Genome-Wide Association Study of Listeria monocytogenes Isolates Causing Three Different Clinical Outcomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Genome Collection
2.2. Phenotype Designations
2.3. Whole Genome Sequencing (WGS)
2.4. Lineage Determination and In Silico MLST Assignment
2.5. Pan and Core Genome Analyses and Phylogenetic Reconstruction Based on Core Genome SNPs
2.6. Virulence Gene Screening
2.7. Gene-Based and SNP-Based GWAS
3. Results
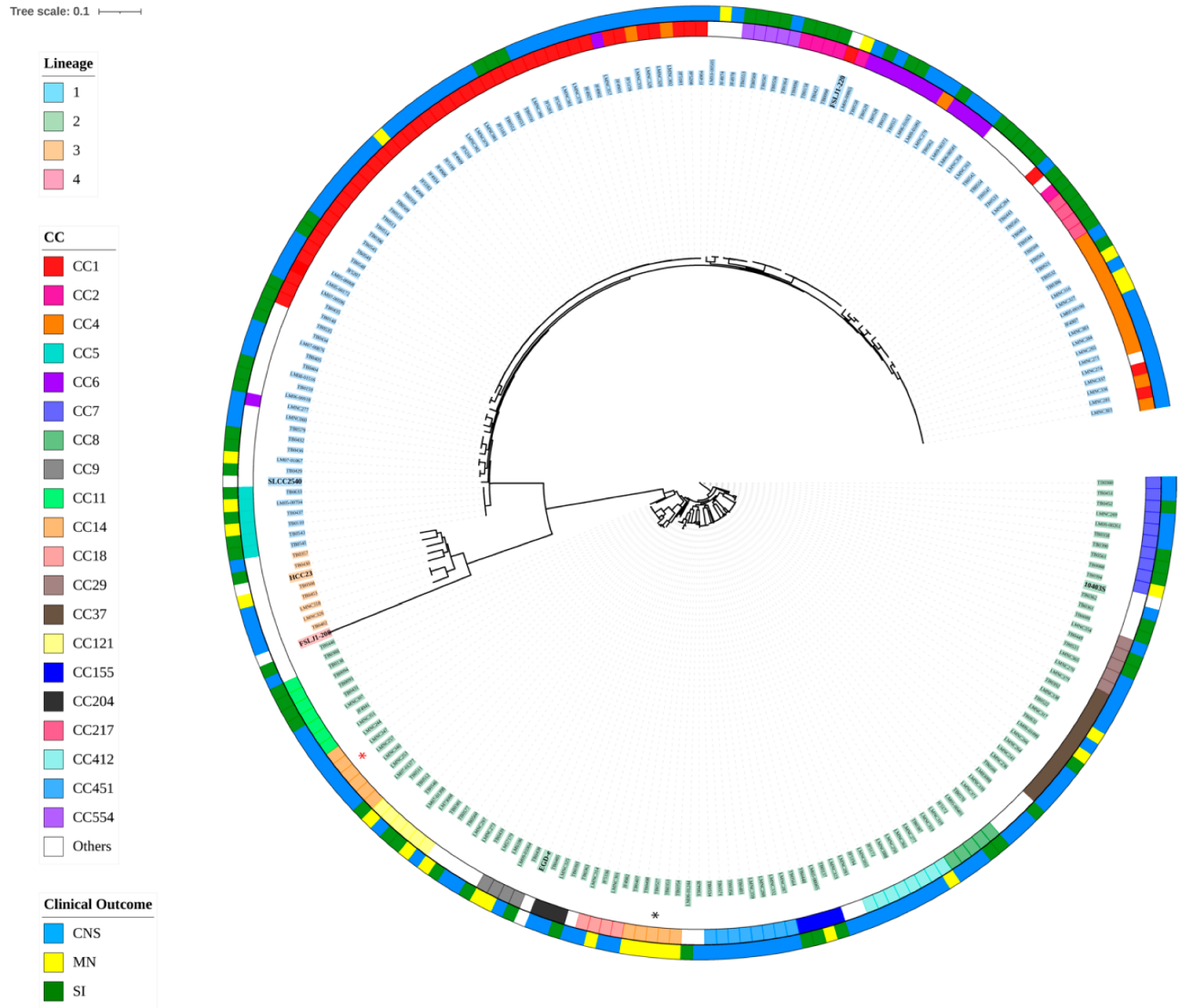
3.1. Variant Calling at the Core Genome Level and Phylogenetic Relationship Determination
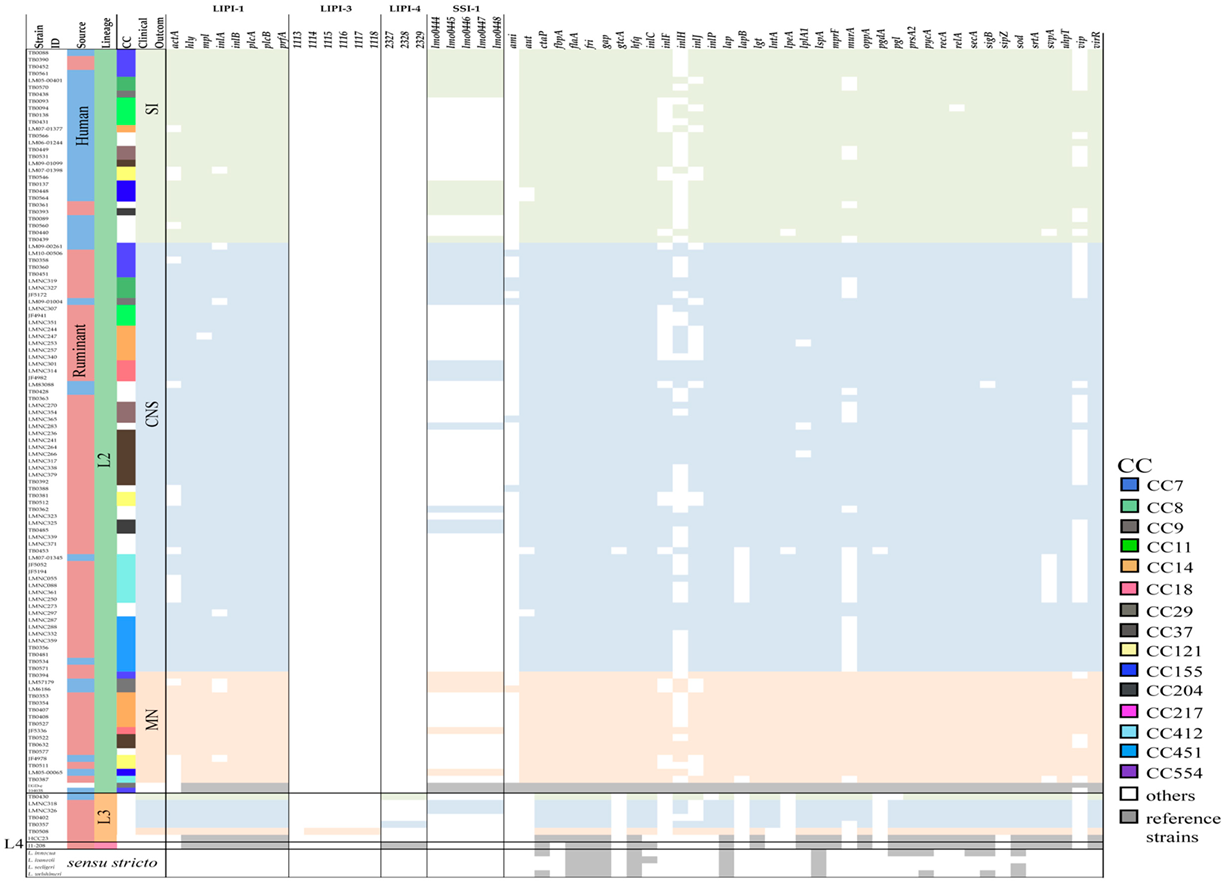
3.2. Distribution of Virulence Genes from Listeriosis Cases Causing CNS, MN, and SI
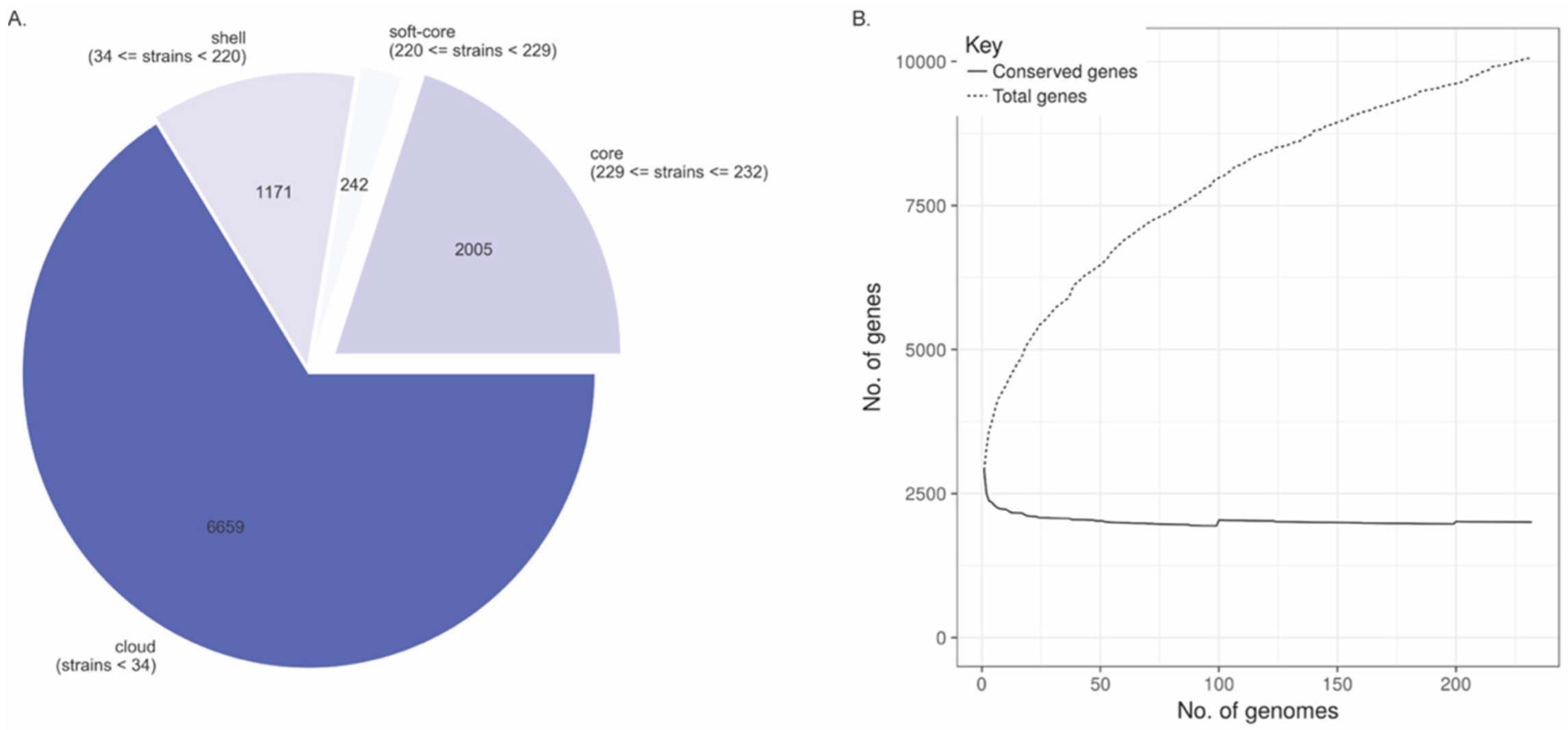
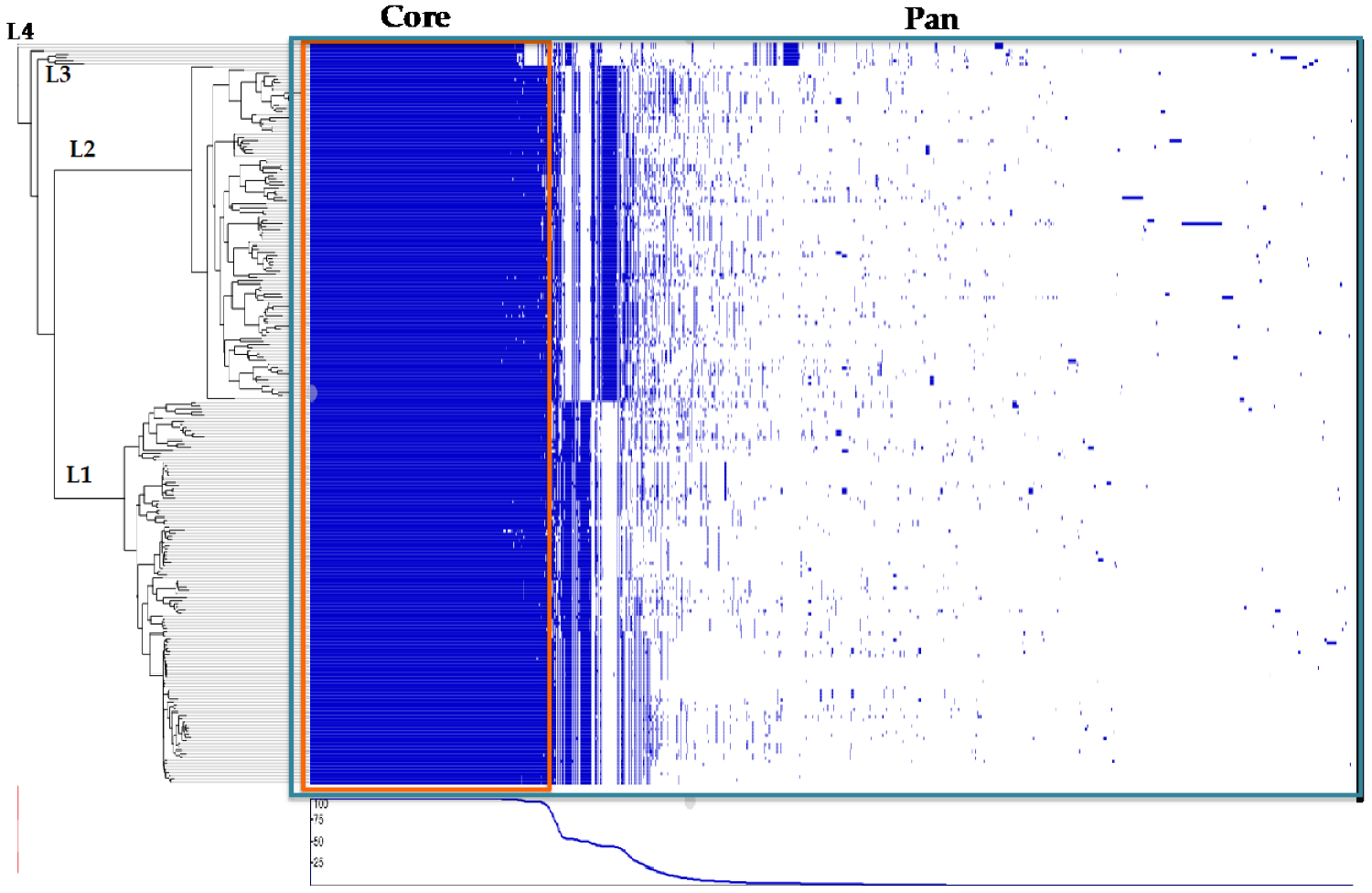
3.3. Pan and Core Genome Analyses
3.4. Gene Association with Clinical Outcomes
3.5. Core-SNPs Association with Clinical Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dreyer, M.; Aguilar-Bultet, L.; Rupp, S.; Guldimann, C.; Stephan, R.; Schock, A.; Otter, A.; Schüpbach, G.; Brisse, S.; Lecuit, M.; et al. Listeria Monocytogenes Sequence Type 1 Is Predominant in Ruminant Rhombencephalitis. Sci. Rep. 2016, 6, 36419. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.-H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria Monocytogenes Hypervirulence by Harnessing Its Biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Rupp, S.; Bärtschi, M.; Frey, J.; Oevermann, A. Hyperinvasiveness and Increased Intercellular Spread of Listeria Monocytogenes Sequence Type 1 Are Independent of Listeriolysin S, Internalin F and Internalin J1. J. Med. Microbiol. 2017, 66, 1053–1062. [Google Scholar] [CrossRef]
- Papić, B.; Pate, M.; Félix, B.; Kušar, D. Genetic Diversity of Listeria Monocytogenes Strains in Ruminant Abortion and Rhombencephalitis Cases in Comparison with the Natural Environment. BMC Microbiol. 2019, 19, 299. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria Monocytogenes Clones’ Adaption to Mammalian Gut Accounts for Their Association with Dairy Products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef]
- Aguilar-Bultet, L.; Nicholson, P.; Rychener, L.; Dreyer, M.; Gözel, B.; Origgi, F.C.; Oevermann, A.; Frey, J.; Falquet, L. Genetic Separation of Listeria Monocytogenes Causing Central Nervous System Infections in Animals. Front. Cell. Infect. Microbiol. 2018, 8, 20. [Google Scholar] [CrossRef]
