Opposite Sides of Pantoea agglomerans and Its Associated Commercial Outlook
Abstract
:1. Introduction
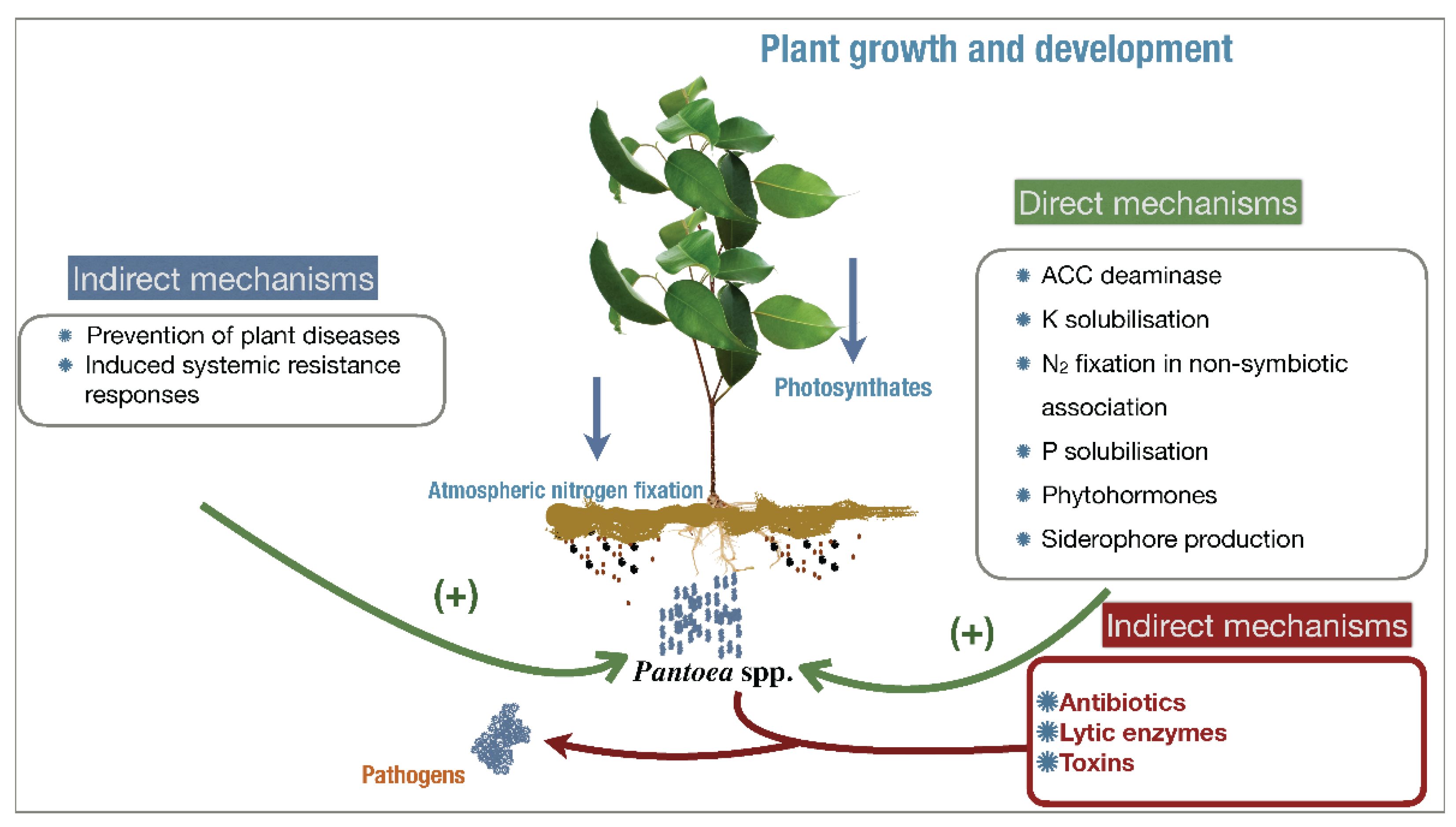
2. Bioprospecting of P. agglomerans and Its Commercial Outlook
2.1. Agricultural Outlook of P. agglomerans as a Biofertilizer
2.1.1. Potassium and Phosphate Solubilization
2.1.2. Nitrogen Fixation, IAA, and Siderophore Production
2.2. Agricultural Outlook of P. agglomerans as a Biocontroller
Genetic Engineering of an Isolate of P. agglomerans for Biocontrol
2.3. Commercial Outlook of P. agglomerans for Human and Environmental Health
3. Pathogenic Potential of P. agglomerans and Its Associated Biological Risks
3.1. Parasitic Colonization of Plants
3.2. Host Association with Humans and Animals
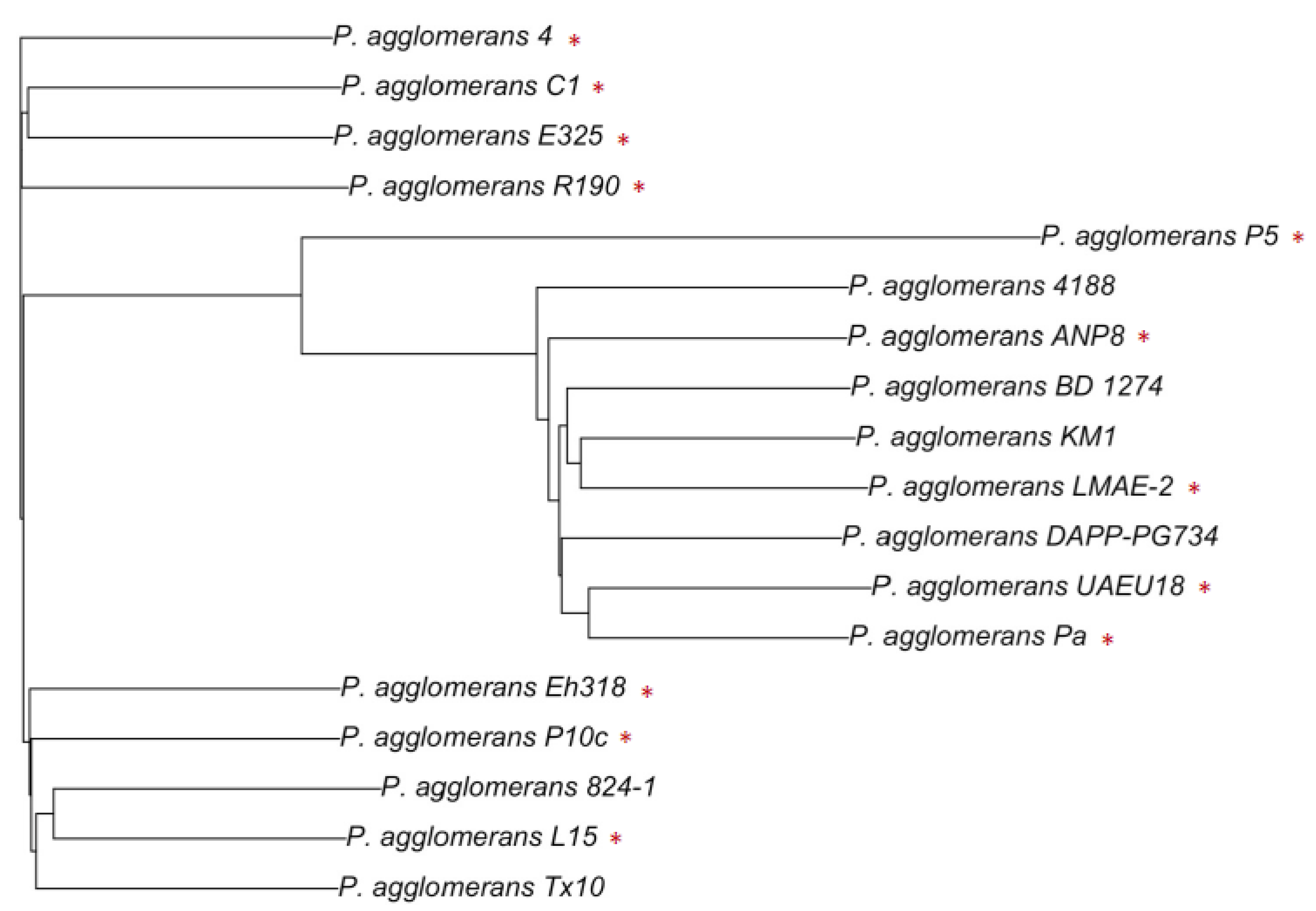
4. Genomics as a Tool to Explore the Potential and Hazards of P. agglomerans
4.1. Genomic Potential of the Beneficial Strains of P. agglomerans: From Plant to Environment
4.2. Exploring the Hazards of P. agglomerans Strains: What Genomic Data Can Tell Us
| Strain | RefSeq | Function | Isolation Source | Location | Genome Size (bp) | GC (%) | Contigs/Scaffolds | Plasmids/Scaffolds of Plasmids | CDS | rRNA | tRNA | tmRNA | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beneficial P. agglomerans strains | |||||||||||||
| L15 | GCF_003860325.1 | Biocontrol | Hypericum perphoratum phyllosphere | Poland | 4,858,869 | 55.10 | 1 | 3 | 4455 | 22 | 81 | 1 | [128] |
| UAEU18 | GCF_010523255.1 | PGPB | Date palm rhizosphere soil | United Arab Emirates | 4,825,350 | 55.20 | 1 | 3 | 4386 | 22 | 80 | 1 | [129] |
| P5 | GCF_002157425.2 | PGPB | Farmland soil | Iran | 5,074,260 | 54.87 | 127 | - | 4684 | 8 | 61 | 1 | [108] |
| C1 | GCF_009759885.1 | PGPB | Lettuce phyllosphere | Italy | 4,846,162 | 55.16 | 21 | - | 4465 | 12 | 72 | 1 | [107] |
| P10c | GCF_001288285.1 | Biocontrol | Pear blossoms | New Zealand | 4,775,916 | 55.06 | 18 | 2 | 4381 | 6 | 67 | 1 | [115] |
| ANP8 | GCF_017315165.1 | PGPB | Alfalfa root nodules | Iran | 5,035,017 | 55.00 | 1390 | - | 4404 | 14 | 75 | 1 | [106] |
| Pa | GCF_002082355.1 | PGPB | Durum wheat rhizosphere | Algeria | 4,795,157 | 55.02 | 45 | - | 4358 | 13 | 70 | 1 | [6] |
| R190 | GCF_000731125.1 | Biocontrol | Apple orchard | South Korea | 5,002,566 | 55.05 | 2 | 3 | 4592 | 24 | 78 | 1 | [117] |
| LMAE-2 | GCF_000814075.1 | Bioremediation | Marine sediment | Chile | 4,981,165 | 55.15 | 155 | - | 4562 | 19 | 80 | 1 | [119] |
| 4 | GCF_000743785.2 | Biocontrol | Wheat seed | Canada | 4,827,890 | 55.17 | 4 | - | 4399 | 19 | 74 | 1 | [130] |
| E325 | GCF_014353865.1 | Biocontrol | Apple flowers | United States | 4,786,783 | 55.21 | 162 | - | 4365 | 14 | 74 | 1 | [131] |
| Eh318 | GCF_000687245.1 | Biocontrol | Stem from an asymptomatic apple | United States | 5,035,839 | 54.77 | 34 | - | 4597 | 21 | 74 | 1 | [116] |
| Harmful P. agglomerans strains | |||||||||||||
