Effects of Allelochemicals, Soil Enzyme Activities, and Environmental Factors on Rhizosphere Soil Microbial Community of Stellera chamaejasme L. along a Growth-Coverage Gradient
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Location Information
2.2. Soil Chemical Analysis
2.3. Quantification of Chemical Substances in Rhizosphere Soil
2.4. High-Throughput Sequencing of Soil Microorganisms
2.5. Data Processing and Analysis
2.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. Soil Physical and Chemical Properties and Enzyme Activity
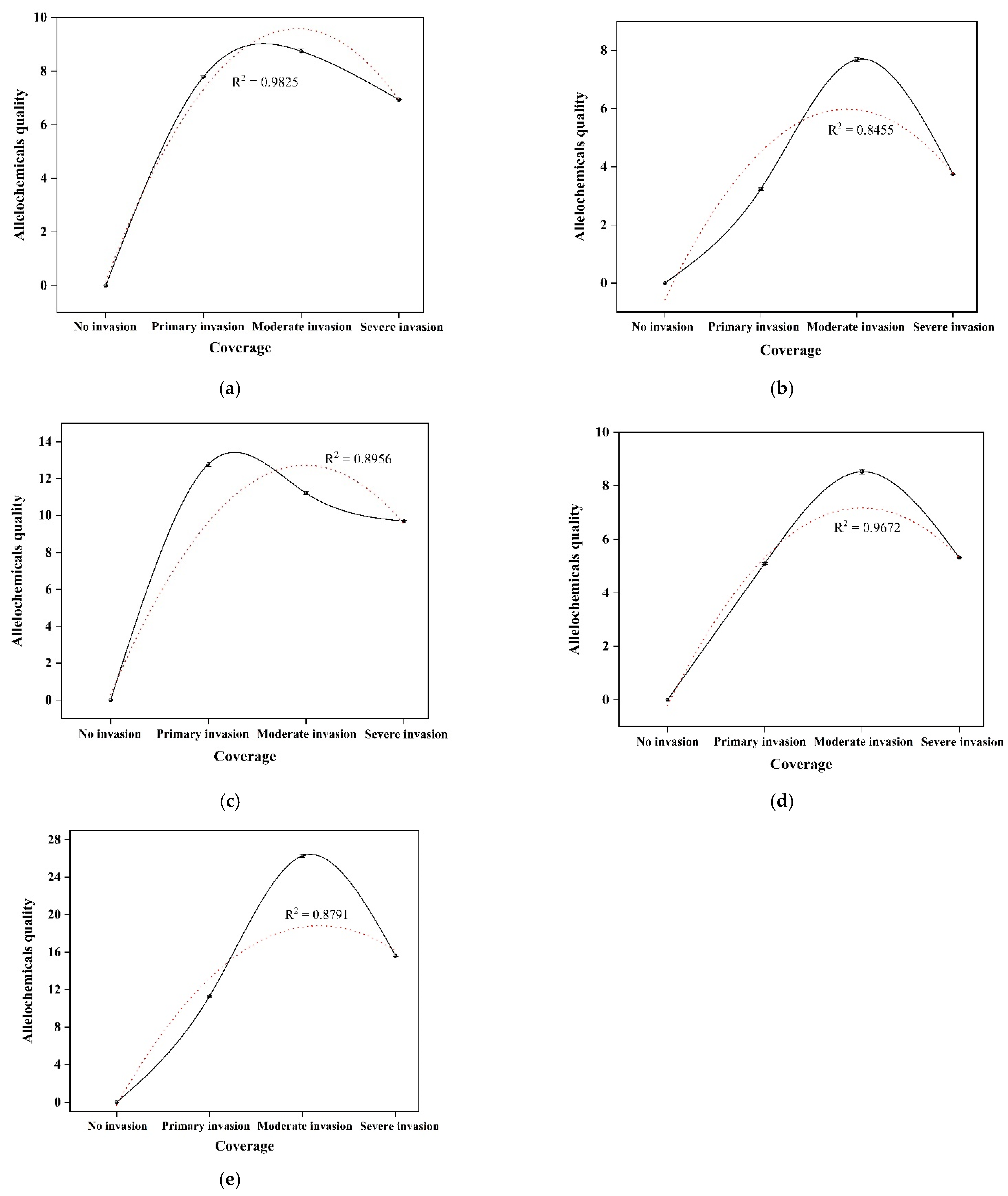
3.2. Quantitative Analysis of Allelochemicals in Rhizosphere Soil
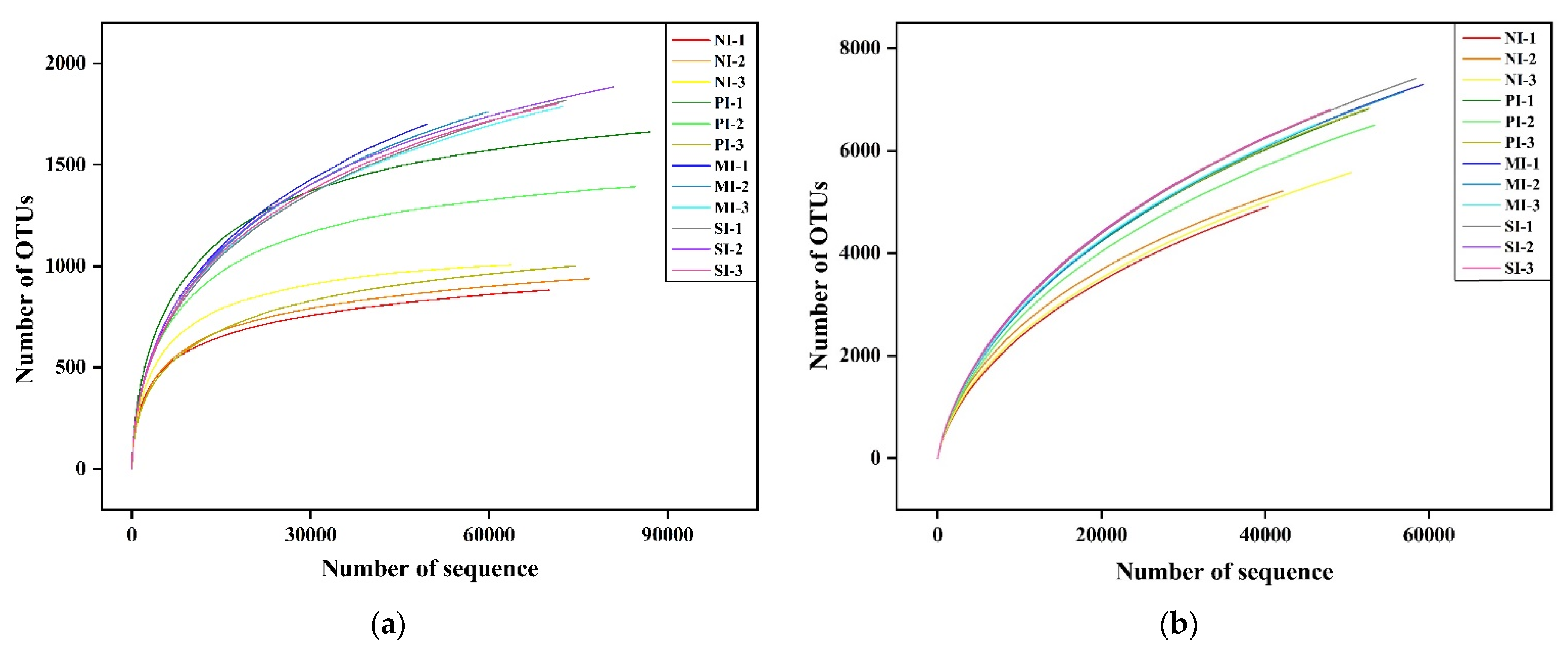
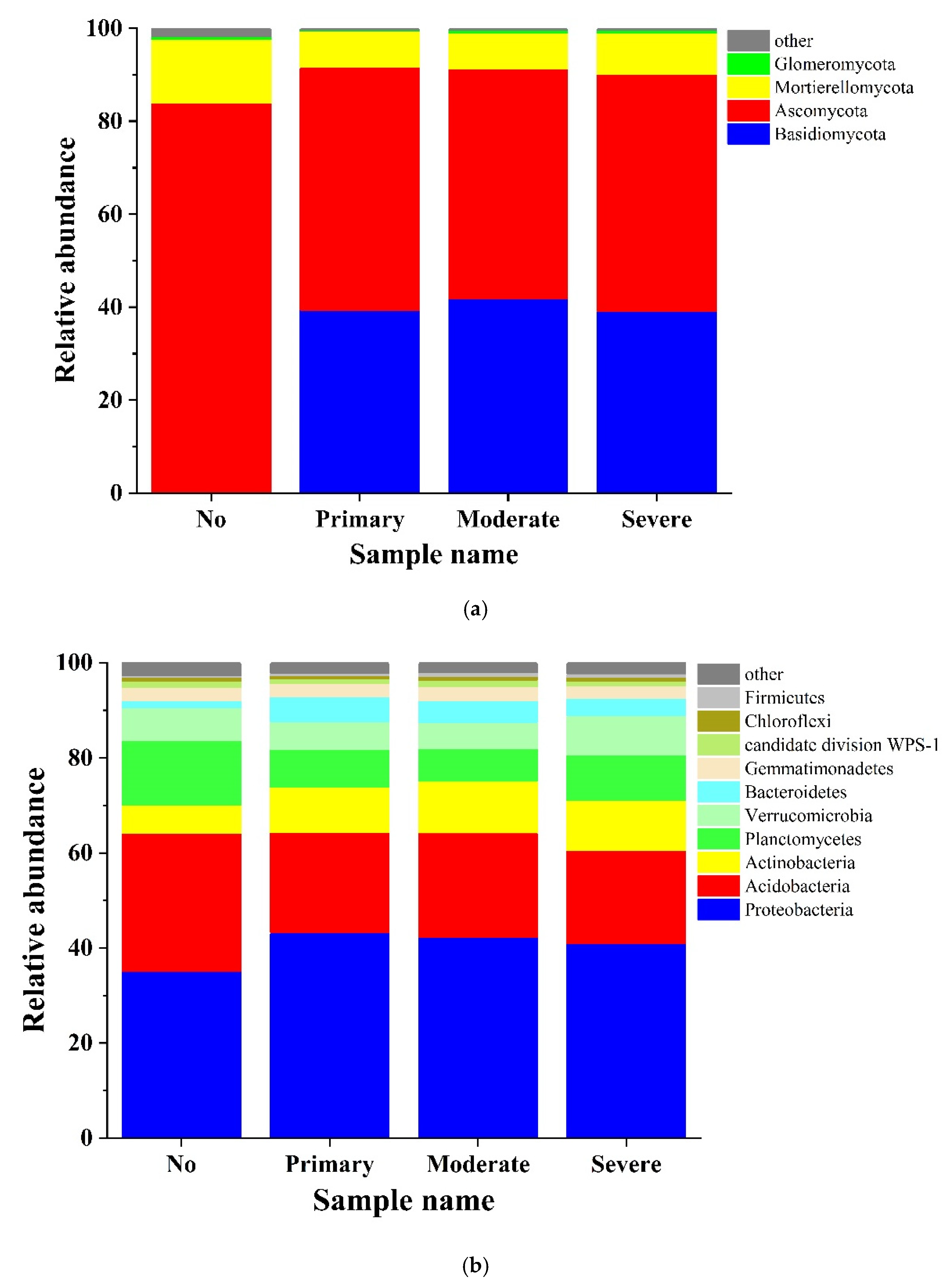
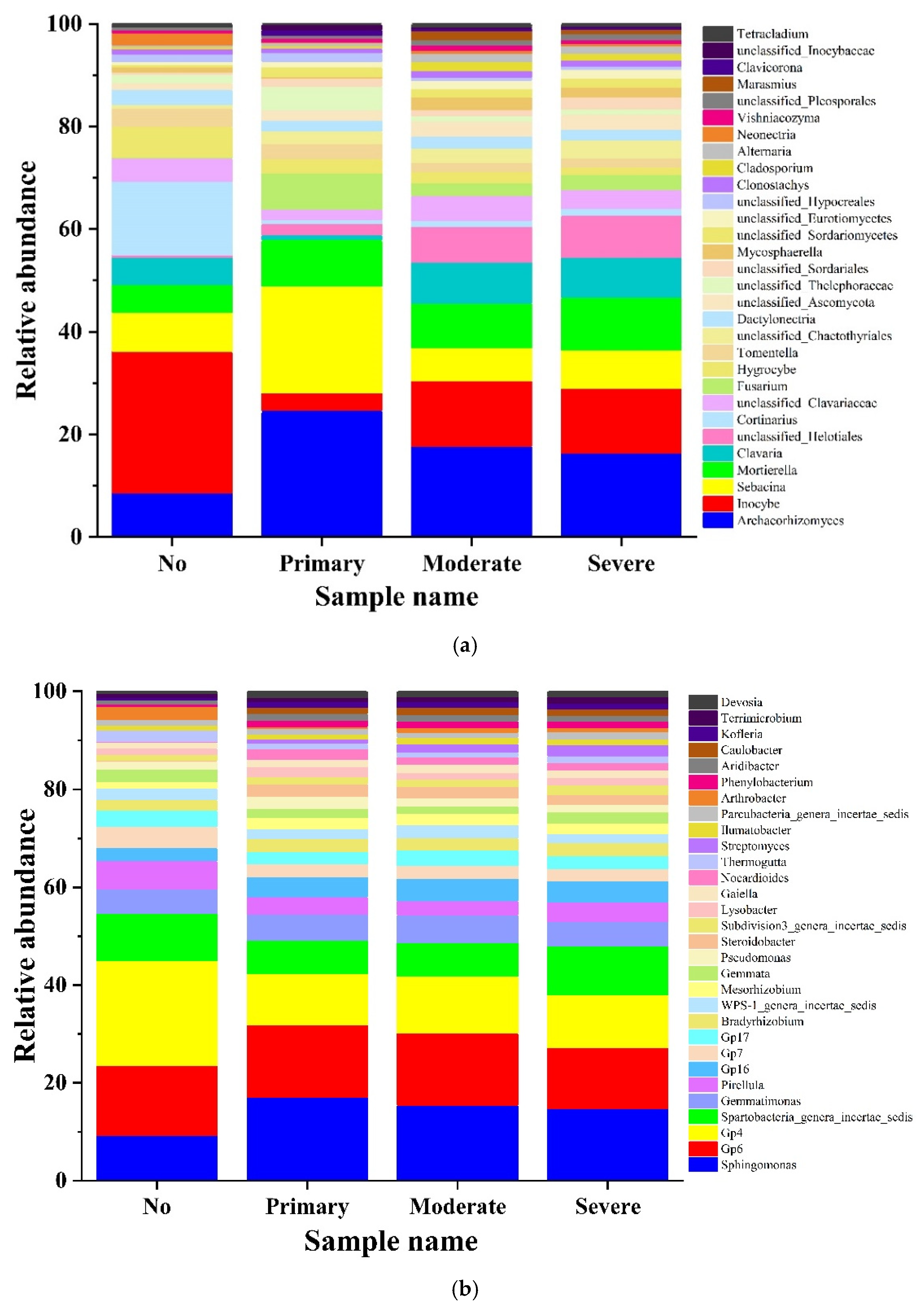
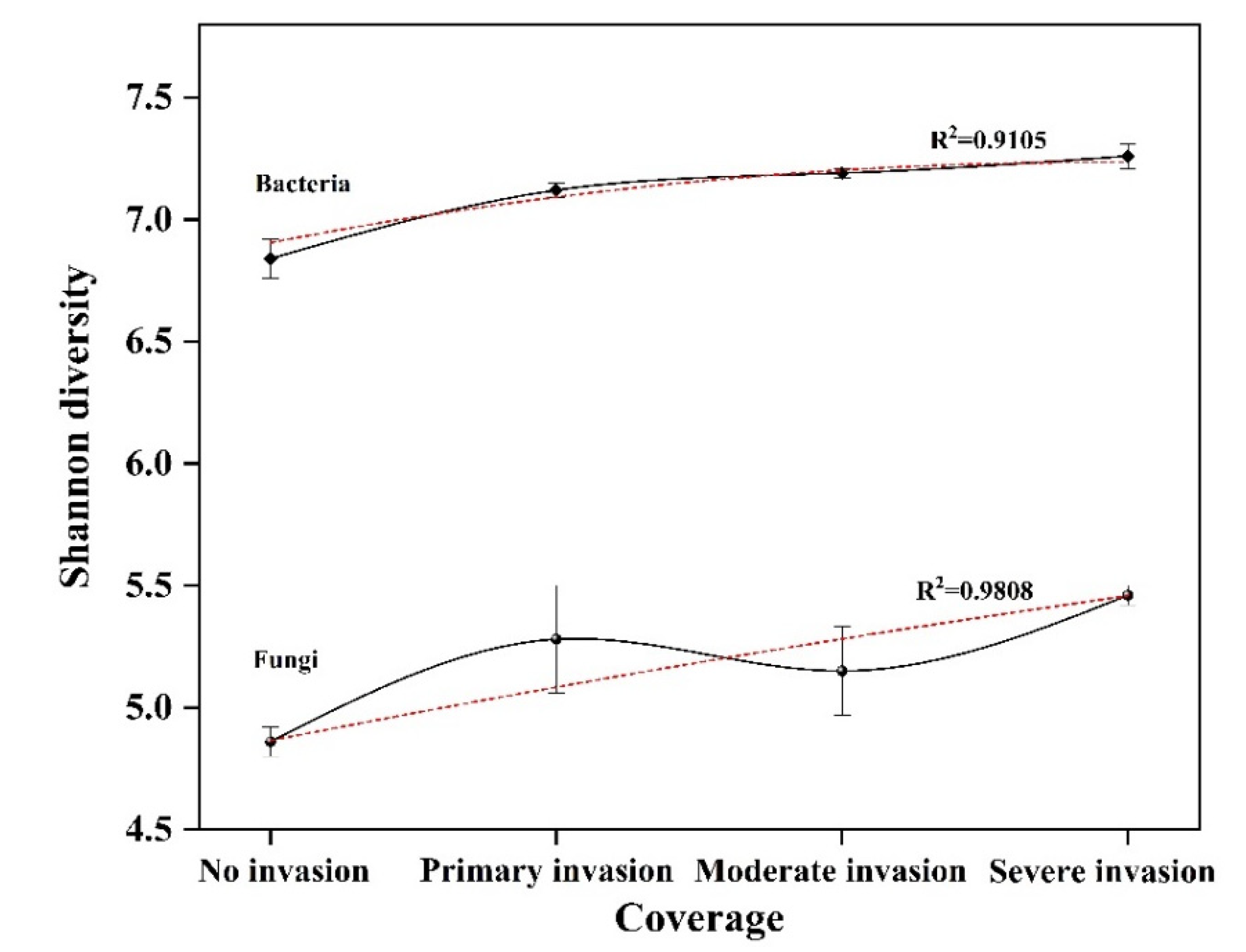
3.3. Analysis of High-Throughput Sequencing Data
3.4. Relationship between Soil Environmental Factors, Soil Enzyme Activity, and Allelopathic Substances and Bacterial Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de la Fuente Cantó, C.; Simonin, M.; King, E.; Moulin, L.; Bennett, M.J.; Castrillo, G.; Laplaze, L. An extended root phenotype: The rhizosphere, its formation and impacts on plant fitness. Plant. J. 2020, 103, 951–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizo-sphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant. Soil. 2009, 321, 341–361. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Li, C.; Wen, L.; Li, W.; Shi, L.; Cheng, K.; Xiao, X. Microbial carbon source utilization in rice rhizosphere and non-rhizosphere soils in a 34-year fertilized paddy field. J. Basic Microb. 2020, 60, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Razavi, B.S.; Zarebanadkouki, M.; Blagodatskaya, E.; Kuzyakov, Y. Rhizosphere shape of lentil and maize: Spatial distribution of enzyme activities. Soil Biol. Biochem. 2016, 96, 229–237. [Google Scholar] [CrossRef]
- Inderjit; Seastedt, T.R.; Callaway, R.M.; Pollock, J.L.; Kaur, J. Allelopathy and plant invasions: Traditional, congeneric, and bio-geographical approaches. Biol. Invasions 2008, 10, 875–890. [Google Scholar] [CrossRef]
