Molecular Epidemiology of mcr-1-Positive Escherichia coli and Klebsiella pneumoniae Isolates: Results from Russian Sentinel Surveillance (2013–2018)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Species Identification, Antimicrobial Susceptibility Testing and PCR Screening of mcr Genes
2.3. Whole-Genome Sequencing
2.4. Bioinformatics Analysis
2.5. Data Availability
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falagas, M.E.; Rafailidis, P.I. Nephrotoxicity of Colistin: New Insight into an Old Antibiotic. Clin. Infect. Dis. 2009, 48, 1729–1731. [Google Scholar] [CrossRef] [Green Version]
- Olaitan, A.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A microbiological and Molecular Biological Study. Lancet Infect Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Shen, Y.; Shen, J.; Wu, C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 2016, 16, 293. [Google Scholar] [CrossRef] [Green Version]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.-M. Epidemiology of Mobile Colistin Resistance (Mcr) Genes in Aquatic Environments. J Glob Antimicrob Resist 2021, 27, 51–62. [Google Scholar] [CrossRef]
- Li, X.-P.; Sun, R.-Y.; Song, J.-Q.; Fang, L.-X.; Zhang, R.-M.; Lian, X.-L.; Liao, X.-P.; Liu, Y.-H.; Lin, J.; Sun, J. Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int. J. Antimicrob. Agents 2020, 55, 105806. [Google Scholar] [CrossRef]
- Martiny, H.-M.; Munk, P.; Brinch, C.; Szarvas, J.; Aarestrup, F.M.; Petersen, T.N. Global Distribution of mcr Gene Variants in 214K Metagenomic Samples. mSystems 2022, 7, 2. [Google Scholar] [CrossRef]
- Matamoros, S.; van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Hoa, N.T.; Bootsma, M.C.J.; van Genderen, P.J.; et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. GLASS Whole-Genome Sequencing for Surveillance of Antimicrobial Resistance, Electronic version; World Health Organization: Geneva, Switzerland, 2020; ISBN 9789240011007. [Google Scholar]
- Smelikova, E.; Tkadlec, J.; Krutova, M. How to: Screening for mcr-mediated resistance to colistin. Clin. Microbiol. Infect. 2022, 28, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Tarabai, H.; Valcek, A.; Jamborova, I.; Vazhov, S.V.; Karyakin, I.V.; Raab, R.; Literak, I.; Dolejska, M. Plasmid-Mediated mcr-1 Colistin Resistance in Escherichia coli from a Black Kite in Russia. Antimicrob. Agents Chemother. 2019, 63, e01266-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ageevets, V.; Lazareva, I.; Mrugova, T.; Gostev, V.; Lobzin, Y.; Sidorenko, S. IncX4 plasmids harbouring mcr-1 genes: Further dissemination. J. Glob. Antimicrob. Resist. 2019, 18, 166–167. [Google Scholar] [CrossRef]
- Sulian, O.; Ageevets, V.; Lazareva, I.; Gostev, V.; Popov, D.; Vostrikova, T.; Sukhinin, A.; Lobzin, Y.; Sidorenko, S. Co-production of MCR-1 and NDM-1 by Escherichia coli sequence type 31 isolated from a newborn in Moscow, Russia. Int. J. Infect. Dis. 2020, 101, 4–5. [Google Scholar] [CrossRef]
- Kuleshov, K.V.; Pavlova, A.S.; Shedko, E.D.; Mikhaylova, Y.V.; Margos, G.; Hepner, S.; Chebotar, I.V.; Korneenko, E.V.; Podkolzin, A.T.; Akimkin, V.G. Mobile Colistin Resistance Genetic Determinants of Non-Typhoid Salmonella enterica Isolates from Russia. Microorganisms 2021, 9, 2515. [Google Scholar] [CrossRef]
- Kuzmenkov, A.Y.; Trushin, I.V.; Vinogradova, A.G.; Avramenko, A.A.; Sukhorukova, M.V.; Malhotra-Kumar, S.; Dekhnich, A.V.; Edelstein, M.V.; Kozlov, R.S. AMRmap: An Interactive Web Platform for Analysis of Antimicrobial Resistance Surveillance Data in Russia. Front. Microbiol. 2021, 12, 620002. [Google Scholar] [CrossRef]
- ISO. Available online: https://www.iso.org/standard/70464.html (accessed on 20 September 2022).
- EUCAST. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2022_manuals/ReadinR_guide_BMD_v_4.0_2022.pdf (accessed on 20 September 2022).
- EUCAST Clinical Breakpoints. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 20 September 2022).