| KM1 | GCF_012241415.1 | Opportunistic pathogen | Short-term fermented homemade kimchi | South Korea | 4,995,756 | 55.07 | 34 | 12 | 4568 | 2 | 64 | 1 | [122] |
| BD 1274 | GCF_003369505.1 | Pathogen of onion | Onion seeds | South Africa | 4,968,508 | 54.96 | 246 | - | 4612 | 11 | 48 | 1 | [125] |
| 824-1 | GCF_001661985.1 | Gall-forming | Gypsophila | United States | 4,989,068 | 54.81 | 55 | - | 4648 | 7 | 73 | 1 | [87] |
| 4188 | GCF_001662025.1 | Gall-forming | Beet | United States | 5,001,997 | 54.96 | 79 | - | 4629 | 10 | 74 | 1 | [87] |
| Tx10 | GCF_000475055.1 | Cystic fibrosis | Sputum of a cystic fibrosis patient | United States | 4,856,993 | 55.10 | 22 | - | 4446 | 16 | 73 | 1 | [71] |
| DAPP-PG 734 | GCF_000710215.1 | Related to olive knot | Olive knots | Italy | 5,365,929 | 54.75 | 195 | - | 5000 | 11 | 71 | 1 | [124] |
5. Final Considerations
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Gavini, F.; Mergaert, J.; Beji, A.; Mielcarek, C.; Izard, D.; Kersters, K.; De Ley, J. Transfer of Enterobacter agglomerans (Beijerinck 1988) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int. J. Syst. Bacteriol. 1989, 39, 337–345. [Google Scholar] [CrossRef]
- Asai, N.; Koizumi, Y.; Yamada, A.; Sakanashi, D.; Watanabe, H.; Kato, H.; Shiota, A.; Hagihara, M.; Suematsu, H.; Yamagishi, Y.; et al. Pantoea dispersa bacteremia in an immunocompetent patient: A case report and review of the literature. J. Med. Case Rep. 2019, 13, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, C.; Cleenwerck, I.; Venter, S.; Engelbeen, K.; De Vos, P.; Coutinho, T.A. Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Micr. 2010, 60, 2430–2440. [Google Scholar]
- Kim, Y.C.; Leveau, J.; Gardener, B.B.M.; Pierson, E.A.; Pierson, L.S.; Ryu, C.-M. The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microbiol. 2011, 77, 1548–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quecine, M.C.; Araújo, W.L.; Rossetto, P.B.; Ferreira, A.; Tsui, S.; Lacava, P.T.; Mondin, M.; Azevedo, J.L.; Pizzirani-Kleiner, A.A. Sugarcane growth promotion by the endophytic bacterium Pantoea agglomerans 33.1. Appl. Environ. Microbiol. 2012, 78, 7511–7518. [Google Scholar] [CrossRef] [Green Version]
- Cherif-Silini, H.; Thissera, B.; Bouket, A.C.; Saadaoui, N.; Silini, A.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Yahiaoui, B.; et al. Durum wheat stress tolerance induced by endophyte Pantoea agglomerans with genes contributing to plant functions and secondary metabolite arsenal. Int. J. Mol. Sci. 2019, 20, 3989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thissera, B.; Alhadrami, H.A.; Hassan, M.H.A.; Hassan, H.M.; Bawazeer, M.; Yaseen, M.; Belbahri, L.; Rateb, M.E.; Behery, F.A. Induction of cryptic antifungal pulicatin derivatives from Pantoea agglomerans by microbial co-culture. Biomolecules 2020, 10, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, A.; Kohchi, C.; Inagawa, H.; Nishizawa, T.; Soma, G.-I. Improvement of allergic dermatitis via regulation of the Th1/Th2 immune system balance by macrophages activated with lipopolysaccharide derived from Pantoea agglomerans (IP-PA1). Anticancer. Res. 2009, 29, 4867–4870. [Google Scholar]
