Learning from the Invaders: What Viruses Teach Us about RNA-Based Regulation in Microbes
Abstract
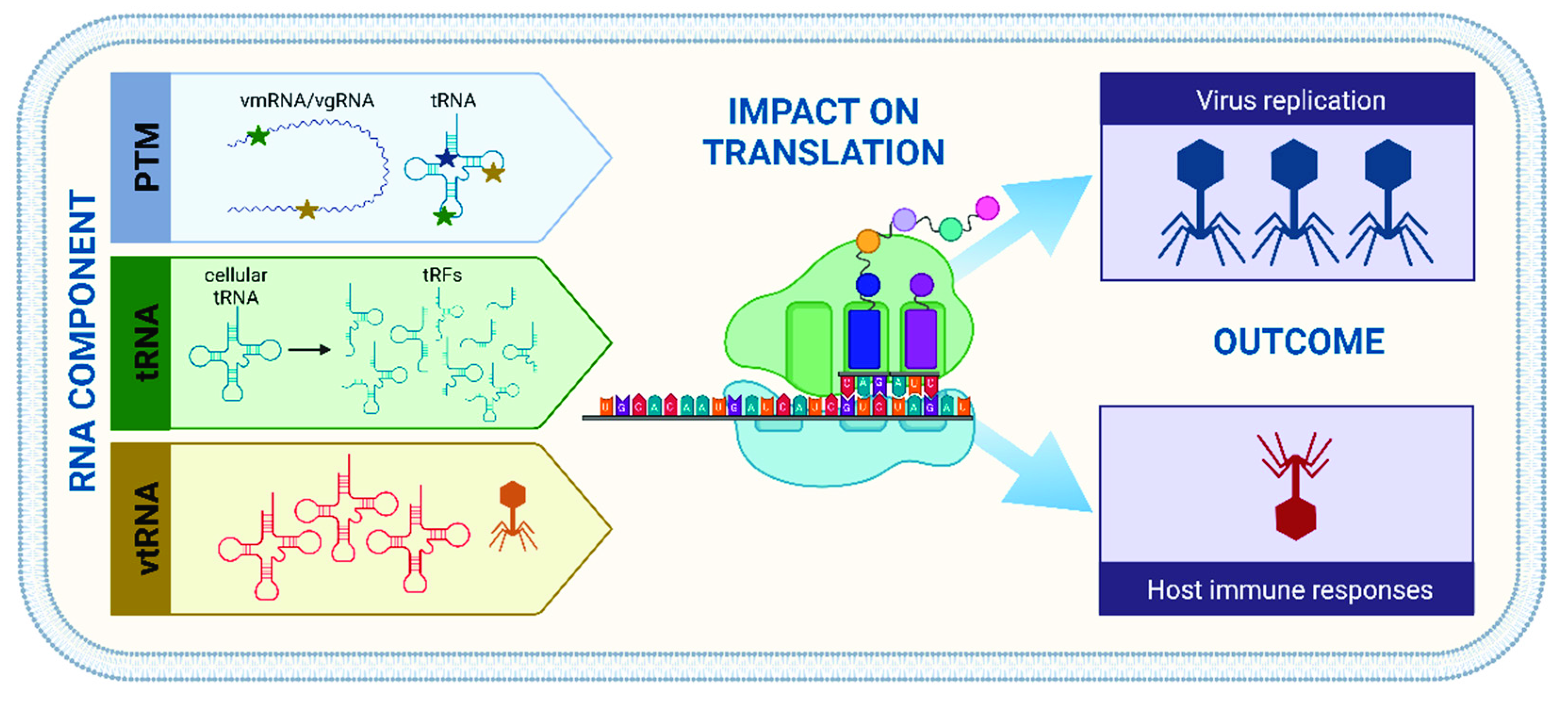
1. Introduction
2. Tools of the Trade—RNA in the Viroscope
2.1. How Modifications Modulate Cellular Functions
2.2. Translation Potluck—Bring Your Own tRNAs
3. Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Rowlands, D.J. A Brief History of Virology. In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 1, pp. 3–13. [Google Scholar]
- Li, N.; Rana, T.M. Regulation of antiviral innate immunity by chemical modification of viral RNA. Wiley Interdiscipl. Rev. RNA 2022, e1720. [Google Scholar] [CrossRef]
- Jin, D.; Musier-Forsyth, K. Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. J. Biol. Chem. 2019, 294, 5352–5364. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Xie, Y.; Zhang, S.; Song, X.; Xiao, B.; Yan, Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 2021, 11, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W. Viral tRNAs and tRNA-like structures. Wiley Interdiscip. Rev. RNA 2010, 1, 402–414. [Google Scholar] [CrossRef]
- Nunes, A.; Ribeiro, D.R.; Marques, M.; Santos, M.A.S.; Ribeiro, D.; Soares, A.R. Emerging Roles of tRNAs in RNA Virus Infections. Trends. Biochem. Sci. 2020, 45, 794–805. [Google Scholar] [CrossRef]
- Wu, S.; Li, X.; Wang, G. tRNA-like structures and their functions. FEBS J. 2022, 289, 5089–5099. [Google Scholar] [CrossRef]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef]
- Koh, C.S.; Sarin, L.P. Transfer RNA modification and infection—Implications for pathogenicity and host responses. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 419–432. [Google Scholar] [CrossRef]
- Zinshteyn, B.; Gilbert, W.V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013, 9, e1003675. [Google Scholar] [CrossRef]
- Alings, F.; Sarin, L.P.; Fufezan, C.; Drexler, H.C.; Leidel, S.A. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 2015, 21, 202–212. [Google Scholar] [CrossRef]
- Damon, J.R.; Pincus, D.; Ploegh, H.L. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol. Biol. Cell 2015, 26, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Shigi, N.; Sakaguchi, Y.; Suzuki, T.; Watanabe, K. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 2006, 281, 14296–14306. [Google Scholar] [CrossRef] [PubMed]
- Chavarria, N.E.; Hwang, S.; Cao, S.; Fu, X.; Holman, M.; Elbanna, D.; Rodriguez, S.; Arrington, D.; Englert, M.; Uthandi, S.; et al. Archaeal Tuc1/Ncs6 homolog required for wobble uridine tRNA thiolation is associated with ubiquitin-proteasome, translation, and RNA processing system homologs. PLoS ONE 2014, 9, e99104. [Google Scholar] [CrossRef]
- Shippy, D.C.; Eakley, N.M.; Bochsler, P.N.; Chopra, A.K.; Fadl, A.A. Biological and virulence characteristics of Salmonella enterica serovar Typhimurium following deletion of glucose-inhibited division (gidA) gene. Microb. Pathog. 2011, 50, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Sinha, H.; David, L.; Pascon, R.C.; Clauder-Munster, S.; Krishnakumar, S.; Nguyen, M.; Shi, G.; Dean, J.; Davis, R.W.; Oefner, P.J.; et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 2008, 180, 1661–1670. [Google Scholar] [CrossRef]
- Cho, K.H.; Caparon, M.G. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: A new strategy to make live attenuated strains. Infect. Immun. 2008, 76, 3176–3186. [Google Scholar] [CrossRef]