- Daniels, J.B.; Campbell, D.; Boyd, S.; Ansari, U.; Lutgring, J.; Rasheed, J.K.; Halpin, A.L.; Sjölund-Karlsson, M. Development and Validation of a Clinical Laboratory Improvement Amendments-Compliant Multiplex Real-Time PCR Assay for Detection of mcr Genes. Microb. Drug Resist. 2019, 25, 991–996. [Google Scholar] [CrossRef]
- Simon Andrews FastQC. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 June 2022).
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglass, A.P.; O’Brien, C.E.; Offei, B.; Coughlan, A.Y.; Ortiz-Merino, R.A.; Butler, G.; Byrne, K.P.; Wolfe, K.H. Coverage-Versus-Length Plots, a Simple Quality Control Step for de Novo Yeast Genome Sequence Assemblies. G3 Genes Genomes Genet. 2019, 9, 879–887. [Google Scholar] [CrossRef] [Green Version]
- Antipov, D.; Raiko, M.; Lapidus, A.; Pevzner, P.A. Plasmid detection and assembly in genomic and metagenomic data sets. Genome Res. 2019, 29, 961–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanfear, R.; Schalamun, M.; Kainer, D.; Wang, W.; Schwessinger, B. MinIONQC: Fast and simple quality control for MinION sequencing data. Bioinformatics 2019, 35, 523–525. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Méric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus long-read assemblies for bacterial genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Ryan Wick Filtlong. Available online: https://github.com/rrwick/Filtlong (accessed on 12 June 2022).
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Holt, K.E. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Research 2019, 8, 2138. [Google Scholar] [CrossRef]
- Vaser, R.; Šikić, M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 2021, 1, 332–336. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies. Medaka. Available online: https://github.com/nanoporetech/medaka (accessed on 12 June 2022).