- Hebishima, T.; Matsumoto, Y.; Watanabe, G.; Soma, G.-I.; Kohchi, C.; Taya, K.; Hayashi, Y.; Hirota, Y. Oral administration of immunopotentiator from Pantoea agglomerans 1 (IP-PA1) improves the survival of B16 melanoma-inoculated model mice. Exp. Anim. 2011, 60, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Nakata, K.; Inagawa, H.; Soma, G. Lipopolysaccharide IPPA1 from Pantoea agglomerans prevents suppression of macrophage function in stress-induced diseases. Anticancer. Res. 2011, 31, 2437–2440. [Google Scholar] [PubMed]
- Thissera, B.; Hallyburton, I.; Ngwa, C.J.; Cherif-Silini, H.; Hassane, A.S.I.; Anderson, M.; Campbell, L.A.; Mutter, N.; Eshelli, M.; Abdelmohsen, U.R.; et al. Potent antiplasmodial alkaloids from the rhizobacterium Pantoea agglomerans as hemozoin modulators. Bioorg. Chem. 2021, 115, 105215. [Google Scholar] [CrossRef] [PubMed]
- Pidot, S.J.; Coyne, S.; Kloss, F.; Hertweck, C. Antibiotics from neglected bacterial sources. Int. J. Med. Microbiol. 2014, 304, 14–22. [Google Scholar] [CrossRef]
- Muraschi, T.; Friend, M.; Bolles, D. Erwinia-like microorganisms isolated from animal and human hosts. Appl. Microbiol. 1965, 13, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Ewing, W.; Fife, M. Enterobacter agglomerans (Beijerinck) comb. nov. (the Herbicola-Lathyri Bacteria). Int. J. Syst. Bacteriol. 1972, 22, 4–11. [Google Scholar] [CrossRef] [Green Version]
- Brady, C.; Cleenwerck, I.; Venter, S.; Vancanneyt, M.; Swings, J.; Coutinho, T. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 2008, 31, 447–460. [Google Scholar] [CrossRef] [Green Version]
- Völksch, B.; Thon, S.; Jacobsen, I.; Gube, M. Polyphasic study of plant- and clinic-associated Pantoea agglomerans strains reveals indistinguishable virulence potential. Infect. Genet. Evol. 2009, 9, 1381–1391. [Google Scholar] [CrossRef]
- Nadarasah, G.; Stavrinides, J. Quantitative evaluation of the host-colonizing capabilities of the enteric bacterium Pantoea using plant and insect hosts. Microbiology 2014, 160, 602–615. [Google Scholar] [CrossRef]
- Slotnick, I.; Tulman, L. A human infection caused by an Erwinia species. Am. J. Med. 1967, 43, 147–150. [Google Scholar] [CrossRef]
- Cruz, A.T.; Cazacu, A.C.; Allen, C.H. Pantoea agglomerans, a plant pathogen causing human disease. J. Clin. Microbiol. 2007, 45, 1989–1992. [Google Scholar] [CrossRef] [Green Version]
- GMBl. No. 68-80 of 06.12.2010. 2010, pp. 1428–1667. Available online: https://www.baua.de/EN/Service/Legislative-texts-and-technical-rules/Rules/TRBA/TRBA-466.html (accessed on 8 July 2022).