- Kalisz, S.; Kivlin, S.N.; Bialic-Murphy, L. Allelopathy is pervasive in invasive plants. Biol. Invasions 2020, 1–5. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Mishra, M.; Jain, S.; Singh, J.; Sharma, S. Exploring the role of plant-microbe interactions in improving soil structure and function through root exudation: A key to sustainable agriculture. Plant.-Microbe Agro-Ecol. Perspect. 2017, 467–487. [Google Scholar] [CrossRef]
- Xiao, Z.X.; Lu, S.G.; Xu, Z.H. Biochemistry of allelopathic plant residues in soil. Ekoloji 2019, 28, 2997–3006. [Google Scholar]
- Dawkins, K.; Esiobu, N. Emerging Insights on Brazilian Pepper Tree (Schinus terebinthifolius) Invasion: The Potential Role of Soil Microorganisms. Front. Plant. Sci. 2016, 7, 712. [Google Scholar] [CrossRef] [Green Version]
- Lankau, R.A. Intraspecific variation in allelochemistry determines an invasive species’ impact on soil microbial communities. Oecologia 2011, 165, 453–463. [Google Scholar] [CrossRef]
- Bressan, M.; Roncato, M.-A.; Bellvert, F.; Comte, G.; Haichar, F.Z.; Achouak, W.; Berge, O. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 2009, 3, 1243–1257. [Google Scholar] [CrossRef]
- Li, X.G.; Ding, C.F.; Hua, K.; Zhang, T.L.; Zhang, Y.N.; Zhao, L.; Yang, Y.R.; Liu, J.G.; Wang, X.X. Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 2014, 78, 149–159. [Google Scholar] [CrossRef]
- Cheng, F.; Cheng, Z.H. Corrigendum: Research Progress on the use of Plant Allelopathy in Agriculture and the Physiological and Ecological Mechanisms of Allelopathy. Front. Plant. Sci. 2016, 7, 1697. [Google Scholar] [CrossRef] [Green Version]
- Lu, F.L.; Zheng, L.L.; Chen, Y.Y.; Li, D.P.; Zeng, R.S.; Li, H.S. Soil microorganisms alleviate the allelopathic effect of Eucalyptus grandis × E. urophylla leachates on Brassica chinensis. J. For. Res. 2017, 28, 1203–1207. [Google Scholar] [CrossRef]
- Błońska, E.; Lasota, J.; Zwydak, M. The relationship between soil properties, enzyme activity and land use. For. Res. Pap. 2017, 78, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Guo, K.Y.; Zhao, Y.Z.; Liu, Y.; Chen, J.H.; Wu, Q.F.; Ruan, Y.F.; Li, S.H.; Shi, J.; Zhao, L.; Sun, X.; et al. Pyrolysis temperature of biochar affects ecoenzymatic stoichiometry and microbial nutrient-use efficiency in a bamboo forest soil. Geoderma 2020, 363, 114162. [Google Scholar] [CrossRef]