- Wick, R.R.; Holt, K.E. Polypolish: Short-read polishing of long-read bacterial genome assemblies. PLoS Comput. Biol. 2022, 18, e1009802. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Salzberg, S.L. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS Comput. Biol. 2020, 16, e1007981. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing Genomic Data Quality and Beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef] [PubMed]
- Torsten Seemann Mlst. Available online: https://github.com/tseemann/mlst (accessed on 17 June 2022).
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [Green Version]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roer, L.; Tchesnokova, V.; Allesøe, R.; Muradova, M.; Chattopadhyay, S.; Ahrenfeldt, J.; Thomsen, M.C.F.; Lund, O.; Hansen, F.; Hammerum, A.M.; et al. Development of a Web Tool for Escherichia coli Subtyping Based on fimH Alleles. J. Clin. Microbiol. 2017, 55, 2538–2543. [Google Scholar] [CrossRef] [Green Version]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Torsten Seemann Abricate. Available online: https://github.com/tseemann/abricate (accessed on 17 June 2022).
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Has-man, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typ-ing. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wise, M.G.; Estabrook, M.A.; Sahm, D.F.; Stone, G.G.; Kazmierczak, K.M. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PLoS ONE 2018, 13, e0195281. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Wang, Y.; Xiao, Y. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosaf. Health 2020, 2, 71–78. [Google Scholar] [CrossRef]