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016, 7, 13302. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Jenner, R.G.; Young, R.A. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 2005, 3, 281–294. [Google Scholar] [CrossRef]
- Lemaitre, B.; Girardin, S.E. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nat. Rev. Microbiol. 2013, 11, 365–369. [Google Scholar] [CrossRef]
- Netzband, R.; Pager, C.T. Epitranscriptomic marks: Emerging modulators of RNA virus gene expression. Wiley Interdiscipl. Rev. RNA 2020, 11, e1576. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, N.S.; Horner, S.M. RNA modifications go viral. PLoS Pathog. 2017, 13, e1006188. [Google Scholar] [CrossRef]
- Hao, H.; Liu, W.; Miao, Y.; Ma, L.; Yu, B.; Liu, L.; Yang, C.; Zhang, K.; Chen, Z.; Yang, J.; et al. N4-acetylcytidine regulates the replication and pathogenicity of enterovirus 71. Nucleic Acids Res. 2022, 50, 9339–9354. [Google Scholar] [CrossRef]
- Korniy, N.; Samatova, E.; Anokhina, M.M.; Peske, F.; Rodnina, M.V. Mechanisms and biomedical implications of -1 programmed ribosome frameshifting on viral and bacterial mRNAs. FEBS Lett. 2019, 593, 1468–1482. [Google Scholar] [CrossRef] [PubMed]
- Jungfleisch, J.; Bottcher, R.; Tallo-Parra, M.; Perez-Vilaro, G.; Merits, A.; Novoa, E.M.; Diez, J. CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nat. Commun. 2022, 13, 4725. [Google Scholar] [CrossRef] [PubMed]
- Maynard, N.D.; Macklin, D.N.; Kirkegaard, K.; Covert, M.W. Competing pathways control host resistance to virus via tRNA modification and programmed ribosomal frameshifting. Mol. Syst. Biol. 2012, 8, 567. [Google Scholar] [CrossRef] [PubMed]
- Maynard, N.D.; Birch, E.W.; Sanghvi, J.C.; Chen, L.; Gutschow, M.V.; Covert, M.W. A forward-genetic screen and dynamic analysis of lambda phage host-dependencies reveals an extensive interaction network and a new anti-viral strategy. PLoS Genet. 2010, 6, e1001017. [Google Scholar] [CrossRef]
- Goldfarb, A.; Daniel, V. Transcriptional control of two gene subclusters in the tRNA operon of bacteriophage T4. Nature 1980, 286, 418–420. [Google Scholar] [CrossRef]
- Daniel, V.; Sarid, S.; Littauer, U.Z. Amino acid acceptor activity of bacteriophage T4 transfer RNA. FEBS Lett. 1968, 2, 39–41. [Google Scholar] [CrossRef]
- McClain, W.H.; Guthrie, C.; Barrell, B.G. Eight transfer RNAs induced by infection of Escherichia coli with bacteriophage T4. Proc. Natl. Acad. Sci. USA 1972, 69, 3703–3707. [Google Scholar] [CrossRef]
- Morgado, S.; Vicente, A.C. Global In-Silico Scenario of tRNA Genes and Their Organization in Virus Genomes. Viruses 2019, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Fang, W.; Miranda-Sanchez, F.; Brown, J.M.; Kauffman, K.M.; Acevero, C.M.; Bartel, D.P.; Polz, M.F.; Kelly, L. Degradation of host translational machinery drives tRNA acquisition in viruses. Cell Syst. 2021, 12, 771–779.e775. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, G.; Askora, A.; Blanc-Mathieu, R.; Kawasaki, T.; Li, Y.; Nakano, M.; Ogata, H.; Yamada, T. Xanthomonas citri jumbo phage XacN1 exhibits a wide host range and high complement of tRNA genes. Sci. Rep. 2018, 8, 4486. [Google Scholar] [CrossRef] [PubMed]
