, F.R.; Fischer, P.; Heiler, M.; Richter, D.; Spielman, A. Hosts on which nymphal Ixodes ricinus most abundantly feed. Am. J. Trop. Med. Hyg. 1991, 44, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Matuschka, F.R.; Fischer, P.; Heiler, M.; Richter, D.; Spielman, A. Capacity of European animals as reservoir hosts for the Lyme disease spirochete. J. Infect. Dis. 1992, 156, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Ostfeld, K.A.; Keesing, R.S. Biodiversity and the dilution effect in disease ecology. ESA 2000, 82, 609–619. [Google Scholar]
- Bastos, A.D.S.; Mohammed, O.B.; Bennett, N.C.; Petevinos, C.; Alagaili, A.N. Molecular detection of novel Anaplasmataceae closely related to Anaplasma platys and Ehrlichia canis in the dromedary camel (Camelus dromedarius). Vet. Microbiol. 2015, 4, 310–314. [Google Scholar] [CrossRef]
- Feliz, M.L.; Munoz-Leal, S.; Carvalho, L.A.; Queirolo, D.; Remesar, S.; Armua-Fernandez, M.T.; Venzal, J.M. Characterization of ‘Candidatus Ehrlichia pampenana’ in Haemaphysalis juxtakochi ticks and gray brocket deer (Mazama gouazoubira) from Uruguay. Microorganisms 2021, 9, 2165. [Google Scholar] [CrossRef]
- Vieira, T.S.W.J.; Collere, F.C.M.; Ferrari, L.D.R.; Baggio, R.A.; Lange, R.R.; Ferrari, M.V.; Duque, J.C.M.; Sanches, G.S.; Pereira, N.A.; Aguiar, D.M.; et al. Novel Anaplasmataceae agents Candidatus Ehrlichia hydrochoerus and Anaplasma spp. Infecting Capybaras, Brazil. Emerg. Infect. Dis. 2022, 28, 480–482. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).