- Doumith, M.; Ellington, M.J.; Livermore, D.M.; Woodford, N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 2009, 63, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Dadashi, M.; Sameni, F.; Bostanshirin, N.; Yaslianifard, S.; Khosravi-Dehaghi, N.; Nasiri, M.J.; Goudarzi, M.; Hashemi, A.; Hajikhani, B. Global prevalence and molecular epidemiology of mcr-mediated colistin resistance in Escherichia coli clinical isolates: A systematic review. J. Glob. Antimicrob. Resist. 2022, 29, 444–461. [Google Scholar] [CrossRef] [PubMed]
- Lev, A.I.; Astashkin, E.I.; Kislichkina, A.A.; Solovieva, E.V.; Kombarova, T.I.; Korobova, O.V.; Ershova, O.N.; Alexandrova, I.A.; Malikov, V.E.; Bogun, A.G.; et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012–2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog. Glob. Health 2018, 112, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Shamina, O.; Kryzhanovskaya, O.; Lazareva, A.; Alyabieva, N.; Polikarpova, S.; Karaseva, O.; Mayanskiy, N. Emergence of a ST307 clone carrying a novel insertion element MITEKpn1 in the mgrB gene among carbapenem-resistant Klebsiella pneumoniae from Moscow, Russia. Int. J. Antimicrob. Agents 2020, 55, 105850. [Google Scholar] [CrossRef]
- Shaidullina, E.; Shelenkov, A.; Yanushevich, Y.; Mikhaylova, Y.; Shagin, D.; Alexandrova, I.; Ershova, O.; Akimkin, V.; Kozlov, R.; Edelstein, M. Antimicrobial Resistance and Genomic Characterization of OXA-48- and CTX-M-15-co-Producing Hypervirulent Klebsiella pneumoniae ST23 Recovered from Nosocomial Outbreak. Antibiotics 2020, 9, 862. [Google Scholar] [CrossRef] [PubMed]
- Fursova, N.K.; Astashkin, E.I.; Gabrielyan, N.I.; Novikova, T.S.; Fedyukina, G.N.; Kubanova, M.K.; Esenova, N.M.; Sharapchenko, S.O.; Volozhantsev, N.V. Emergence of Five Genetic Lines ST395NDM-1, ST13OXA-48, ST3346OXA-48, ST39CTX-M-14, and Novel ST3551OXA-48 of Multidrug-Resistant Clinical Klebsiella pneumoniae in Russia. Microb. Drug Resist. 