Effect of Growth Media on the Diversity of Neocallimastigomycetes from Non-Rumen Habitats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Media Preparation and Enrichment of AF
2.3. DNA Extraction, Library Construction, and Illumina Sequencing
2.4. QIIME2 Pipeline
3. Results
3.1. AF Enrichment Cultures in Different Media
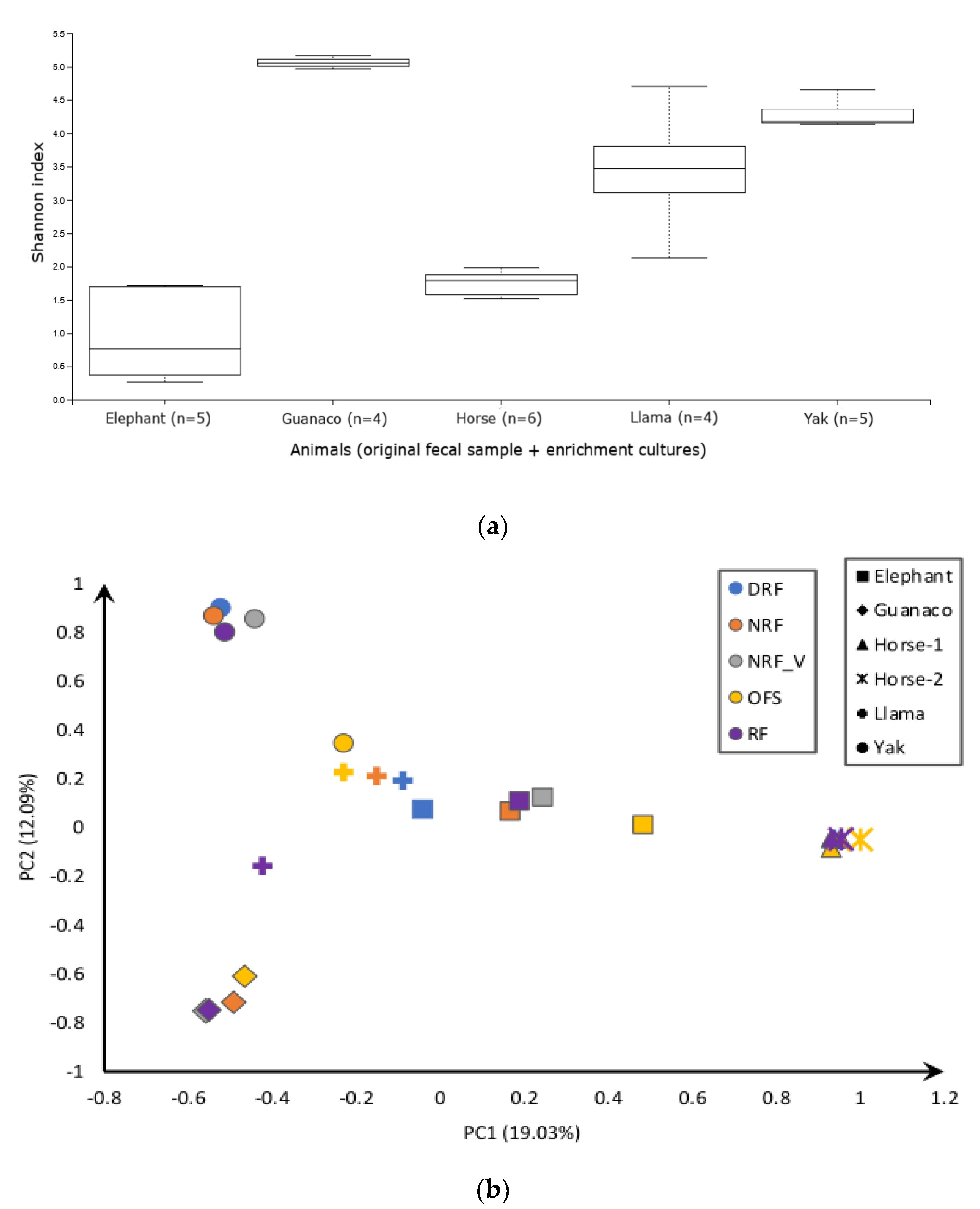
3.2. Sequencing Results and Diversity Analysis
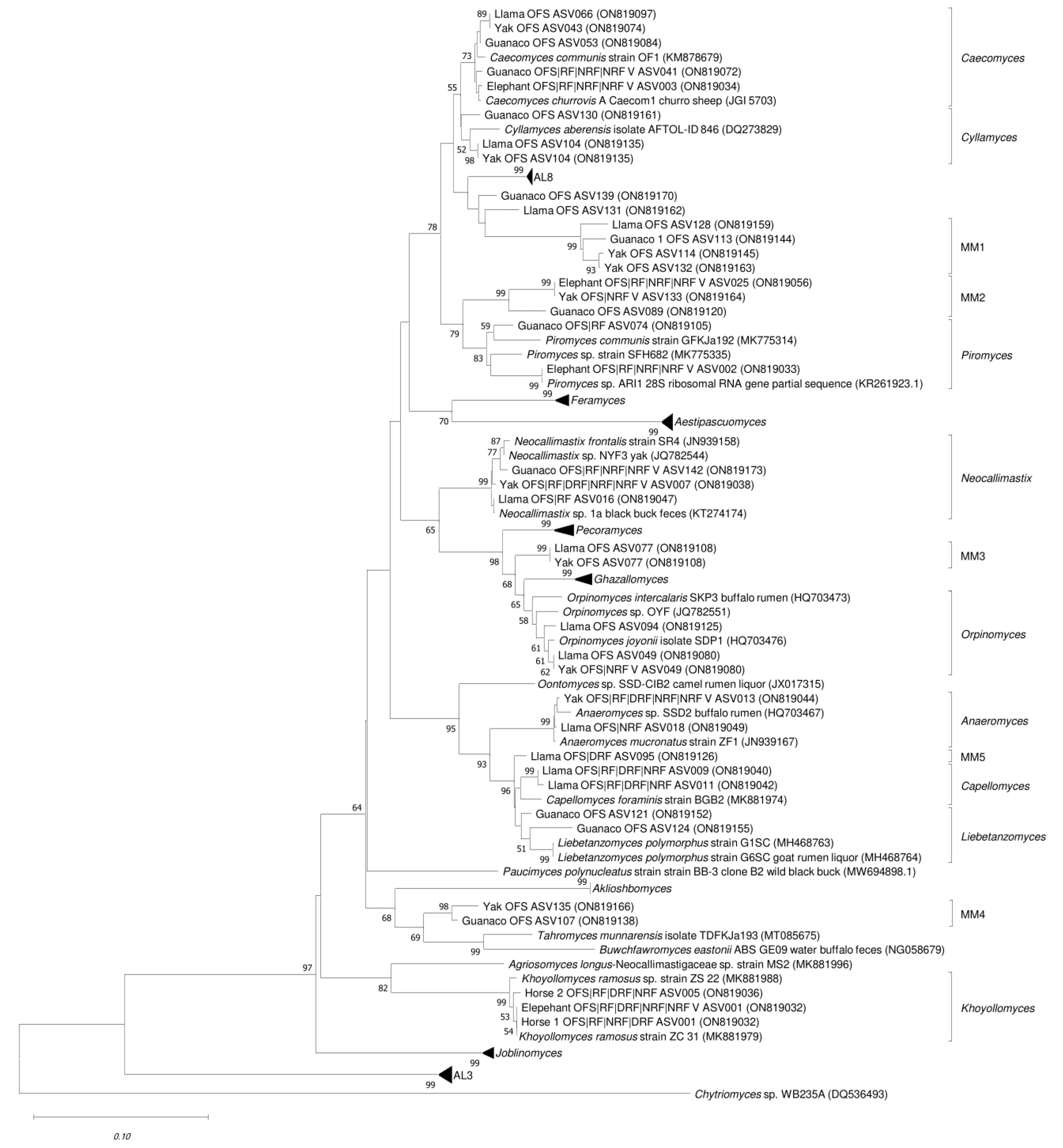
3.3. Phylogenetic Assignment
4. Discussion
4.1. Diversity of AF in Rumen and Non-Rumen Origin Samples
4.2. Influence of the Medium on the Observed Diversity of AF
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gruninger, R.J.; Puniya, A.K.; Callaghan, T.M.; Edwards, J.E.; Youssef, N.; Dagar, S.S.; Fliegerova, K.; Griffith, G.W.; Forster, R.; Tsang, A.; et al. Anaerobic Fungi (Phylum Neocallimastigomycota): Advances in Understanding Their Taxonomy, Life Cycle, Ecology, Role and Biotechnological Potential. FEMS Microbiol. Ecol. 2014, 90, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, I.B.; Bauchop, T.; Skipp, R.A. Assignment of the Rumen Anaerobe Neocallimastix frontalis to the Spizellomycetales (Chytridiomycetes) on the Basis of Its Polyflagellate Zoospore Ultrastructure. Can. J. Bot. 1983, 61, 295–307. [Google Scholar] [CrossRef]
- Orpin, C.G. Studies on the Rumen Flagellate Neocallimastix frontalis. Microbiology 1975, 91, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Hanafy, R.A.; Dagar, S.S.; Griffith, G.W.; Pratt, C.J.; Youssef, N.H.; Elshahed, M.S. Taxonomy of the Anaerobic Gut Fungi (Neocallimastigomycota): A Review of Classification Criteria and Description of Current Taxa. Int. J. Syst. Evol. Microbiol. 2022, 72, 005322. [Google Scholar] [CrossRef]
- Hess, M.; Paul, S.S.; Puniya, A.K.; van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerová, K. Anaerobic Fungi: Past, Present, and Future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef]
- Schulz, D.; Pšenková-Profousová, I.; Červená, B.; Procter, M.; Neba, T.F.; Modrý, D.; Petrželková, K.J.; Qablan, M.A. Occurrence and Diversity of Anaerobic Gut Fungi in Wild Forest Elephants and Buffaloes Inhabiting Two Separated Forest Ecosystems in Central West Africa. J. Vertebr. Biol. 2021, 71, 21033.1–13. [Google Scholar] [CrossRef]