- Bian, X.B.; Xiao, S.Y.; Zhao, Y.; Xu, Y.H.; Yang, H.; Zhang, L.X. Comparative analysis of rhizosphere soil physiochemical characteristics and microbial communities between rusty and healthy ginseng root. Sci. Rep.-UK 2020, 10, 15756. [Google Scholar] [CrossRef]
- Batista, É.R.; Carneiro, J.J.; Pinto, F.A.; Santos, J.V.D.; Carneiro, M.A.C. Environmental drivers of shifts on microbial traits in sites disturbed by a large-scale tailing dam collapse. Sci. Total Environ. 2020, 738, 139453. [Google Scholar] [CrossRef]
- Peng, X.Q.; Wang, W. Stoichiometry of soil extracellular enzyme activity along a climatic transect in temperate grasslands of northern China. Soil Biol. Biochem. 2016, 98, 74–84. [Google Scholar] [CrossRef]
- Jin, H.; Yang, X.Y.; Liu, R.T.; Yan, Z.Q.; Li, X.D.; Li, X.Z.; Su, A.X.; Zhao, Y.H.; Qin, B. Bacterial community structure associated with the rhizosphere soils and roots of S. chamaejasme along a Tibetan elevation gradient. Ann. Microbiol. 2018, 68, 273–286. [Google Scholar] [CrossRef]
- Guo, H.R.; Cui, H.Y.; Jin, H.; Yan, Z.Q.; Ding, L.; Qin, B. Potential allelochemicals in root zone soils of S. chamaejasme and variations at different geographical growing sites. Plant. Growth Regul. 2015, 77, 335–342. [Google Scholar] [CrossRef]
- Guo, H.R.; Zeng, L.M.; Yan, Z.Q.; Jin, H.; Li, X.Z.; Guan, J.F.; Qin, B. Allelochemical from the root exudates of S. chamaejasme and its degradation. Allelopath. J. 2016, 38, 103–112. [Google Scholar]
- Yan, Z.Q.; Guo, H.R.; Yang, J.Y.; Liu, Q.; Jin, H.; Xu, R.; Cui, H.Y.; Qin, B. Phytotoxic flavonoids from roots of S. chamaejasme (Thymelaeaceae). Phytochemistry 2014, 106, 61–68. [Google Scholar] [CrossRef]
- Huang, X.Q.; Chen, Y.; Zhang, X.; Li, F.M.; Ye, H.T. Extract of S. chamaejasme inhibits the progression of hepatocellular carcinoma by regulating miR-134-5p and JAK1/STAT3 pathway. Cancer Biother. Radiopharm. 2020, 35, 586–595. [Google Scholar] [CrossRef]
- Jing, C.X.; Guo, J.J.; Yang, B.J.; Fan, S.R.; Wang, Y.T.; Chen, D.Z.; Hao, X.J. Stelleraguaianone B and C, two new sesquiterpenoids from Stellera chamaejasme L. Fitoterapia 2019, 134, 443–446. [Google Scholar] [CrossRef]