2020, 26, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Fursova, N.K.; Astashkin, E.I.; Ershova, O.N.; Aleksandrova, I.A.; Savin, I.A.; Novikova, T.S.; Fedyukina, G.N.; Kislichkina, A.A.; Fursov, M.V.; Kuzina, E.S.; et al. Multidrug-Resistant Klebsiella pneumoniae Causing Severe Infections in the Neuro-ICU. Antibiotics 2021, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Khrulnova, S.; Fedorova, A.; Frolova, I.; Tandilova, K.; Likold, E.; Klyasova, G. Distribution of virulence genes and capsule types in Klebsiella pneumoniae among bloodstream isolates from patients with hematological malignancies. Diagn. Microbiol. Infect. Dis. 2022, 104, 115744. [Google Scholar] [CrossRef] [PubMed]
- Mmatli, M.; Marylucy Mbelle, N.; Sekyere, J.O. Global Epidemiology and Genetic Environment of Mcr Genes: A One Health Systematic Review of Current and Emerging Trends. medRxiv 2022. [Google Scholar] [CrossRef]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 117. [Google Scholar] [CrossRef] [PubMed]
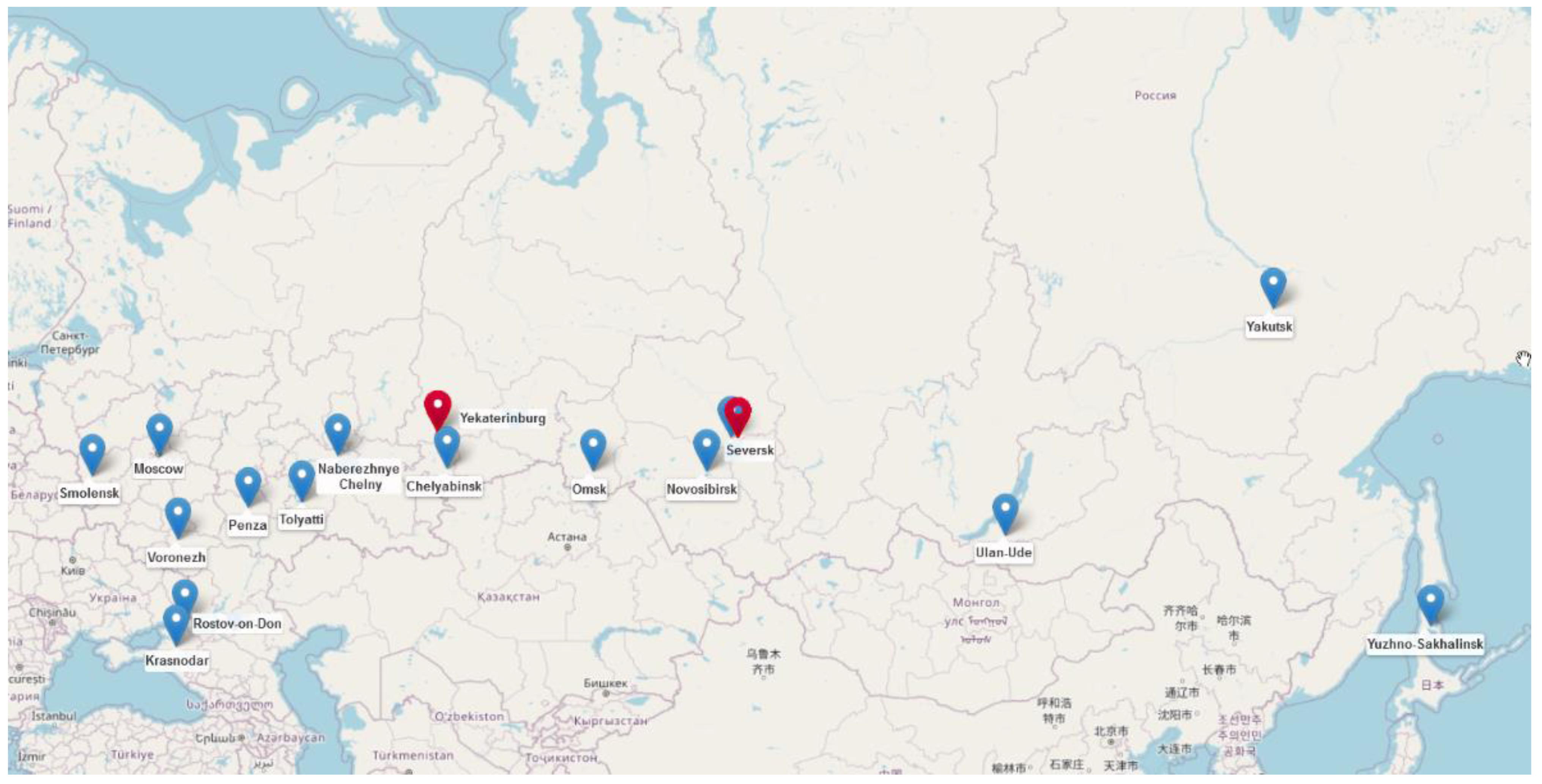
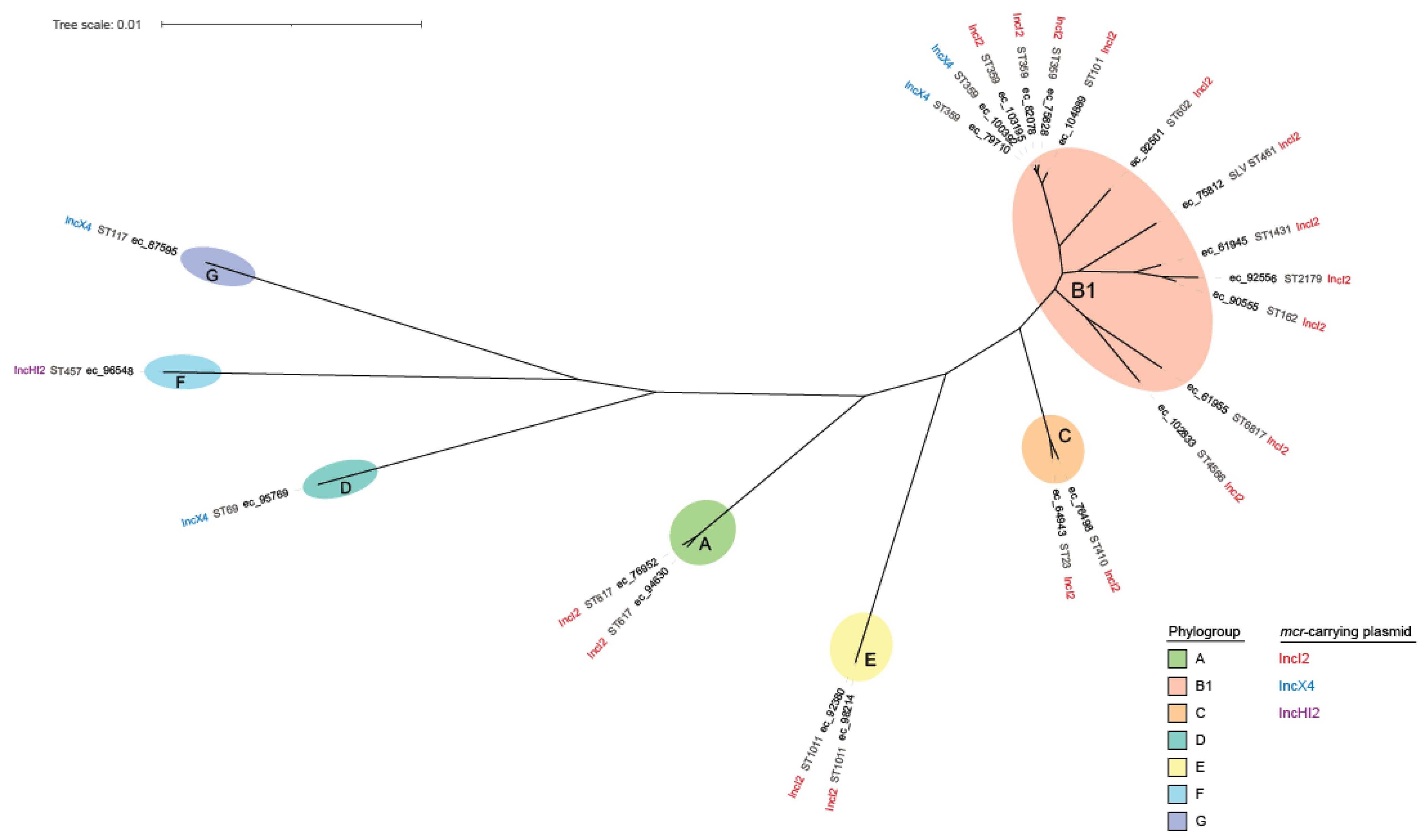
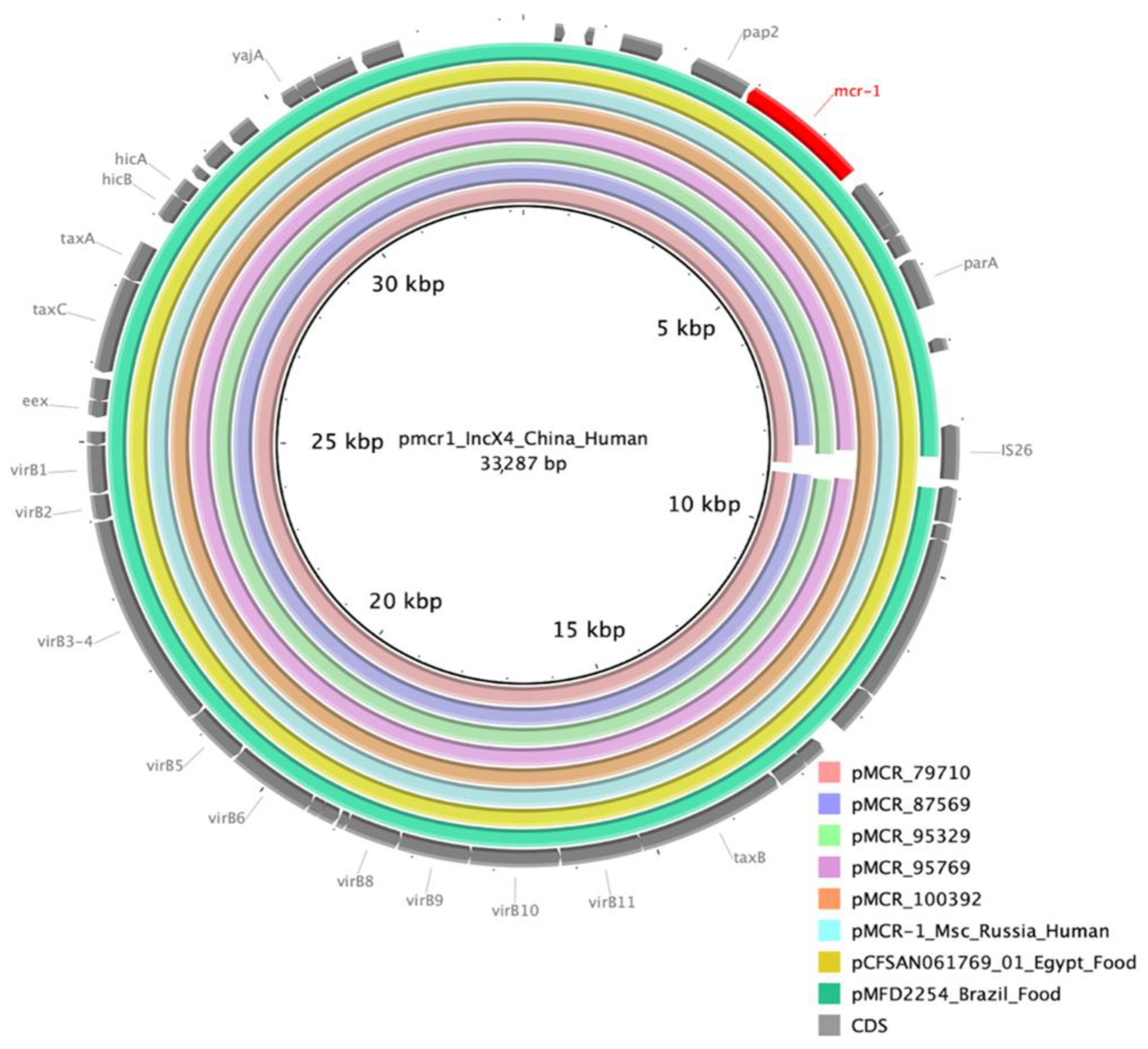
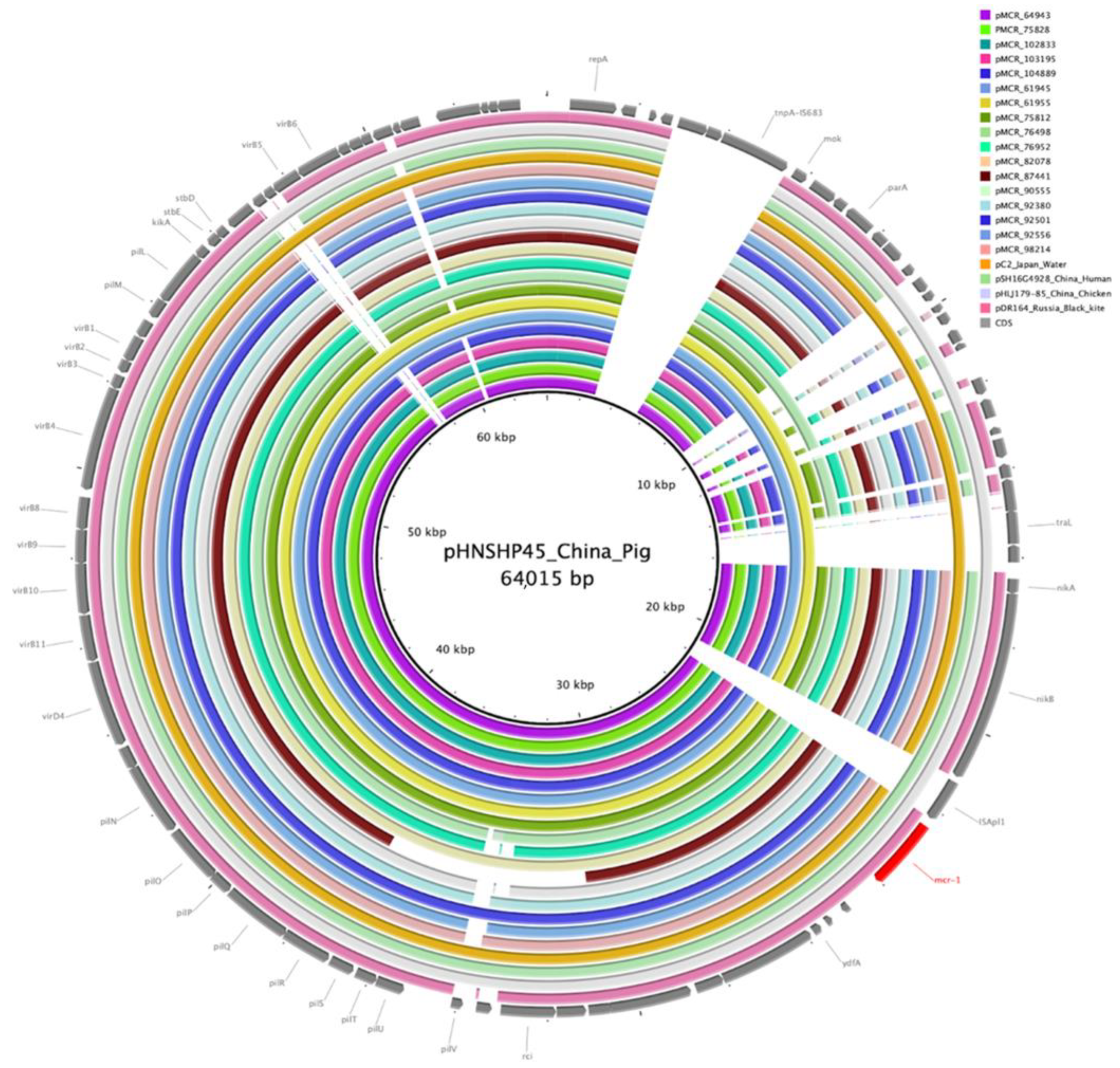
| ID | Collection Date | Phylo-Group | ST | Serotype | fimH Type | Plasmid Type (Length of mcr-1.1 Contig) | ARGs |
|---|---|---|---|---|---|---|---|
| ec_61945 | 1 January 2013 | B1 | ST1431 | O8:H19 | fimH32 | IncI2 (60859 bp) | mcr-1.1, blaCTX-M-1, aac(6′)-Ib-cr, arr-3, dfrA27, aadA16, qacEdelta1, sul1, sul2, tet(A) |
| ec_61955 | 5 August 2013 | B1 | ST6817 | O85:H23 | fimH31 | IncI2 (60860 bp) | mcr-1.1, blaTEM-1, blaCTX-M-15, dfrA12, aadA2, cmlA1, aadA1, qacL, tet(A), tet(M), sul3, aph(3′)-Ia |
| ec_64943 | 30 May 2014 | C | ST23 | O78:H4 | fimH35 | IncI2 (61030 bp) | mcr-1.1, catA1, dfrA17, aadA5, tet(B), blaTEM-1, sul2, aph(3″)-Ib, aph(6)-Id, aph(4)-Ia, aac(3)-IVa, qnrB19 |
| ec_75812 | 1 June 2015 | B1 | new (SLV ST461) | O178:H28 | fimH31 | IncI2 (60903 bp) | mcr-1.1, sul1, qacEdelta1, aac(3)-VIa, aadA1, qnrS1, blaCTX-M-15 |