- Hanafy, R.A.; Johnson, B.; Youssef, N.H.; Elshahed, M.S. Assessing Anaerobic Gut Fungal Diversity in Herbivores Using D1/D2 Large Ribosomal Subunit Sequencing and Multi-Year Isolation. Environ. Microbiol. 2020, 22, 3883–3908. [Google Scholar] [CrossRef] [PubMed]
- Young, D.; Joshi, A.; Huang, L.; Munk, B.; Wurzbacher, C.; Youssef, N.; Elshahed, M.; Moon, C.; Ochsenreither, K.; Griffith, G.W.; et al. Simultaneous Metabarcoding and Quantification of Neocallimastigomycetes from Environmental Samples: Insights into Community Composition and Novel Lineages. Microorganisms 2022, 10, 1749. [Google Scholar] [CrossRef]
- Trinci, A.P.J.; Davies, D.R.; Gull, K.; Lawrence, M.I.; Bonde Nielsen, B.; Rickers, A.; Theodorou, M.K. Anaerobic Fungi in Herbivorous Animals. Mycol. Res. 1994, 98, 129–152. [Google Scholar] [CrossRef]
- Li, J.; Heath, I.B.; Bauchop, T. Piromyces Mae and Piromyces Dumbonica, Two New Species of Uniflagellate Anaerobic Chytridiomycete Fungi from the Hindgut of the Horse and Elephant. Can. J. Bot. 1990, 68, 1021–1033. [Google Scholar] [CrossRef]
- Bauchop, T. Rumen Anaerobic Fungi of Cattle and Sheep. Appl. Environ. Microbiol. 1979, 38, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Joshi, A.; Lanjekar, V.B.; Dhakephalkar, P.K.; Callaghan, T.M.; Griffith, G.W.; Dagar, S.S. Liebetanzomyces polymorphus Gen. et Sp. Nov., a New Anaerobic Fungus (Neocallimastigomycota) Isolated from the Rumen of a Goat. MycoKeys 2018, 40, 89–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elshahed, M.S.; Hanafy, R.A.; Cheng, Y.; Dagar, S.S.; Edwards, J.E.; Flad, V.; Fliegerová, K.O.; Griffith, G.W.; Kittelmann, S.; Lebuhn, M.; et al. Characterization and Rank Assignment Criteria for the Anaerobic Fungi (Neocallimastigomycota). Int. J. Syst. Evol. Microbiol. 2022, 72, 005449. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, R.A.; Elshahed, M.S.; Liggenstoffer, A.S.; Griffith, G.W.; Youssef, N.H. Pecoramyces ruminantium, Gen. Nov., Sp. Nov., an Anaerobic Gut Fungus from the Feces of Cattle and Sheep. Mycologia 2017, 109, 231–243. [Google Scholar] [CrossRef]