- Fermin, G. Host Range, Host–Virus Interactions, and Virus Transmission. In Viruses; Tennant, P., Fermin, G., Foster, J.E., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 101–134. [Google Scholar]
- Chen, Y.; Bao, X. Respiratory Syncytial Virus Induces a Functional tRNA-derived Fragment to Promote Infection by Targeting SYNE2. J. Immunol. 2020, 93, 1. [Google Scholar]
- Deng, J.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.; Phan, T.; Deng, X.; Zhou, J.; Lee, I.; Lee, Y.S.; et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Selitsky, S.R.; Baran-Gale, J.; Honda, M.; Yamane, D.; Masaki, T.; Fannin, E.E.; Guerra, B.; Shirasaki, T.; Shimakami, T.; Kaneko, S.; et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci. Rep. 2015, 5, 7675. [Google Scholar] [CrossRef]
- Avcilar-Kucukgoze, I.; Kashina, A. Hijacking tRNAs From Translation: Regulatory Functions of tRNAs in Mammalian Cell Physiology. Front. Mol. Biosci. 2020, 7, 610617. [Google Scholar] [CrossRef]
- Fricker, R.; Brogli, R.; Luidalepp, H.; Wyss, L.; Fasnacht, M.; Joss, O.; Zywicki, M.; Helm, M.; Schneider, A.; Cristodero, M.; et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun. 2019, 10, 118. [Google Scholar] [CrossRef]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Ruger, W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. MMBR 2003, 67, 86–156, table of contents. [Google Scholar] [CrossRef]
- Hofer, K.; Jaschke, A. Epitranscriptomics: RNA Modifications in Bacteria and Archaea. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Pilotto, S.; Werner, F. How to Shut Down Transcription in Archaea during Virus Infection. Microorganisms 2022, 10, 1824. [Google Scholar] [CrossRef] [PubMed]
- Gregorova, P.; Sipari, N.H.; Sarin, L.P. Broad-range RNA modification analysis of complex biological samples using rapid C18-UPLC-MS. RNA Biol. 2021, 18, 1382–1389. [Google Scholar] [CrossRef] [PubMed]
- Hagelskamp, F.; Borland, K.; Ramos, J.; Hendrick, A.G.; Fu, D.; Kellner, S. Broadly applicable oligonucleotide mass spectrometry for the analysis of RNA writers and erasers in vitro. Nucleic Acids Res. 2020, 48, e41. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Hussmann, J.A.; Weissman, J.S. Ribosome Profiling: Global Views of Translation. Cold Spring Harb. Perspect. Biol. 2019, 11, a032698. [Google Scholar] [CrossRef] [PubMed]
- Gelsinger, D.R.; Dallon, E.; Reddy, R.; Mohammad, F.; Buskirk, A.R.; DiRuggiero, J. Ribosome profiling in archaea reveals leaderless translation, novel translational initiation sites, and ribosome pausing at single codon resolution. Nucleic Acids Res. 2020, 48, 5201–5216. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarin, L.P. Learning from the Invaders: What Viruses Teach Us about RNA-Based Regulation in Microbes. Microorganisms 2022, 10, 2106. https://doi.org/10.3390/microorganisms10112106
Sarin LP. Learning from the Invaders: What Viruses Teach Us about RNA-Based Regulation in Microbes. Microorganisms. 2022; 10(11):2106. https://doi.org/10.3390/microorganisms10112106
Chicago/Turabian StyleSarin, L. Peter. 2022. "Learning from the Invaders: What Viruses Teach Us about RNA-Based Regulation in Microbes" Microorganisms 10, no. 11: 2106. https://doi.org/10.3390/microorganisms10112106
APA StyleSarin, L. P. (2022). Learning from the Invaders: What Viruses Teach Us about RNA-Based Regulation in Microbes. Microorganisms, 10(11), 2106. https://doi.org/10.3390/microorganisms10112106