| ec_75828 | 2 June 2015 | B1 | ST359 | O23:H21 | fimH35 | IncI2 (61036 bp) | mcr-1.1, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib, tet(B), blaTEM-1, sul2 |
| ec_76498 | 16 June 2015 | C | ST410 | O66:H45 | fimH24 | IncI2 (62806 bp) | mcr-1.1, blaCTX-M-27, floR, aac(6′)-Ib-cr, arr-3, dfrA27, aadA16, qacEdelta1, sul1, mph(A), blaTEM-1, aac(3)-IId, bleO, dfrA17, catA1, tet(B), blaTEM-1, sul2, aph(3″)-Ib, aph(6)-Id, aph(3′)-Ia, blaCMY-2 |
| ec_76952 | 22 October 2015 | A | ST617 | O101:H10 | fimH29 | IncI2 (59391 bp) | mcr-1.1, mph(A), catB3, blaOXA-1, aac(6′)-Ib-cr, aph(4)-Ia, aac(3)-IVa, aph(6)-Id, aph(3″)-Ib, tet(B), blaCTX-M-15, sul1, qacEdelta1, aadA5, dfrA17 |
| ec_79710 | 16 March 2016 | B1 | ST359 | O36:H31 | fimH35 | IncX4 (33106 bp) | mcr-1.1, blaCTX-M-1, tet(A), sul1, qacEdelta1, aac(3)-VIa, aadA1, aac(3)-IId, blaTEM-1, mph(A) |
| ec_82078 | 9 June 2016 | B1 | ST359 | O29:H31 | fimH35 | IncI2 (61024 bp) | mcr-1.1, tet(A), dfrA12, mph(A), sul1, qacEdelta1, cmlA1, aadA1, qacL, sul3, aph(4)-Ia, aac(3)-IVa, aph(6)-Id, aph(3″)-Ib, aadA2 |
| ec_87595 | 15 March 2017 | G | ST117 | O45:H51 | fimH97 | IncX4 (32744 bp) | mcr-1.1, blaCMY-2, mph(A), sul1, qacEdelta1, aadA2, dfrA12, tet(A), sul2, aph(3″)-Ib, aph(6)-Id |
| ec_90555 | 7 March 2017 | B1 | ST162 | O88:H21 | fimH32 | IncI2 (60006 bp) | mcr-1.1, blaTEM-1, blaCTX-M-65, mph(A), sul1, qacEdelta1, aadA5, dfrA17, aac(3)-IId, floR, fosA3, aph(3″)-Ib, aph(6)-Id, tet(A), sul2 |
| ec_92380 | 17 July 2017 | E | ST1011 | O188:H28 | fimH31 | IncI2 (60934 bp) | mcr-1.1, blaTEM-1, blaTEM-1, armA, msr(E), mph(E), blaCTX-M-15, blaCTX-M-15 |
| ec_92501 | 14 August 2017 | B1 | ST602 | O-*:H21 | fimH86 | IncI2 (61163 bp) | mcr-1.1, floR, blaTEM-1, aph(6)-Id, aph(3″)-Ib, dfrA17, aac(3)-IId, fosA7.5, sul2, catA1, blaCTX-M-169 |