- Liggenstoffer, A.S.; Youssef, N.H.; Couger, M.B.; Elshahed, M.S. Phylogenetic Diversity and Community Structure of Anaerobic Gut Fungi (Phylum Neocallimastigomycota) in Ruminant and Non-Ruminant Herbivores. ISME J. 2010, 4, 1225–1235. [Google Scholar] [CrossRef]
- Schulz, D.; Qablan, M.A.; Profousova-Psenkova, I.; Vallo, P.; Fuh, T.; Modry, D.; Piel, A.K.; Stewart, F.; Petrzelkova, K.J.; Fliegerová, K. Anaerobic Fungi in Gorilla (Gorilla gorilla gorilla) Feces: An Adaptation to a High-Fiber Diet? Int. J. Primatol. 2018, 39, 567–580. [Google Scholar] [CrossRef]
- Paul, S.S.; Bu, D.; Xu, J.; Hyde, K.D.; Yu, Z. A Phylogenetic Census of Global Diversity of Gut Anaerobic Fungi and a New Taxonomic Framework. Fungal Divers. 2018, 89, 253–266. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Groenewald, J.Z. Phylogeny of Anaerobic Fungi (Phylum Neocallimastigomycota), with Contributions from Yak in China. Antonie Leeuwenhoek 2017, 110, 87–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orpin, C.G. Isolation of Cellulolytic Phycomycete Fungi from the Caecum of the Horse. J. Gen. Microbiol. 1981, 123, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Orpin, C.G. Studies on the Rumen Flagellate Sphaeromonas communis. Microbiology 1976, 94, 270–280. [Google Scholar] [CrossRef]
- Orpin, C.G. The Rumen Flagellate Piromonas communis: Its Life-History and Invasion of Plant Material in the Rumen. Microbiology 1977, 99, 107–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, D.R.; Theodorou, M.K.; Lawrence, M.I.; Trinci, A.P. Distribution of Anaerobic Fungi in the Digestive Tract of Cattle and Their Survival in Faeces. J. Gen. Microbiol. 1993, 139 Pt 6, 1395–1400. [Google Scholar] [CrossRef] [Green Version]
- Gold, J.J.; Brent Heath, I.; Bauchop, T. Ultrastructural Description of a New Chytrid Genus of Caecum Anaerobe, Caecomyces equi Gen. Nov., Sp. Nov., Assigned to the Neocallimasticaceae. Biosystems 1988, 21, 403–415. [Google Scholar] [CrossRef]