- Faralla, C.; Rizzuto, G.A.; Lowe, D.E.; Kim, B.; Cooke, C.; Shiow, L.R.; Bakardjiev, A.I. InlP, a New Virulence Factor with Strong Placental Tropism. Infect. Immun. 2016, 84, 3584–3596. [Google Scholar] [CrossRef]
- Hingston, P.; Chen, J.; Dhillon, B.K.; Laing, C.; Bertelli, C.; Gannon, V.; Tasara, T.; Allen, K.; Brinkman, F.S.L.; Truelstrup Hansen, L.; et al. Genotypes Associated with Listeria Monocytogenes Isolates Displaying Impaired or Enhanced Tolerances to Cold, Salt, Acid, or Desiccation Stress. Front. Microbiol. 2017, 8, 369. [Google Scholar] [CrossRef]
- Haley, B.J.; Sonnier, J.; Schukken, Y.H.; Karns, J.S.; Van Kessel, J.A.S. Diversity of Listeria Monocytogenes Within a U.S. Dairy Herd, 2004–2010. Foodborne Pathog. Dis. 2015, 12, 844–850. [Google Scholar] [CrossRef]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a Novel Peptide Haemolysin Associated with a Subset of Lineage I Listeria Monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef]
- Bagatella, S.; Tavares-Gomes, L.; Oevermann, A. Listeria Monocytogenes at the Interface between Ruminants and Humans: A Comparative Pathology and Pathogenesis Review. Vet. Pathol. 2022, 59, 186–210. [Google Scholar] [CrossRef]
- Hilliard, A.; Leong, D.; O’Callaghan, A.; Culligan, E.; Morgan, C.; DeLappe, N.; Hill, C.; Jordan, K.; Cormican, M.; Gahan, C. Genomic Characterization of Listeria Monocytogenes Isolates Associated with Clinical Listeriosis and the Food Production Environment in Ireland. Genes 2018, 9, 171. [Google Scholar] [CrossRef]
- Kathariou, S.; Evans, P.; Dutta, V. Strain-Specific Virulence Differences in Listeria Monocytogenes: Current Perspectives in Addressing an Old and Vexing Issue. In Foodborne Pathogens; Gurtler, J.B., Doyle, M.P., Kornacki, J.L., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 61–92. ISBN 978-3-319-56834-8. [Google Scholar]
- Quereda, J.J.; Meza-Torres, J.; Cossart, P.; Pizarro-Cerdá, J. Listeriolysin S: A Bacteriocin from Epidemic Listeria Monocytogenes Strains That Targets the Gut Microbiota. Gut Microbes 2017, 8, 384–391. [Google Scholar] [CrossRef]