- Guo, L.Z.; Li, J.H.; He, W.; Liu, L.; Huang, D.; Wang, K. High nutrient uptake efficiency and high water use efficiency facilitate the spread of S. chamaejasme in degraded grasslands. BMC Ecol. 2019, 19, 50. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Yang, X.Y.; Yan, Z.Q.; Liu, Q.; Li, X.Z.; Chen, J.X.; Zhang, D.H.; Zeng, L.M.; Qin, B. Characterization of rhizosphere and endophytic bacterial communities from leaves, stems and roots of medicinal S. chamaejasme. Syst. App. Microbiol. 2014, 37, 376–385. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics - ScienceDirect. PCR Protoc. 1990, 315–322. [Google Scholar] [CrossRef]
- Sofie, T.; Michiel, O.D.B.; Bram, B.; Sascha, T.; Vincent, S.; Van, H.J.D.; Nele, W.; Jaco, V. Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal database project: Tools and data for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.C.; Yang, C.Q.; Zhang, J.; Zhang, Q.; Yang, F.; Yang, W.Y.; Liu, J. Organ-specific differential NMR-based metabonomic analysis of soybean [Glycine max (L.) Merr.] fruit reveals the metabolic shifts and potential protection mechanisms involved in field mold infection. Front. Plant. Sci. 2017, 8, 508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.H.; Liu, Y.X.; Pu, Z.E.; Wang, J.R.; Zheng, Y.L.; Li, Y.H.; Wei, Y.M. Regulation, evolution, and functionality of flavonoids in cereal crops. Biotechnol. Lett. 2013, 35, 1765–1780. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant. Physiol. Bioch. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weston, L.A.; Mathesius, U. Flavonoids: Their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 2013, 39, 283. [Google Scholar] [CrossRef]
- Jin, H.; Yang, X.Y.; Lu, D.X.; Li, C.J.; Yan, Z.Q.; Li, X.Z.; Zeng, L.M.; Qin, B. Phylogenic diversity and tissue specificity of fungal endophytes associated with the pharmaceutical plant, S. chamaejasme revealed by a cultivation-independent approach. Anton. Leeuw. 2015, 108, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.; Doddapaneni, H.; Subramanian, V.; Lam, Y.W.; Yadav, J.S. Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs). Biochem. Bioph. Res. Co. 2010, 399, 492–497. [Google Scholar] [CrossRef] [Green Version]