- Teunissen, M.J.; Op den Camp, H.J.; Orpin, C.G.; Huis in’t Veld, J.H.; Vogels, G.D. Comparison of Growth Characteristics of Anaerobic Fungi Isolated from Ruminant and Non-Ruminant Herbivores during Cultivation in a Defined Medium. J. Gen. Microbiol. 1991, 137, 1401–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calkins, S.; Elledge, N.C.; Hanafy, R.A.; Elshahed, M.S.; Youssef, N. A Fast and Reliable Procedure for Spore Collection from Anaerobic Fungi: Application for RNA Uptake and Long-Term Storage of Isolates. J. Microbiol. Methods 2016, 127, 206–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, S.E.; Theodorou, M.K.; Trinci, A.P.J.; Hespell, R.B. Growth of Anaerobic Rumen Fungi on Defined and Semi-Defined Media Lacking Rumen Fluid. Microbiology 1985, 131, 2225–2229. [Google Scholar] [CrossRef] [Green Version]
- Caldwell, D.R.; Bryant, M.P. Medium Without Rumen Fluid for Nonselective Enumeration and Isolation of Rumen Bacteria. Appl. Microbiol. 1966, 14, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Dehority, B.A.; Grubb, J.A. Basal Medium for the Selective Enumeration of Rumen Bacteria Utilizing Specific Energy Sources. Appl. Environ. Microbiol. 1976, 32, 703–710. [Google Scholar] [CrossRef] [Green Version]
- Haitjema, C.H.; Solomon, K.V.; Henske, J.K.; Theodorou, M.K.; O’Malley, M.A. Anaerobic Gut Fungi: Advances in Isolation, Culture, and Cellulolytic Enzyme Discovery for Biofuel Production. Biotechnol. Bioeng. 2014, 111, 1471–1482. [Google Scholar] [CrossRef]
- Hungate, R.E.; Bryant, M.P.; Mah, R.A. The Rumen Bacteria and Protozoa. Annu. Rev. Microbiol. 1964, 18, 131–166. [Google Scholar] [CrossRef] [PubMed]
- Stabel, M.; Hanafy, R.A.; Schweitzer, T.; Greif, M.; Aliyu, H.; Flad, V.; Young, D.; Lebuhn, M.; Elshahed, M.S.; Ochsenreither, K.; et al. Aestipascuomyces dupliciliberans Gen. Nov, Sp. Nov., the First Cultured Representative of the Uncultured SK4 Clade from Aoudad Sheep and Alpaca. Microorganisms 2020, 8, 1734. [Google Scholar] [CrossRef] [PubMed]
- McGranaghan, P.; Davies, J.C.; Griffith, G.W.; Davies, D.R.; Theodorou, M.K. The Survival of Anaerobic Fungi in Cattle Faeces. FEMS Microbiol. Ecol. 1999, 29, 293–300. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. A Serum Bottle Modification of the Hungate Technique for Cultivating Obligate Anaerobes. Appl. Microbiol. 1974, 27, 985–987. [Google Scholar] [CrossRef] [PubMed]