- Parsons, C.; Lee, S.; Kathariou, S. Dissemination and Conservation of Cadmium and Arsenic Resistance Determinants in Listeria and Other Gram-positive Bacteria. Mol. Microbiol. 2020, 113, 560–569. [Google Scholar] [CrossRef]
- Rupp, S.; Aguilar-Bultet, L.; Jagannathan, V.; Guldimann, C.; Drögemüller, C.; Pfarrer, C.; Vidondo, B.; Seuberlich, T.; Frey, J.; Oevermann, A. A Naturally Occurring PrfA Truncation in a Listeria Monocytogenes Field Strain Contributes to Reduced Replication and Cell-to-Cell Spread. Vet. Microbiol. 2015, 179, 91–101. [Google Scholar] [CrossRef]
- Camejo, A.; Carvalho, F.; Reis, O.; Leitão, E.; Sousa, S.; Cabanes, D. The Arsenal of Virulence Factors Deployed by Listeria Monocytogenes to Promote Its Cell Infection Cycle. Virulence 2011, 2, 379–394. [Google Scholar] [CrossRef]
- Berthenet, E.; Yahara, K.; Thorell, K.; Pascoe, B.; Meric, G.; Mikhail, J.M.; Engstrand, L.; Enroth, H.; Burette, A.; Megraud, F.; et al. A GWAS on Helicobacter Pylori Strains Points to Genetic Variants Associated with Gastric Cancer Risk. BMC Biol. 2018, 16, 84. [Google Scholar] [CrossRef]
- Collins, C.; Didelot, X. A Phylogenetic Method to Perform Genome-Wide Association Studies in Microbes That Accounts for Population Structure and Recombination. PLoS Comput. Biol. 2018, 14, e1005958. [Google Scholar] [CrossRef]
- Fritsch, L.; Felten, A.; Palma, F.; Mariet, J.-F.; Radomski, N.; Mistou, M.-Y.; Augustin, J.-C.; Guillier, L. Insights from Genome-Wide Approaches to Identify Variants Associated to Phenotypes at Pan-Genome Scale: Application to L. Monocytogenes’ Ability to Grow in Cold Conditions. Int. J. Food Microbiol. 2019, 291, 181–188. [Google Scholar] [CrossRef]
- Melnyk, R.A.; Hossain, S.S.; Haney, C.H. Convergent Gain and Loss of Genomic Islands Drive Lifestyle Changes in Plant-Associated Pseudomonas. ISME J. 2019, 13, 1575–1588. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Didelot, X.; Meric, G.; Torralbo, A.; Jolley, K.A.; Kelly, D.J.; Bentley, S.D.; Maiden, M.C.J.; Parkhill, J.; Falush, D. Genome-Wide Association Study Identifies Vitamin B 5 Biosynthesis as a Host Specificity Factor in Campylobacter. Proc. Natl. Acad. Sci. USA 2013, 110, 11923–11927. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, T.M.; Shah, M.K.; Burall, L.S.; Rakic-Martinez, M.; Datta, A.R. Genomic and Phenotypic Diversity of Listeria Monocytogenes Clonal Complexes Associated with Human Listeriosis. Appl. Microbiol. Biotechnol. 