| ec_92556 | 23 September 2017 | B1 | ST2179 | O9/09a:H9 | fimH32 | IncI2 (59951 bp) | mcr-1.1, blaTEM-1, blaCTX-M-65, tet(A), aph(6)-Id, aph(3″)-Ib, sul2, mph(A), floR |
| ec_94630 | 10 October 2017 | A | ST617 | O101/O9:H9 | * | IncI2 ** (23072 bp) | mcr-1.1, blaCTX-M-55, blaTEM, aph(3′)-IIa, ble, dfrA14, aac(6′)-Ib-cr, arr-3, dfrA27, aadA16, qacEdelta1, sul1, qnrB6, qacEdelta1, sul1, aph(6)-Id, aph(3″)-Ib, sul2, floR |
| ec_95769 | 28 November 2017 | D | ST69 | O15:H18 | fimH27 | IncX4 (32819 bp) | mcr-1.1, blaCTX-M-15, qnrS1, blaTEM-1, aph(6)-Id, aph(3″)-Ib, sul2, tet(A), dfrA14 |
| ec_96548 | 1 November 2017 | F | ST457 | O11:H25 | fimH145 | IncHI2 * (2325 bp) | mcr-1.1, blaTEM-1, aadA2, tet(A), sul3, qacL, aadA1, cmlA1, floR, sul1, qacEdelta1, lnu(F), dfrA14, mph(A) |
| ec_98214 | 22 March 2018 | E | ST1011 | O188:H28 | fimH31 | IncI2 (60456 bp) | mcr-1.1, blaTEM-1 |
| ec_100392 | 3 April 2018 | B1 | ST359 | O36:H31 | fimH35 | IncX4 (33437 bp) | mcr-1.1, blaCTX-M-55 |
| ec_102833 | 28 April 2018 | B1 | ST4566 | O163:H23 | fimH60 | IncI2 (61680 bp) | mcr-1.1, tet(A), sul2, blaCTX-M-1 |
| ec_103195 | 27 September 2018 | B1 | ST359 | O36:H31 | fimH35 | IncI2 (60820 bp) | mcr-1.1, sul2, blaCTX-M-1, tet(A), mph(A), dfrA17, aac(3)-IId, blaTEM-1 |
| ec_104889 | 31 October 2018 | B1 | ST101 | O115:H31 | fimH86 | IncI2 (61030 bp) | mcr-1.1, aph(3″)-Ib, aph(6)-Id, blaCTX-M-55, catA1, dfrA14, mph(A), tet(B) |
| ID | Collection Date | ST | K_Locus | O_Locus | Plasmid Type (Length of mcr-1.1 Contig) | ARGs |
|---|---|---|---|---|---|---|
| kp_87441 | 9 February 2017 | 307 | KL102 | O2v2 | IncI2 (55661 bp) | mcr-1.1, oqxB19, oqxA, blaSHV-28, fosA, sul3, qacL, aadA1, cmlA1, aadA2, sul2, aph(3″)-Ib, aph(6)-Id, blaTEM-1, blaCTX-M-15, dfrA14, aac(3)-IIe, qnrB1, tet(A), catB3, blaOXA-1, aac(6′)-Ib-cr, tet(A), catB3, blaOXA-1, aac(6′)-Ib-cr, tet(A), catB3, blaOXA-1, aac(6′)-Ib-cr |