- Leedle, J.A.; Hespell, R.B. Differential Carbohydrate Media and Anaerobic Replica Plating Techniques in Delineating Carbohydrate-Utilizing Subgroups in Rumen Bacterial Populations. Appl. Environ. Microbiol. 1980, 39, 709–719. [Google Scholar] [CrossRef] [Green Version]
- Tophinke, A.H.; Joshi, A.; Baier, U.; Hufenus, R.; Mitrano, D.M. Systematic Development of Extraction Methods for Quantitative Microplastics Analysis in Soils Using Metal-Doped Plastics. Environ. Pollut. 2022, 311, 119933. [Google Scholar] [CrossRef]
- Lebuhn, M.; Effenberger, M.; Gronauer, A.; Wilderer, P.A.; Wuertz, S. Using Quantitative Real-Time PCR to Determine the Hygienic Status of Cattle Manure. Water Sci. Technol. 2003, 48, 97–103. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Reitmeier, S.; Hitch, T.C.A.; Treichel, N.; Fikas, N.; Hausmann, B.; Ramer-Tait, A.E.; Neuhaus, K.; Berry, D.; Haller, D.; Lagkouvardos, I.; et al. Handling of Spurious Sequences Affects the Outcome of High-Throughput 16S rRNA Gene Amplicon Profiling. ISME Commun. 2021, 1, 31. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Fliegerova, K.O.; Podmirseg, S.M.; Vinzelj, J.; Grilli, D.J.; Kvasnová, S.; Schierová, D.; Sechovcová, H.; Mrázek, J.; Siddi, G.; Arenas, G.N.; et al. The Effect of a High-Grain Diet on the Rumen Microbiome of Goats with a Special Focus on Anaerobic Fungi. Microorganisms 2021, 9, 157. [Google Scholar] [CrossRef] [PubMed]
- Mura, E.; Edwards, J.; Kittelmann, S.; Kaerger, K.; Voigt, K.; Mrázek, J.; Moniello, G.; Fliegerova, K. Anaerobic Fungal Communities Differ along the Horse Digestive Tract. Fungal Biol. 2019, 123, 240–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, M.J.; McSweeney, C.S.; Mackie, R.I.; Brookman, J.L.; Theodorou, M.K. Diversity of Anaerobic Gut Fungal Populations Analysed Using Ribosomal ITS1 Sequences in Faeces of Wild and Domesticated Herbivores. Anaerobe 2010, 16, 66–73. [Google Scholar] [CrossRef]