- Godoy, P.; Reina, R.; Calderón, A.; Wittich, R.M.; García-Romera, I.; Aranda, E. Exploring the potential of fungi isolated from PAH-polluted soil as a source of xenobiotics-degrading fungi. Environ. Sci. Pollut. R. 2016, 23, 20985–20996. [Google Scholar] [CrossRef]
- Lv, X.F.; Yu, J.B.; Fu, Y.Q.; Ma, B.; Qu, F.Z.; Ning, K.; Wu, H.F. A Meta-Analysis of the bacterial and archaeal diversity observed in wetland soils. Sci. World J. 2014, 437684. [Google Scholar] [CrossRef] [Green Version]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microb. 2011, 77, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.J.; Cho, H.J.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [Green Version]
- McLeod, M.L.; Cleveland, C.C.; Lekberg, Y.; Maron, J.L.; Philippot, L.; Bru, D.; Callaway, R.M. Exotic invasive plants increase productivity, abundance of ammonia-oxidizing bacteria and nitrogen availability in intermountain grasslands. J. Ecol. 2016, 104, 994–1002. [Google Scholar] [CrossRef] [Green Version]
- Ni, G.Y.; Song, L.Y.; Zhang, J.L.; Peng, S.L. Effects of root extracts of Mikania micrantha H.B.K. on soil microbial community. Alleloapthy J. 2006, 17, 247–254. [Google Scholar]
- Shaw, L.J.; Morris, P.; Hooker, J.E. Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ. Microbiol. 2006, 8, 1867–1880. [Google Scholar] [CrossRef]
- Vivanco, J.M.; Bais, H.P.; Stermitz, F.R.; Thelen, G.C.; Callaway, R.M. Biogeographical variation in community response to root allelochemistry: Novel weapons and exotic invasion. Ecol. Lett. 2004, 7, 285–292. [Google Scholar] [CrossRef]
- Ma, S.H.; Chen, G.P.; Tang, W.G.; Xing, A.J.; Chen, X.; Xiao, W.; Zhou, L.H.; Zhu, J.L.; Li, Y.D.; Zhu, B.; et al. Inconsistent responses of soil microbial community structure and enzyme activity to nitrogen and phosphorus additions in two tropical forests. Plant. Soil. 2021, 460, 1–16. [Google Scholar] [CrossRef]
- Chu, H.Y.; Lin, X.G.; Fujii, T.; Morimoto, S.; Yagi, K.; Hu, J.L.; Zhang, J.B. Soil microbial biomass, dehydrogenase activity, bacterial community structure in response to long-term fertilizer management. Soil Biol. Biochem. 2007, 39, 2971–2976. [Google Scholar] [CrossRef]