2018, 102, 3475–3485. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole Genome-Based Population Biology and Epidemiological Surveillance of Listeria Monocytogenes. Nat. Microbiol. 2017, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Marder, E.P.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Hurd, S.; Jervis, R.; Lathrop, S.; Muse, A.; Ryan, P.; Smith, K.; et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2017. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Steckler, A.J.; Cardenas-Alvarez, M.X.; Townsend Ramsett, M.K.; Dyer, N.; Bergholz, T.M. Genetic Characterization of Listeria Monocytogenes from Ruminant Listeriosis from Different Geographical Regions in the U.S. Vet. Microbiol. 2018, 215, 93–97. [Google Scholar] [CrossRef]
- Cardenas-Alvarez, M.X.; Zeng, H.; Webb, B.T.; Mani, R.; Munoz, M.; Bergholz, T.M. Comparative Genomics of Listeria Monocytogenes Isolated from Ruminant Listeriosis Cases in the Midwest United States. Microbiol. Spectr. 2022, in press. [Google Scholar]
- Flamm, R.K.; Hinrichs, D.J.; Thomashow, M.F. Introduction of PAM Beta 1 into Listeria Monocytogenes by Conjugation and Homology between Native L. Monocytogenes Plasmids. Infect. Immun. 1984, 44, 157–161. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Wolfgang, W.J.; Balkey, M.; Venkata, S.L.G.; Randolph, R.; Allard, M.; Strain, E. Optimizing Open Data to Support One Health: Best Practices to Ensure Interoperability of Genomic Data from Bacterial Pathogens. One Health Outlook 2020, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Gardner, S.N.; Slezak, T.; Hall, B.G. KSNP3.0: SNP Detection and Phylogenetic Analysis of Genomes without Genome Alignment or Reference Genome: Table 1. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An Interactive Viewer for Bacterial Population Genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Didelot, X.; Wilson, D.J. ClonalFrameML: Efficient Inference of Recombination in Whole Bacterial Genomes. PLoS Comput. Biol. 2015, 11, e1004041. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- San, J.E.; Baichoo, S.; Kanzi, A.; Moosa, Y.; Lessells, R.; Fonseca, V.; Mogaka, J.; Power, R.; de Oliveira, T. Current Affairs of Microbial Genome-Wide Association Studies: Approaches, Bottlenecks and Analytical Pitfalls. Front. Microbiol. 2020, 10, 3119. [Google Scholar] [CrossRef] [PubMed]