- Rabee, A.E.; Forster, R.J.; Elekwachi, C.O.; Kewan, K.Z.; Sabra, E.A.; Shawket, S.M.; Mahrous, H.A.; Khamiss, O.A. Community Structure and Fibrolytic Activities of Anaerobic Rumen Fungi in Dromedary Camels. J. Basic Microbiol. 2019, 59, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, P.; Liu, X.; Zhang, C.; Lu, Q.; Xi, D.; Yang, R.; Wang, S.; Bai, W.; Yang, Z.; et al. The Composition of Fungal Communities in the Rumen of Gayals (Bos frontalis), Yaks (Bos grunniens), and Yunnan and Tibetan Yellow Cattle (Bos taurs). Pol. J. Microbiol. 2019, 68, 505–514. [Google Scholar] [CrossRef] [Green Version]
- Sijtsma, L.; Tan, B. Degradation and Utilization of Grass Cell Walls by Anaerobic Fungi Isolated from Yak, Llama and Sheep. Anim. Feed Sci. Technol. 1993, 44, 221–236. [Google Scholar] [CrossRef]
- Marvin-Sikkema, F.D.; Lahpor, G.A.; Kraak, M.N.; Gottschal, J.C.; Prins, R.A. Characterization of an Anaerobic Fungus from Ilama Faeces. Microbiology 1992, 138, 2235–2241. [Google Scholar] [CrossRef] [Green Version]
- Dijkerman, R.; Ledeboer, J.; Op den Camp, H.J.M.; Prins, R.A.; van der Drift, C. The Anaerobic Fungus Neocallimastix Sp. Strain L2: Growth and Production of (Hemi)Cellulolytic Enzymes on a Range of Carbohydrate Substrates. Curr. Microbiol. 1997, 34, 91–96. [Google Scholar] [CrossRef]
- Sirohi, S.K.; Choudhury, P.K.; Dagar, S.S.; Puniya, A.K.; Singh, D. Isolation, Characterization and Fibre Degradation Potential of Anaerobic Rumen Fungi from Cattle. Ann. Microbiol. 2013, 63, 1187–1194. [Google Scholar] [CrossRef]
- Theodorou, M.K.; Gill, M.; King-Spooner, C.; Beever, D.E. Enumeration of Anaerobic Chytridiomycetes as Thallus-Forming Units: Novel Method for Quantification of Fibrolytic Fungal Populations from the Digestive Tract Ecosystem. Appl. Environ. Microbiol. 1990, 56, 1073–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wubah, D.A.; Fuller, M.S.; Akin, D.E. Resistant Body Formation in Neocallimastix Sp., An Anaerobic Fungus from the Rumen of a Cow. Mycologia 1991, 83, 40–47. [Google Scholar] [CrossRef]
- Minich, J.J.; Sanders, J.G.; Amir, A.; Humphrey, G.; Gilbert, J.A.; Knight, R. Quantifying and Understanding Well-to-Well Contamination in Microbiome Research. mSystems 2019, 4, e00186-19. [Google Scholar] [CrossRef] [Green Version]
- De Aguiar, S.C.; Zeoula, L.M.; do Prado, O.P.P.; Arcuri, P.B.; Forano, E. Characterization of Rumen Bacterial Strains Isolated from Enrichments of Rumen Content in the Presence of Propolis. World J. Microbiol. Biotechnol. 2014, 30, 2917–2926. [Google Scholar] [CrossRef] [PubMed]