- Liu, J.G.; Li, Y.B.; Jiang, G.Y.; Bian, X.M.; Geng, W. Allelopathic effects of cotton in continuous cropping. Allelopathy J. 2008, 21, 299–305. [Google Scholar]
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z.; Yang, S.H.; Herrera-Estrella, L.; Xu, G.H.; Chao, D.Y.; Li, J.R.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sc. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Mulderij, G.; Mooij, W.M.; Smolders, A.J.P.; Donk, E.V. Allelopathic inhibition of phytoplankton by exudates from Stratiotes aloides. Aquat. Bot. 2005, 82, 284–296. [Google Scholar] [CrossRef]
- Yu, X.J.; Yu, D.; Lu, Z.J.; Keping, M.A. A new mechanism of invader success: Ex-otic plant inhibits natural vegetation restoration by changing soil mi-crobe community. Chin. Sci. Bull. 2005, 50, 1105–1112. [Google Scholar] [CrossRef]

| Sample Number | Altitude (m) | Northern Latitude | Eastern Longitude | Population Coverage | Coverage Gradient (%) |
|---|---|---|---|---|---|
| 1 | 2960 | 37°7′37″ | 102°50′3″ | 0.00 | 0 |
| 2 | 2970 | 37°7′36″ | 102°50′3″ | 0.00 | |
| 3 | 2970 | 37°7′33″ | 102°49′38″ | 0.00 | |
| 4 | 2970 | 37°7′33″ | 102°50′0″ | 0.00 | |
| 5 | 2960 | 37°7′36″ | 102°50′4″ | 25.50 | 25.13 ± 1.26 |
| 6 | 2890 | 37°7′33″ | 102°49′56″ | 26.75 | |
| 7 | 2930 | 37°7′37″ | 102°50′17″ | 25.00 | |
| 8 | 2940 | 37°7′36″ | 102°50′17″ | 23.25 | |
| 9 | 2940 | 37°7′34″ | 102°50′1″ | 50.50 | 52.63 ± 2.76 |
| 10 | 2920 | 37°7′42″ | 102°50′14″ | 57.25 | |
| 11 | 2930 | 37°7′48″ | 102°50′16″ | 50.50 | |
| 12 | 2950 | 37°7′42″ | 102°50′5″ | 52.25 | |
| 13 | 2960 | 37°7′35″ | 102°50′2″ | 85.00 | 89.69 ± 4.81 |
| 14 | 2970 | 37°7′35″ | 102°49′59″ | 85.75 | |
| 15 | 2950 | 37°7′43″ | 102°50′12″ | 97.00 | |
| 16 | 2950 | 37°7′45″ | 102°50′16″ | 91.00 |
| Name | No Invasion | Primary Invasion | Moderate Invasion | Severe Invasion |
|---|---|---|---|---|
| pH | 7.46 ± 0.01 b | 7.60 ± 0.01 a | 7.60 ± 0.02 a | 7.61 ± 0.01 a |
| At (m) | 2968 ± 2.50 a | 2930 ± 14.72 b | 2935 ± 6.45 b | 2958 ± 4.79 a |
| ST (°C) | 4.80 ± 0.15 a | 4.93 ± 0.09 a | 5.03 ± 0.09 a | 5.17 ± 0.09 a |
| SH (%) | 39.39 ± 2.56 a | 30.48 ± 0.34 b | 29.63 ± 0.18 b | 29.18 ± 0.37 b |
| SOM (g/kg) | 131.41 ± 0.94 b | 113.83 ± 0.29 c | 132.11 ± 0.52 b | 135.45 ± 0.85 a |