- Liu, L.; Kloepper, J.W.; Tuzun, S. Induction of systemic resistance in cucumber against Fusarium wilt by plant growth-promoting rhizobacteria. Phytopathology 1995, 85, 695–698. [Google Scholar] [CrossRef]
- Ongena, M.; Daayf, F.; Jacques, P.; Thonart, P.; Benhamou, N.; Paulitz, T.C.; Belanger, R.R. Systemic induction of phytoalexins in cucumber in response to treatments with fluorescent pseudomonads. Plant Pathol. 2000, 49, 523–530. [Google Scholar] [CrossRef]
- Bonaterra, A.; Mari, M.; Casalini, L.; Montesinos, E. Biological control of Monilinia laxa and Rhizopus stolonifer in postharvest of stone fruit by Pantoea agglomerans EPS125 and putative mechanisms of antagonism. Int. J. Food Microbiol. 2003, 84, 93–104. [Google Scholar] [CrossRef]
- Quecine, M.C.; Araújo, W.L.; Tsui, S.; Parra, J.R.P.; Azevedo, J.L.; Pizzirani-Kleiner, A.A. Control of Diatraea saccharalis by the endophytic Pantoea agglomerans 33.1 expressing cry1Ac7. Arch. Microbiol. 2014, 196, 227–234. [Google Scholar] [CrossRef]
- Torres, A.R.; Araújo, W.L.; Cursino, L.; Rossetto, P.B.; Mondin, M.; Hungria, M.; Azevedo, J.L. Colonization of Madagascar periwinkle (Catharanthus roseus), by endophytes encoding gfp marker. Arch. Microbiol. 2013, 195, 483–489. [Google Scholar] [CrossRef]
- Dong, Z.; Canny, M.J.; McCully, M.E.; Roboredo, M.G.; Cabadilla, C.F.; Ortega, E.; Rodés, R. A nitrogen-fixing endophyte of sugarcane stems (A new role for the apoplast). Plant Physiol. 1994, 105, 1139–1147. [Google Scholar] [CrossRef]
- Silini-Chérif, H.; Silini, A.; Ghoul, M.; Yadav, S. Isolation and characterization of plant growth promoting traits of a rhizobacteria: Pantoea agglomerans lma2. Pak. J. Biol. Sci. 2012, 15, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part I. Deleterious effects: Dust-borne endotoxins and allergens—Focus on cotton dust. Ann. Agric. Environ. Med. 2015, 22, 576–588. [Google Scholar] [CrossRef]
- Berni, R.J.; DeLucca, A.J.; Shafer, G.P. Statistical and biological analysis of epiphytic bacteria and endotoxin on cotton. In Cotton Dust, Proceedings of the 12th Cotton Dust Research Conference, New Orleans, LA, USA, 6–7 January 1988; Jacobs, R.R., Wakelyn, P.J., Eds.; National Cotton Council: Memphis, TN, USA, 1988; pp. 20–22. [Google Scholar]
- Loncaric, I.; Heigl, H.; Licek, E.; Moosbeckhofer, R.; Busse, H.-J.; Rosengarten, R. Typing of Pantoea agglomerans isolated from colonies of honey bees (Apis mellifera) and culturability of selected strains from honey. Apidologie 2009, 40, 40–54. [Google Scholar] [CrossRef]
- Feng, Y.; Shen, D.; Song, W. Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J. Appl. Microbiol. 2006, 100, 938–945. [Google Scholar] [CrossRef]
- Kamran, S.; Shahid, I.; Baig, D.N.; Rizwan, M.; Malik, K.A.; Mehnaz, S. Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front. Microbiol. 2017, 8, 2593. [Google Scholar] [CrossRef] [Green Version]
- Da Luz, W.C. Evaluation of plant growth-promoting and bioprotecting rhizobacteria on wheat crop. Fitopatol. Bras. 2001, 26, 597–600. [Google Scholar] [CrossRef]
- Zakria, K.U.; Ogawa, T.; Yamamoto, A.; Saeki, Y.; Akao, S. Influence of inoculation technique on the endophytic colonization of rice by Pantoea sp. isolated from sweet potato and by Enterobacter sp. isolated from sugarcane Muhammad. Soil Sci. Plant Nutr. 2008, 54, 224–236. [Google Scholar] [CrossRef]
- Bakhshandeh, E.; Pirdashti, H.; Gilani, Z. Application of mathematical models to describe rice growth and nutrients uptake in the presence of plant growth promoting microorganisms. Appl. Soil. Ecol. 2017, 124, 171–184. [Google Scholar] [CrossRef]
- Khanghahi, M.Y.; Pirdashti, H.; Rahimian, H.; Nematzadeh, G.A.; Ghajar Sepanlou, M. Potassium solubilising bacteria (KSB) isolated from rice paddy soil: From isolation, identification to K use efficiency. Symbiosis 2017, 77, 13–23. [Google Scholar]
- De Souza, R.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Yaghoubi, M.; Rahimian, H.; Pirdashti, H.; Nematzadeh, G. The role of potassium solubilizing bacteria (KSB) inoculations on grain yield, dry matter remobilization and translocation in rice (Oryza sativa L.). J. Plant Nutr. 2019, 42, 1165–1179. [Google Scholar] [CrossRef]
- Saadouli, I.; Mosbah, A.; Ferjani, R.; Stathopoulou, P.; Galiatsatos, I.; Asimakis, E.; Marasco, R.; Daffonchio, D.; Tsiamis, G.; Ouzari, H.-I. The impact of the inoculation of phosphate-solubilizing bacteria Pantoea agglomerans on phosphorus availability and bacterial community dynamics of a semi-arid soil. Microorganisms 2021, 9, 1661. [Google Scholar] [CrossRef]
- Li, L.; Chen, R.; Zuo, Z.; Lv, Z.; Yang, Z.; Mao, W.; Liu, Y.; Zhou, Y.; Huang, J.; Song, Z. Evaluation and improvement of phosphate solubilization by an isolated bacterium Pantoea agglomerans ZB. World J. Microbiol. Biotechnol. 2020, 36, 27. [Google Scholar] [CrossRef]
- Malboobi, M.A.; Owlia, P.; Behbahani, M.; Sarokhani, E.; Moradi, S.; Yakhchali, B. Solubilization of organic an inorganic phosphates by three highly efficient soil bacterial isolates. World J. Microbiol. Biotechnol. 2009, 25, 1471–1477. [Google Scholar] [CrossRef]