- Ziemer, C.J. Newly Cultured Bacteria with Broad Diversity Isolated from Eight-Week Continuous Culture Enrichments of Cow Feces on Complex Polysaccharides. Appl. Environ. Microbiol. 2014, 80, 574–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swift, C.L.; Louie, K.B.; Bowen, B.P.; Hooker, C.A.; Solomon, K.V.; Singan, V.; Daum, C.; Pennacchio, C.P.; Barry, K.; Shutthanandan, V.; et al. Cocultivation of Anaerobic Fungi with Rumen Bacteria Establishes an Antagonistic Relationship. mBio 2021, 12, e0144221. [Google Scholar] [CrossRef] [PubMed]
- Stabel, M.; Schweitzer, T.; Haack, K.; Gorenflo, P.; Aliyu, H.; Ochsenreither, K. Isolation and Biochemical Characterization of Six Anaerobic Fungal Strains from Zoo Animal Feces. Microorganisms 2021, 9, 1655. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, A.; Young, D.; Huang, L.; Mosberger, L.; Munk, B.; Vinzelj, J.; Flad, V.; Sczyrba, A.; Griffith, G.W.; Podmirseg, S.M.; et al. Effect of Growth Media on the Diversity of Neocallimastigomycetes from Non-Rumen Habitats. Microorganisms 2022, 10, 1972. https://doi.org/10.3390/microorganisms10101972
Joshi A, Young D, Huang L, Mosberger L, Munk B, Vinzelj J, Flad V, Sczyrba A, Griffith GW, Podmirseg SM, et al. Effect of Growth Media on the Diversity of Neocallimastigomycetes from Non-Rumen Habitats. Microorganisms. 2022; 10(10):1972. https://doi.org/10.3390/microorganisms10101972
Chicago/Turabian StyleJoshi, Akshay, Diana Young, Liren Huang, Lona Mosberger, Bernhard Munk, Julia Vinzelj, Veronika Flad, Alexander Sczyrba, Gareth W. Griffith, Sabine Marie Podmirseg, and et al. 2022. "Effect of Growth Media on the Diversity of Neocallimastigomycetes from Non-Rumen Habitats" Microorganisms 10, no. 10: 1972. https://doi.org/10.3390/microorganisms10101972
APA StyleJoshi, A., Young, D., Huang, L., Mosberger, L., Munk, B., Vinzelj, J., Flad, V., Sczyrba, A., Griffith, G. W., Podmirseg, S. M., Warthmann, R., Lebuhn, M., & Insam, H. (2022). Effect of Growth Media on the Diversity of Neocallimastigomycetes from Non-Rumen Habitats. Microorganisms, 10(10), 1972. https://doi.org/10.3390/microorganisms10101972