| TN (g/kg) | 6.83 ± 0.04 c | 8.24 ± 0.01 a | 6.66 ± 0.01 d | 7.45 ± 0.01 b |
| AN (mg/kg) | 586.23 ± 0.96 c | 690.35 ± 2.56 a | 582.06 ± 1.91 c | 634.12 ± 2.74 b |
| AK (mg/kg) | 255.4 ± 0.68 c | 368.1 ± 1.36 a | 223.42 ± 1.99 d | 310.28 ± 1.88 b |
| AP (mg/kg) | 38.4 ± 0.21 d | 58.94 ± 0.17 a | 49.97 ± 0.41 c | 56.91 ± 0.23 b |
| PPO (mg/d/g) | 13.53 ± 0.12 c | 13.74 ± 0.12 c | 16.03 ± 0.10 b | 17.61 ± 0.05 a |
| POD (mg/d/g) | 30.44 ± 0.17 c | 45.59 ± 0.26 b | 35.79 ± 0.06 a | 35.59 ± 0.46 b |
| UE (μg/d/g) | 860.93 ± 1.41 a | 756.62 ± 0.29 c | 758.08 ± 0.41 c | 776.10 ± 0.48 b |
| DHA (μg/d/g) | 6.09 ± 0.08 d | 23.03 ± 0.16 a | 10.85 ± 0.13 c | 17.33 ± 0.15 b |
| SC (mg/d/g) | 62.81 ± 0.17 a | 59.79 ± 0.16 d | 61.56 ± 0.13 b | 60.74 ± 0.31 c |
| AKP (umol/d/g) | 8.11 ± 0.13 a | 4.25 ± 0.06 b | 4.55 ± 0.06 b | 4.41 ± 0.14 b |
| ACP (umol/d/g) | 15.63 ± 0.10 a | 13.99 ± 0.10 b | 12.99 ± 0.20 c | 14.45 ± 0.15 b |
| SYT (mg/kg) | -- | 7.79 ± 0.06 b | 8.74 ± 0.08 a | 6.93 ± 0.02 c |
| YA (mg/kg) | -- | 3.24 ± 0.06 c | 7.69 ± 0.07 a | 3.75 ± 0.01 b |
| XB (mg/kg) | -- | 12.78 ± 0.13 a | 11.21 ± 0.09 b | 9.69 ± 0.05 c |
| QB (mg/kg) | -- | 5.10 ± 0.04 b | 8.52 ± 0.10 a | 5.32 ± 0.01 c |
| JA (mg/kg) | -- | 11.31 ± 0.10 c | 26.28 ± 0.20 a | 15.62 ± 0.09 b |
| Sample Name | Effective Tags | OTU | Shannon Index | Simpson Index | Chao1 Index | ACE Index | Coverage (%) | |
|---|---|---|---|---|---|---|---|---|
| Fungi | F-0 | 70,240 ± 3823 b | 938 ± 40 b | 4.86 ± 0.06 b | 0.026 ± 0.003 a | 1039.00 ± 38.65 b | 1028.29 ± 34.01 b | 99.80 |
| F-1 | 81,977 ± 3836 a | 1387 ± 201 a | 5.28 ± 0.22 a | 0.013 ± 0.003 a | 1553.82 ± 208.18 a | 1525.87 ± 203.69 a | 99.08 | |
| F-2 | 89,715 ± 13232 a | 1429 ± 57 a | 5.15 ± 0.18 a | 0.017 ± 0.007 a | 1678.31 ± 33.62 a | 1665.56 ± 53.87 a | 99.10 | |
| F-3 | 77,406 ± 8489 a | 1634 ± 92 a | 5.46 ± 0.04 a | 0.010 ± 0.000 b | 1841.96 ± 90.65 a | 1844.77 ± 97.90 a | 99.30 | |
| Bacteria | B-0 | 44,335 ± 3130 a | 5235 ± 191 b | 6.84 ± 0.08 b | 0.005 ± 0.000 b | 8413.62 ± 309.63 b | 10,702.66 ± 442.60 c | 94.60 |
| B-1 | 52,847 ± 271 a | 6713 ± 181 a | 7.12 ± 0.03 a | 0.004 ± 0.000 a | 10,251.66 ± 116.19 a | 12,729.96 ± 125.85 b | 94.40 | |