- Goljanian-Tabrizi, S.; Amiri, S.; Nikaein, D.; Motesharrei, Z. The comparison of five low cost liquid formulations to preserve two phosphate solubilizing bacteria from the genera Pseudomonas and Pantoea. Iran J. Microbiol. 2016, 8, 377–382. [Google Scholar]
- Weaver, R.; Frederick, L. Effect of inoculum rate on competitive nodulation of Glycine max L. Merril. II field studies. Agron. J. 1974, 66, 233–236. [Google Scholar] [CrossRef]
- Hume, D.; Blair, D. Effect of number of Bradyrhizobium japonicum applied in commercial inoculants in soybean seed yield in Ontario. Can. J. Microbiol. 1992, 38, 588–593. [Google Scholar] [CrossRef]
- Lupwayi, N.Z.; Olsen, P.E.; Sande, E.S.; Keyser, H.H.; Collins, M.M.; Singleton, P.W. Inoculant quality and its evaluation. F. Crops Res. 2000, 65, 259–2170. [Google Scholar] [CrossRef]
- Deaker, R.; Roughley, R.J.; Kennedy, I.R. Legume seed inoculation technology—A review. Soil Biol. Biochem. 2004, 36, 1275–1288. [Google Scholar] [CrossRef]
- De Gregorio, P.R.; Michavila, G.; Muller, L.R.; Borges, C.S.; Pomares, M.F.; de Sá, E.L.S.; Pereira, C.; Vincent1, P.A. Beneficial rhizobacteria immobilized in nanofibers for potential application as soybean seed bioinoculants. PLoS ONE 2017, 12, e0176930. [Google Scholar] [CrossRef] [Green Version]
- Viñas, I.; Usall, J.; Nunes, C.; Teixidó, N. Nueva cepa Bacteriana Pantoea agglomerans, Beijerinck (1888) Gavini, Mergaert, Beji, Mielcareck, Izard, Kerstersy, De Ley (1989) y su Utilización como Agente de Control Biológico de las Enfermedades Fúngicas de Fruta. Spanish Patent P9900612, 26 March 1999. [Google Scholar]
- Costa, E.; Teixidó, N.; Usall, J.; Atarés, E.; Viñas, I. Production of the biocontrol agent Pantoea agglomerans strain CPA-2 using commercial products and by-products. Appl. Microbiol. Biotechnol. 2001, 56, 367–371. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 2016, 23, 206–222. [Google Scholar] [CrossRef]
- Haidar, R.; Amira, Y.; Roudet, J.; Marc, F.; Patrice, R. Application methods and modes of action of Pantoea agglomerans and Paenibacillus sp. to control the grapevine trunk disease-pathogen, Neofusicoccum parvum. OENO One 2021, 55, 1–16. [Google Scholar] [CrossRef]
- Giddens, S.R.; Feng, Y.; Mahanty, H.K. Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol. Microbiol. 2002, 45, 769–783. [Google Scholar] [CrossRef] [Green Version]
- Wright, S.; Jin, M.; Clardy, J.; Beer, S. The biosynthetic genes of pantocin A and pantocin B of Pantoea agglomerans Eh318. Acta Hortic. 2006, 704, 313–319. [Google Scholar] [CrossRef]
- Vanneste, J.; Yu, J.; Cornish, D.A. Presence of genes homologous to those necessary for synthesis of microcin MccEh252 in strains of Pantoea agglomerans. Acta Hortic. 2008, 793, 391–396. [Google Scholar] [CrossRef]
- Jin, M.; Fischbach, M.A.; Clardy, J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J. Am. Chem. Soc. 2006, 128, 10660–10661. [Google Scholar] [CrossRef] [Green Version]
- Greiner, M.; Winkelmann, G. Fermentation and isolation of herbicolin A, a peptide antibiotic produced by Erwinia herbicola strain A 111. Appl. Microbiol. Biotechnol. 1991, 34, 565–569. [Google Scholar] [CrossRef]
- Kamber, T.; Lansdell, T.A.; Stockwell, V.O.; Ishimaru, C.A.; Smits, T.H.; Duffy, B. Characterization of the biosynthetic operon for the antibacterial peptide herbicolin in Pantoea vagans biocontrol strain C9–1 and incidence in Pantoea species. Appl. Environ. Microbiol. 2012, 78, 4412–4419. [Google Scholar] [CrossRef] [Green Version]
- Shoji, J.; Sakazaki, R.; Hattori, T.; Matsumoto, K.; Uotani, N.; Yoshida, T. Isolation and characterization of agglomerins A, B, C and D. J. Antibiot. 1989, 42, 1729–1733. [Google Scholar] [CrossRef] [Green Version]
- Nunez, W.J.; Colmer, A.R. Differentiation of Aerobacter-Klebsiella isolated from sugarcane. Appl. Microbiol. 1968, 16, 1875–1878. [Google Scholar] [CrossRef]
- Baldani, J.I.; Baldani, V.L.D.; Seldin, L.; Döbereiner, J. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. Int. J. Syst. Bacteriol. 1986, 36, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Cavalcante, V.A.; Döbereiner, J. A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 1988, 108, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Loiret, F.G.; Ortega, E.; Kleiner, D.; Ortega-Rode, P.; Rode’s, R.; Dong, Z. A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J. Appl. Microbiol. 2004, 97, 504–511. [Google Scholar] [CrossRef]
- Dean, D.H. Biochemical genetics of the bacterial insect-control agent Bacillus thuringiensis: Basic principles and prospect for genetic engineering. Biotechnol. Genet. Eng. 1984, 2, 341–363. [Google Scholar] [CrossRef]
- Wei, J.Z.; Hale, K.; Carta, L.; Platzer, E.; Wong, C.; Fang, S.C.; Aroian, R.V. Bacillus thuringiensis crystal proteins that target nematodes. Proc. Natl. Acad. Sci. USA 2003, 100, 2760–2765. [Google Scholar] [CrossRef] [Green Version]
- Kotze, A.C.; O’Grady, J.; Gough, J.M.; Pearson, R.; Bagnall, N.H.; Kemp, D.H.; Akhurst, R.J. Toxicity of Bacillus thuringiensis to parasitic and free-living life-stages of nematode parasites of livestock. Int. J. Parasitol. 2005, 35, 1013–1022. [Google Scholar] [CrossRef]