- Oren, Y.; Smith, M.B.; Johns, N.I.; Kaplan Zeevi, M.; Biran, D.; Ron, E.Z.; Corander, J.; Wang, H.H.; Alm, E.J.; Pupko, T. Transfer of Noncoding DNA Drives Regulatory Rewiring in Bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 16112–16117. [Google Scholar] [CrossRef] [PubMed]
- Drebes Dörr, N.C.; Proutière, A.; Jaskólska, M.; Stutzmann, S.; Bader, L.; Blokesch, M. Single Nucleotide Polymorphism Determines Constitutive versus Inducible Type VI Secretion in Vibrio Cholerae. ISME J. 2022, 16, 1868–1872. [Google Scholar] [CrossRef]
- Hammarlöf, D.L.; Kröger, C.; Owen, S.V.; Canals, R.; Lacharme-Lora, L.; Wenner, N.; Schager, A.E.; Wells, T.J.; Henderson, I.R.; Wigley, P.; et al. Role of a Single Noncoding Nucleotide in the Evolution of an Epidemic African Clade of Salmonella. Proc. Natl. Acad. Sci. USA 2018, 115, E2614–E2623. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Makarova, K.S.; Natale, D.A.; Spiridonov, A.N.; Tatusov, R.L.; Wolf, Y.I.; Yin, J.; Koonin, E.V. Congruent Evolution of Different Classes of Non-Coding DNA in Prokaryotic Genomes. Nucleic Acids Res. 2002, 30, 4264–4271. [Google Scholar] [CrossRef]
- Smith, G.A.; Theriot, J.A.; Portnoy, D.A. The Tandem Repeat Domain in the Listeria Monocytogenes ActA Protein Controls the Rate of Actin-Based Motility, the Percentage of Moving Bacteria, and the Localization of Vasodilator-Stimulated Phosphoprotein and Profilin. J. Cell Biol. 1996, 135, 647–660. [Google Scholar] [CrossRef]
- Bakardjiev, A.I.; Stacy, B.A.; Portnoy, D.A. Growth of Listeria Monocytogenes in the Guinea Pig Placenta and Role of Cell-to-Cell Spread in Fetal Infection. J. Infect. Dis. 2005, 191, 1889–1897. [Google Scholar] [CrossRef]
- Le Monnier, A.; Autret, N.; Join-Lambert, O.F.; Jaubert, F.; Charbit, A.; Berche, P.; Kayal, S. ActA Is Required for Crossing of the Fetoplacental Barrier by Listeria monocytogenes. Infect. Immun. 2007, 75, 950–957. [Google Scholar] [CrossRef]
- Lee, S.; Chen, Y.; Gorski, L.; Ward, T.J.; Osborne, J.; Kathariou, S. Listeria Monocytogenes Source Distribution Analysis Indicates Regional Heterogeneity and Ecological Niche Preference among Serotype 4b Clones. mBio 2018, 9, e00396-18. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Perrodeau, É.; Leclercq, A. Cazenave, Benoît; Pilmis, Benoît Clinical Features and Prognostic Factors of Listeriosis: The MONALISA National Prospective Cohort Study. Lancet Infect. Dis. 2017, 17, 510–519. [Google Scholar] [CrossRef]