| kp_95329 | 27 December 2017 | 23 | KL57 | O2v2 | IncX4 (32850 bp) | mcr-1.1, aac(3)-IIe, catB3, blaOXA-1, aac(6′)-Ib-cr, oqxA, oqxB, blaSHV-1, fosA, qnrS1, blaLAP-2, tet(A), sul2, blaCTX-M-55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapovalova, V.; Shaidullina, E.; Azizov, I.; Sheck, E.; Martinovich, A.; Dyachkova, M.; Matsvay, A.; Savochkina, Y.; Khafizov, K.; Kozlov, R.; et al. Molecular Epidemiology of mcr-1-Positive Escherichia coli and Klebsiella pneumoniae Isolates: Results from Russian Sentinel Surveillance (2013–2018). Microorganisms 2022, 10, 2034. https://doi.org/10.3390/microorganisms10102034
Shapovalova V, Shaidullina E, Azizov I, Sheck E, Martinovich A, Dyachkova M, Matsvay A, Savochkina Y, Khafizov K, Kozlov R, et al. Molecular Epidemiology of mcr-1-Positive Escherichia coli and Klebsiella pneumoniae Isolates: Results from Russian Sentinel Surveillance (2013–2018). Microorganisms. 2022; 10(10):2034. https://doi.org/10.3390/microorganisms10102034
Chicago/Turabian StyleShapovalova, Valeria, Elvira Shaidullina, Ilya Azizov, Eugene Sheck, Alexey Martinovich, Marina Dyachkova, Alina Matsvay, Yulia Savochkina, Kamil Khafizov, Roman Kozlov, and et al. 2022. "Molecular Epidemiology of mcr-1-Positive Escherichia coli and Klebsiella pneumoniae Isolates: Results from Russian Sentinel Surveillance (2013–2018)" Microorganisms 10, no. 10: 2034. https://doi.org/10.3390/microorganisms10102034
APA StyleShapovalova, V., Shaidullina, E., Azizov, I., Sheck, E., Martinovich, A., Dyachkova, M., Matsvay, A., Savochkina, Y., Khafizov, K., Kozlov, R., Shipulin, G., & Edelstein, M. (2022). Molecular Epidemiology of mcr-1-Positive Escherichia coli and Klebsiella pneumoniae Isolates: Results from Russian Sentinel Surveillance (2013–2018). Microorganisms, 10(10), 2034. https://doi.org/10.3390/microorganisms10102034