- De Macedo, C.L.; Martins, E.S.; Pepino de Macedo, L.L.; Santos, A.C.; Praça, L.B.; Góis, L.A.B.; Monnerat, R.G. Selection and characterization of Bacillus thuringiensis efficient strains against Diatraea saccharalis (Lepidoptera: Crambidae). Pesq. Agropec. Bras. 2012, 47, 1759–1765. [Google Scholar] [CrossRef]
- Bisi, D.C.; Lampe, D.J. Secretion of anti-Plasmodium effector proteins from a natural Pantoea agglomerans isolate by using PelB and HlyA secretion signals. Appl. Environ. Microbiol. 2011, 77, 4669–4675. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Ghosh, A.K.; Bongio, N.; Stebbings, K.A.; Lampe, D.J.; Jacobs-Lorena, M. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. USA 2012, 109, 12734–12739. [Google Scholar] [CrossRef] [Green Version]
- Stiebler, R.; Hoang, A.N.; Egan, T.J.; Wright, D.W.; Oliveira, M.F. Increase on the initial soluble heme levels in acidic conditions is an important mechanism for spontaneous heme crystallisation in vitro. PLoS ONE 2010, 5, e12694. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.D.; Kirzinger, M.W.; Stavrinides, J. Draft genome sequence of the antibiotic-producing cystic fibrosis isolate Pantoea agglomerans Tx10. Genome Announc. 2013, 1, e00904-13. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Wang, G.; Qiao, H.; Cai, J. Fermentative hydrogen production by the new marine Pantoea agglomerans isolated from the mangrove sludge. Int. J. Hydrog. Energy 2008, 33, 6116–6123. [Google Scholar] [CrossRef]
- Kurlanda-Witek, H.; Ngwenya, B.T.; Butler, I.B. The influence of biofilms on the mobility of bare and capped zinc oxide nanoparticles in saturated sand and glass beads. J. Contam. Hydrol. 2015, 179, 160–170. [Google Scholar] [CrossRef] [Green Version]
- McInroy, J.; Kloepper, J. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 1995, 173, 337–342. [Google Scholar] [CrossRef]
- Elvira-Recuenco, M.; van Vuurde, J. Natural incidence of endophytic bacteria in pea cultivars under field conditions. Can. J. Microbiol. 2000, 46, 1036–1041. [Google Scholar] [CrossRef]
- Asis, C.; Adachi, K. Isolation of endophytic diazotroph Pantoea agglomerans and nondiazotroph Enterobacter asburiae from sweet potato stem in Japan. Lett. Appl. Microbiol. 2004, 38, 19–23. [Google Scholar] [CrossRef]
- Magnani, G.; Cruz, L.; Weber, H.; Bespalhok, J.; Daros, E.; Baura, V.; Yates, M.; Monteiro, R.; Faoro, H.; Pedrosa, F.; et al. Culture-independent analysis of endophytic bacterial communities associated with Brazilian sugarcane. Genet. Mol. Res. 2013, 12, 4549–4558. [Google Scholar] [CrossRef]
- Sood, A.; Nadha, H.; Salwan, R.; Kasana, R.; Anand, M. Identification and elimination of bacterial contamination during in vitro propagation of Guadua angustifolia Kunth. Pharmacogn. Mag. 2012, 8, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Stets, M.I.; Pinto, A.S., Jr.; Huergo, L.F.; de Souza, E.M.; Guimarães, V.F.; Alves, A.C.; Steffens, M.B.; Monteiro, R.A.; de Oliveira Pedrosa, F.; Cruz, L.M. Rapid identification of bacterial isolates from wheat roots by high resolution whole cell MALDI-TOF MS analysis. J. Biotechnol. 2013, 165, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Guanlin, X. First report of palea browning in China and characterization of the causal organism by phenotypic tests and Biolog. Int. Rice Res. Notes. 2001, 26, 25–26. [Google Scholar]
- Burr, T.J.; Katz, B.H.; Abawi, G.S.; Crosier, D.C. Comparison of tumorigenic strains of Erwinia herbicola isolated from table beet with E. h. gypsophilae. Plant Dis. 1991, 75, 855–858. [Google Scholar] [CrossRef]
- Brady, C.; Venter, S.; Cleenwerck, I.; Engelbeen, K.; de Vos, P.; Wingfield, M.J.; Telechea, N.; Coutinho, T.A. Isolation of Enterobacter cowanii from Eucalyptus showing symptoms of bacterial blight and dieback in Uruguay. Lett. Appl. Microbiol. 2009, 49, 461–465. [Google Scholar] [CrossRef]
- Brady, C.; Venter, S.; Cleenwerck, I.; Engelbeen, K.; Vancanneyt, M.; Swings, J.; Coutinho, T.A. Pantoea vagans sp. nov., Pantoea eucalypti sp. nov., Pantoea deleyi sp. nov and Pantoea anthophila sp. nov. Int. J. Syst. Evol. Micr. 2009, 59, 2339–2345. [Google Scholar] [CrossRef] [PubMed]
- Romeiro, R.; Macagnan, D.; Mendonca, H.; Neto, J.R. Bacterial spot of Chinese taro (Alocasia cucullata) in Brazil induced by Pantoea agglomerans. Plant Pathol. 2007, 56, 1038. [Google Scholar] [CrossRef]
- Cooksey, D.A. Galls of Gypsophila paniculata caused by Erwinia herbicola. Plant Dis. 1986, 70, 464–468. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Ann. Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef]
- Nissan, G.; Gershovits, M.; Morozov, M.; Chalupowicz, L.; Sessa, G.; Manulis-Sasson, S.; Barash, I.; Pupko, T. Revealing the inventory of type III effectors in Pantoea agglomerans gall-forming pathovars using draft genome sequences and a machine-learning approach. Mol. Plant Pathol. 2018, 19, 381–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koirala, S.; Zhao, M.; Agarwal, G.; Gitaitis, R.; Stice, S.; Kvitko, B.; Dutta, B. Identification of two novel pathovars of Pantoea stewartii subsp. indologenes affecting Allium sp. and Millets. Phytopathology 2021, 111, 1509–1519. [Google Scholar] [PubMed]