- Cardenas-Alvarez, M.X.; Townsend Ramsett, M.K.; Malekmohammadi, S.; Bergholz, T.M. Evidence of Hypervirulence in Listeria Monocytogenes Clonal Complex 14. J. Med. Microbiol. 2019, 68, 1677–1685. [Google Scholar] [CrossRef]
- Orsi, R.H.; Jagadeesan, B.; Baert, L.; Wiedmann, M. Identification of Closely Related Listeria Monocytogenes Isolates with No Apparent Evidence for a Common Source or Location: A Retrospective Whole Genome Sequencing Analysis. J. Food Prot. 2021, 84, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.A.; Bentley, S.D. Bacterial GWAS: Not Just Gilding the Lily. Nat. Rev. Microbiol. 2016, 14, 406. [Google Scholar] [CrossRef]
- Hain, T.; Chatterjee, S.S.; Ghai, R.; Kuenne, C.T.; Billion, A.; Steinweg, C.; Domann, E.; Kärst, U.; Jänsch, L.; Wehland, J.; et al. Pathogenomics of Listeria spp. Int. J. Med. Microbiol. 2007, 297, 541–557. [Google Scholar] [CrossRef]
- Fox, E.M.; Allnutt, T.; Bradbury, M.I.; Fanning, S.; Chandry, P.S. Comparative Genomics of the Listeria Monocytogenes ST204 Subgroup. Front. Microbiol. 2016, 7, 2057. [Google Scholar] [CrossRef]
- Schmitz-Esser, S.; Müller, A.; Stessl, B.; Wagner, M. Genomes of Sequence Type 121 Listeria Monocytogenes Strains Harbor Highly Conserved Plasmids and Prophages. Front. Microbiol. 2015, 6, 380. [Google Scholar] [CrossRef]
- Hain, T.; Ghai, R.; Billion, A.; Kuenne, C.T.; Steinweg, C.; Izar, B.; Mohamed, W.; Mraheil, M.A.; Domann, E.; Schaffrath, S.; et al. Comparative Genomics and Transcriptomics of Lineages I, II, and III Strains of Listeria Monocytogenes. BMC Genom. 2012, 13, 144. [Google Scholar] [CrossRef]
- Bergholz, T.M.; den Bakker, H.C.; Katz, L.S.; Silk, B.J.; Jackson, K.A.; Kucerova, Z.; Joseph, L.A.; Turnsek, M.; Gladney, L.M.; Halpin, J.L.; et al. Determination of Evolutionary Relationships of Outbreak-Associated Listeria Monocytogenes Strains of Serotypes 1/2a and 1/2b by Whole-Genome Sequencing. Appl. Environ. Microbiol. 2016, 82, 928–938. [Google Scholar] [CrossRef]