- Delétoile, A.; Decré, D.; Courant, S.; Passet, V.; Audo, J.; Grimont, P.; Arlet, G.; Brisse, S. Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. J. Clin. Microbiol. 2009, 47, 300–310. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, B.; Nabin, K.C.; Bastola, C.; Jahir, T.; Risal, R.; Thapa, S.; Enriquez, D.; Schmidt, F. Pantoea agglomerans: An elusive contributor to chronic obstructive pulmonary disease exacerbation. Cureus 2021, 13, e18562. [Google Scholar] [CrossRef]
- Maki, D.G.; Rhame, F.S.; Mackel, D.C.; Bennett, J.V. Nationwide epidemic of septicemia caused by contaminated intravenous products. I. Epidemiologic and clinical features. Am. J. Med. 1976, 60, 471–485. [Google Scholar] [CrossRef]
- Gibson, J.A.; Eaves, L.E.; O’Sullivan, B.M. Equine abortion associated with Enterobacter agglomerans. Equine Vet. J. 1982, 14, 122–125. [Google Scholar] [CrossRef]
- Hong, C.B.; Donahue, J.M.; Giles, R.C.; Petrites-Murphy, M.B.; Poonacha, K.B.; Roberts, A.W.; Smith, B.J.; Tramontin, R.R.; Tuttle, P.A.; Swerczek, T.W. Etiology and pathology of equine placentitis. J. Vet. Diagn. Investig. 1993, 5, 56–63. [Google Scholar] [CrossRef]
- Hansen, G.H.; Raa, J.; Olafsen, J.A. Isolation of Enterobacter agglomerans from dolphin fish, Coryphaena hippurus L. J. Fish Dis. 1990, 13, 93–96. [Google Scholar] [CrossRef]
- Pomorski, Z.; Dutkiewicz, J.; Taszkun, I.; Woźniak, M.; Sitkowski, W.; Skórska, C.; Cholewa, G. Studies on allergic conditioning of cattle pneumopathy, resulting from breathing in pneumoallergens comprised in organic dusts. Ann. Univ. Mariae Curie Sklodowska Med. 1993, 48, 183–193. [Google Scholar]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef] [PubMed]
- Batista, B.D.; Dourado, M.N.; Figueredo, E.F.; Hortencio, R.O.; Marques, J.P.R.; Piotto, F.A.; Bonatelli, M.L.; Settles, M.L.; Azevedo, J.L.; Quecine, M.C. The auxin-producing Bacillus thuringiensis RZ2MS9 promotes the growth and modifies the root architecture of tomato (Solanum lycopersicum cv. Micro-Tom). Arch. Microbiol. 2021, 203, 3869–3882. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef] [Green Version]
- Badet, T.; Croll, D. The rise and fall of genes: Origins and functions of plant pathogen pangenomes. Curr. Opin. Plant Biol. 2020, 56, 65–73. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes (No. e1900v1). Peer J. Prepr. 2016, 4, e1900v1. [Google Scholar]
- Dobrindt, U.; Chowdary, M.G.; Krumbholz, G.; Hacker, J. Genome dynamics and its impact on evolution of Escherichia Coli. Med. Microbiol. Immunol. 2010, 199, 145–154. [Google Scholar] [CrossRef]
- Carroll, L.M.; Cheng, R.A.; Wiedmann, M.; Kovac, J. Keeping up with the Bacillus cereus group: Taxonomy through the genomics era and beyond. Crit. Rev. Food Sci. Nutr. 2021, 62, 7677–7702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Jin, T.; Kumar Sahu, S.; Xu, J.; Shi, Q.; Liu, H.; Wang, Y. The distribution of tryptophan-dependent indole-3-acetic acid synthesis pathways in bacteria unraveled by large-scale genomic analysis. Molecules 2019, 24, 1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Yang, J.; Shen, P.; Li, Q.; Wu, W.; Jiang, X.; Qin, L.; Huang, J.; Cao, X.; Qi, F. High-level production of indole-3-acetic acid in the metabolically engineered Escherichia coli. J. Agric. Food Chem. 2021, 69, 1916–1924. [Google Scholar] [CrossRef]
- Noori, F.; Etesami, H.; Noori, S.; Forouzan, E.; Jouzani, G.S.; Malboobi, M.A. Whole genome sequence of Pantoea agglomerans ANP8, a salinity and drought stress–resistant bacterium isolated from alfalfa (Medicago sativa L.) root nodules. Biotechnol. Rep. 2021, 29, e00600. [Google Scholar] [CrossRef]
- Luziatelli, F.; Ficca, A.G.; Cardarelli, M.; Melini, F.; Cavalieri, A.; Ruzzi, M. Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 2020, 8, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shariati, J.V.; Malboobi, M.A.; Tabrizi, Z.; Tavakol, E.; Owlia, P.; Safari, M. Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 2017, 7, 15610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, S.; Fontanille, P.; Pandey, A.; Larroche, C. Gluconic acid: Properties, applications and microbial production. Food Technol. Biotechnol. 2006, 44, 184–195. [Google Scholar]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Kloepper, J.W.; Paré, P.W. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Li, X.; Ma, L.; Borriss, R.; Wu, Z.; Gao, X. Acetoin and 2, 3-butanediol from Bacillus amyloliquefaciens induce stomatal closure in Arabidopsis thaliana and Nicotiana benthamiana. J. Exp. Bot. 2018, 69, 5625–5635. [Google Scholar] [CrossRef]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef]
- Crosa, J.H.; Walsh, C.T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 223–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef] [Green Version]
- Smits, T.H.; Rezzonico, F.; Blom, J.; Goesmann, A.; Abelli, A.; Morelli, R.K.; Vanneste, J.; Duffy, B. Draft genome sequence of the commercial biocontrol strain Pantoea agglomerans P10c. Genome Announc. 2015, 3, e01448-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, S.A.; Zumoff, C.H.; Schneider, L.; Beer, S.V. Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl. Environ. Microbiol. 2001, 67, 284–292. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.A.; Lee, D.H.; Kim, B.Y.; Heu, S. Draft genome sequence of Pantoea agglomerans R190, a producer of antibiotics against phytopathogens and foodborne pathogens. J. Biotechnol. 2014, 188, 7–8. [Google Scholar] [CrossRef]
- Zhang, H.; Yuan, X.; Xiong, T.; Wang, H.; Jiang, L. Bioremediation of co-contaminated soil with heavy metals and pesticides: Influence factors, mechanisms and evaluation methods. Chem. Eng. Sci. 2020, 398, 125657. [Google Scholar] [CrossRef]
- Corsini, G.; Valdés, N.; Pradel, P.; Tello, M.; Cottet, L.; Muiño, L.; Karahanian, E.; Castillo, A.; Gonzalez, A.R. Draft genome sequence of a copper-resistant marine bacterium, Pantoea agglomerans strain LMAE-2, a bacterial strain with potential use in bioremediation. Genome Announc. 2016, 4, e00525-16. [Google Scholar] [CrossRef] [Green Version]
- Cazorla, F.M.; Arrebola, E.; Sesma, A.; Pérez-García, A.; Codina, J.C.; Murillo, J.; de Vicente, A. Copper resistance in Pseudomonas syringae strains isolated from mango is encoded mainly by plasmids. Phytopathology 2002, 92, 909–916. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.X.; Rainey, P.B. Regulation of copper homeostasis in Pseudomonas fluorescens SBW25. Environ. Microbiol. 2008, 10, 3284–3294. [Google Scholar] [CrossRef]
- Guevarra, R.B.; Magez, S.; Peeters, E.; Chung, M.S.; Kim, K.H.; Radwanska, M. Comprehensive genomic analysis reveals virulence factors and antibiotic resistance genes in Pantoea agglomerans KM1, a potential opportunistic pathogen. PLoS ONE 2021, 16, e0239792. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, C.; Cortese, C.; Passos da Silva, D.; Venturi, V.; Torelli, E.; Firrao, G.; Buonaurio, R. Draft genome sequence of a hypersensitive reaction-inducing Pantoea agglomerans strain isolated from olive knots caused by Pseudomonas savastanoi pv. savastanoi. Genome Announc. 2014, 2, e00774-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moloto, V.M.; Goszczynska, T.; Hassen, A.I.; Pierneef, R.; Coutinho, T. Draft genome sequences of Pantoea agglomerans strains BD1274 and BD1212, isolated from onion seeds, reveal major differences in pathogenicity and functional genes. Microbiol. Resour. Announc. 2020, 9, e01507-19. [Google Scholar] [CrossRef] [PubMed]
- Scortichini, M.; Katsy, E.I. Common themes and specific features in the genomes of phytopathogenic and plant-beneficial bacteria. In Plasticity in Plant-Growth-Promoting and Phytopathogenic Bacteria; Springer: New York, NY, USA, 2014; pp. 1–26. [Google Scholar]
- Misra, B.B.; Langefeld, C.D.; Olivier, M.; Cox, L.A. Integrated Omics: Tools, Advances, and Future Approaches. J. Mol. Endocrinol. 2018, 62, R21–R45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rekosz-Burlaga, H.; Borys, M.; Goryluk-Salmonowicz, A. Cultivable microorganisms inhabiting the aerial parts of Hypericum perforatum. Acta Sci. Pol. Hortorum Cultus. 2014, 13, 117–129. [Google Scholar]
- Alkaabi, A.S.; Sudalaimuthuasari, N.; Kundu, B.; AlMaskari, R.S.; Salha, Y.; Hazzouri, K.M.; El-Tarabily, K.A.; AbuQamar, S.; Amiri, K.M.A. Complete genome sequence of the plant growth-promoting bacterium Pantoea agglomerans strain UAEU18, isolated from date palm rhizosphere soil in the United Arab Emirates. Microbiol. Resour. Announc. 2020, 9, e00174-20. [Google Scholar] [CrossRef]
- Town, J.; Links, M.; Dumonceaux, T.J. High-quality draft genome sequences of Pantoea agglomerans isolates exhibiting antagonistic interactions with wheat seed-associated fungi. Genome Announc. 2016, 4, e00511-16. [Google Scholar] [CrossRef] [Green Version]
- Pusey, P.L.; Stockwell, V.O.; Reardon, C.L.; Smits, T.H.M.; Duffy, B. Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology 2011, 101, 1234–1241. [Google Scholar] [CrossRef] [Green Version]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorenzi, A.S.; Bonatelli, M.L.; Chia, M.A.; Peressim, L.; Quecine, M.C. Opposite Sides of Pantoea agglomerans and Its Associated Commercial Outlook. Microorganisms 2022, 10, 2072. https://doi.org/10.3390/microorganisms10102072
Lorenzi AS, Bonatelli ML, Chia MA, Peressim L, Quecine MC. Opposite Sides of Pantoea agglomerans and Its Associated Commercial Outlook. Microorganisms. 2022; 10(10):2072. https://doi.org/10.3390/microorganisms10102072
Chicago/Turabian StyleLorenzi, Adriana Sturion, Maria Letícia Bonatelli, Mathias Ahii Chia, Leonardo Peressim, and Maria Carolina Quecine. 2022. "Opposite Sides of Pantoea agglomerans and Its Associated Commercial Outlook" Microorganisms 10, no. 10: 2072. https://doi.org/10.3390/microorganisms10102072
APA StyleLorenzi, A. S., Bonatelli, M. L., Chia, M. A., Peressim, L., & Quecine, M. C. (2022). Opposite Sides of Pantoea agglomerans and Its Associated Commercial Outlook. Microorganisms, 10(10), 2072. https://doi.org/10.3390/microorganisms10102072