- Tinsley, C.R.; Bille, E.; Nassif, X. Bacteriophages and Pathogenicity: More than Just Providing a Toxin? Microbes and Infection 2006, 8, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ward, T.J.; Siletzky, R.M.; Kathariou, S. Two Novel Type II Restriction-Modification Systems Occupying Genomically Equivalent Locations on the Chromosomes of Listeria Monocytogenes Strains. Appl. Environ. Microbiol. 2012, 78, 2623–2630. [Google Scholar] [CrossRef] [PubMed]
- Pieta, L.; Guzman, F.; Eguiluz, M.; Margis, R.; Campos, F.S.; Frazzon, A.P.G.; Frazzon, J. Comparative Genomics Suggests Differences Related to Resistance and Virulence between Food-Isolated Listeria Monocytogenes Serotypes 1/2a and 4b. EC Microbiol. 2018, 14, 755–766. [Google Scholar]
- Arous, S.; Buchrieser, C.; Folio, P.; Glaser, P.; Namane, A.; Hébraud, M.; Héchard, Y. Global Analysis of Gene Expression in an RpoN Mutant of Listeria Monocytogenes. Microbiology 2004, 150, 1581–1590. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, S.; Orsi, R.H.; Tang, S.; Zhang, W.; Wiedmann, M.; Boor, K.J. Phosphotransferase System-Dependent Extracellular Growth of Listeria Monocytogenes Is Regulated by Alternative Sigma Factors σ L and σ H. Appl. Environ. Microbiol. 2014, 80, 7673–7682. [Google Scholar] [CrossRef] [PubMed]
- Korsak, D.; Krawczyk-Balska, A. Rifampicin- and Rifabutin-Resistant Listeria Monocytogenes Strains Isolated from Food Products Carry Mutations in RpoB Gene. Foodborne Pathog. Dis. 2016, 13, 363–368. [Google Scholar] [CrossRef]
- Suzuki, M.; Shibayama, K.; Yahara, K. A Genome-Wide Association Study Identifies a Horizontally Transferred Bacterial Surface Adhesin Gene Associated with Antimicrobial Resistant Strains. Sci. Rep. 2016, 6, 37811. [Google Scholar] [CrossRef]

| Locus ID | Gene Product | Gene Length (ncl) | Clinical Outcome | Number of Strains Found (n = 232) a |
|---|---|---|---|---|
| EAL09991 * | tRNA-Phe(gaa) | 74 | CNS | 195 |
| WP_044683321 | hypothetical protein/ phage capsid protein (phiX174) | 117 | MN | 45 |
| ENH11862 | hypothetical protein/minor spike protein H (phiX174) | 987 | MN | 45 |
| NP_040712 | hypothetical protein/major spike protein G (phiX174) | 528 | MN | 45 |
| ABN49622 | hypothetical protein/replication initiation protein gpA (phiX174) | 1406 | MN | 41 |
| WP_016670801 | hypothetical protein/phage protein C (phiX174) | 144 | MN | 46 |
| WP_000084700 | hypothetical protein/phage protein D (phiX174) | 459 | MN | 45 |
| WP_000033471 | hypothetical protein/DNA binding protein J (phiX174) | 117 | MN | 49 |
| EAL09991 * | tRNA-Phe(gaa) | 73 | MN | 195 |
| EAL09991 | tRNA-Tyr(gta) | 83 | MN | 209 |
| BAO93225 | tRNA-Ala(tgc) | 75 | MN | 59 |
| WP_003734550 | type I restriction-modification system subunit M | 2576 | SI | 30 |
| WP_003743526 | type I restriction endonuclease subunit R | 3107 | SI | 30 |
| WP_021496534 | hypothetical protein | 338 | SI | 30 |
| Associated Position | Reference Nucleotide | SNP | Locus Tag | Gene Product | Protein ID | Region Start | Region End | Clinical Outcome |
|---|---|---|---|---|---|---|---|---|
| 77,529 | T | C | LMRG_RS00365 | Phosphoenolpyruvate mutase | WP_014600361.1 | 76,760 | 77,533 | CNS |
| 271,402 | A | G | LMRG_RS01320 | DNA-directed RNA polymerase subunit beta | WP_003723046.1 | 268,638 | 272,243 | CNS |
| 38,820 | C | A | LMRG_RS00170 | PTS sugar transporter subunit IIC | WP_003721657.1 | 38,033 | 39,385 | SI |
| 271,402 | A | G | LMRG_RS01320 | DNA-directed RNA polymerase subunit beta | WP_003723046.1 | 268,638 | 272,243 | SI |
| 298,930 | C | T/A | LMRG_RS01435 | ABC transporter ATP-binding protein | WP_014600437.1 | 298,775 | 299,440 | SI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardenas-Alvarez, M.X.; Restrepo-Montoya, D.; Bergholz, T.M. Genome-Wide Association Study of Listeria monocytogenes Isolates Causing Three Different Clinical Outcomes. Microorganisms 2022, 10, 1934. https://doi.org/10.3390/microorganisms10101934
Cardenas-Alvarez MX, Restrepo-Montoya D, Bergholz TM. Genome-Wide Association Study of Listeria monocytogenes Isolates Causing Three Different Clinical Outcomes. Microorganisms. 2022; 10(10):1934. https://doi.org/10.3390/microorganisms10101934
Chicago/Turabian StyleCardenas-Alvarez, Maria X., Daniel Restrepo-Montoya, and Teresa M. Bergholz. 2022. "Genome-Wide Association Study of Listeria monocytogenes Isolates Causing Three Different Clinical Outcomes" Microorganisms 10, no. 10: 1934. https://doi.org/10.3390/microorganisms10101934
APA StyleCardenas-Alvarez, M. X., Restrepo-Montoya, D., & Bergholz, T. M. (2022). Genome-Wide Association Study of Listeria monocytogenes Isolates Causing Three Different Clinical Outcomes. Microorganisms, 10(10), 1934. https://doi.org/10.3390/microorganisms10101934