| B-2 | 54,133 ± 4029 a | 6989 ± 414 a | 7.19 ± 0.02 a | 0.004 ± 0.000 a | 10,912.19 ± 146.59 a | 14,014.21 ± 220.17 a | 94.10 | |
| B-3 | 50,430 ± 6233 a | 6940 ± 423 a | 7.26 ± 0.05 a | 0.003 ± 0.000 a | 10,914.76 ± 254.76 a | 13,567.63 ± 224.51 a | 93.80 | |
| Basidiomycota | Ascomycota | Mortierellomycota | Glomeromycota | Total Fungi | Diversity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | p | CC | p | CC | p | CC | p | CC | p | CC | p | |
| pH | −0.906 | 0.094 | 0.875 | 0.125 | 0.160 | 0.840 | 0.633 | 0.367 | 0.760 | 0.240 | 0.939 | 0.061 |
| At | 0.821 | 0.179 | −0.827 | 0.173 | −0.021 | 0.979 | 0.100 | 0.900 | −0.414 | 0.586 | −0.602 | 0.398 |
| ST | −0.533 | 0.467 | 0.477 | 0.523 | 0.027 | 0.973 | 0.903 | 0.097 | 0.819 | 0.181 | 0.864 | 0.136 |
| SH | 0.873 | 0.127 | −0.838 | 0.162 | −0.097 | 0.903 | −0.642 | 0.358 | −0.806 | 0.194 | −0.962 | 0.038 |
| SOM | 0.573 | 0.427 | −0.627 | 0.373 | −0.583 | 0.417 | 0.324 | 0.676 | 0.397 | 0.603 | 0.106 | 0.894 |
| TN | −0.729 | 0.271 | 0.764 | 0.236 | 0.915 | 0.085 | 0.298 | 0.702 | −0.256 | 0.744 | 0.097 | 0.903 |
| AN | −0.771 | 0.229 | 0.803 | 0.197 | 0.890 | 0.110 | 0.312 | 0.688 | −0.200 | 0.800 | 0.155 | 0.845 |
| AK | −0.651 | 0.349 | 0.686 | 0.314 | 0.961 | 0.039 | 0.342 | 0.658 | −0.321 | 0.679 | 0.024 | 0.976 |
| AP | −0.962 | 0.038 | 0.952 | 0.048 | 0.538 | 0.462 | 0.650 | 0.350 | 0.445 | 0.555 | 0.727 | 0.273 |
| PPO | −0.258 | 0.742 | 0.194 | 0.806 | −0.183 | 0.817 | 0.847 | 0.153 | 0.821 | 0.179 | 0.761 | 0.239 |
| POD | −0.909 | 0.091 | 0.935 | 0.065 | 0.585 | 0.415 | 0.116 | 0.884 | 0.058 | 0.942 | 0.385 | 0.615 |
| UE | 0.934 | 0.066 | −0.912 | 0.088 | −0.099 | 0.901 | −0.427 | 0.573 | −0.715 | 0.285 | −0.902 | 0.098 |
| DHA | −0.923 | 0.077 | 0.936 | 0.064 | 0.747 | 0.253 | 0.485 | 0.515 | 0.128 | 0.872 | 0.466 | 0.534 |
| SC | 0.968 | 0.032 | −0.975 | 0.025 | −0.644 | 0.356 | −0.489 | 0.511 | −0.255 | 0.745 | −0.577 | 0.423 |
| ALP | 0.941 | 0.059 | −0.915 | 0.085 | −0.202 | 0.798 | −0.581 | 0.419 | −0.710 | 0.290 | −0.911 | 0.089 |
| ACP | 0.687 | 0.313 | −0.657 | 0.343 | 0.355 | 0.645 | −0.127 | 0.873 | −0.822 | 0.178 | −0.872 | 0.128 |
| SYT | −0.89 | 0.110 | 0.861 | 0.139 | 0.001 | 0.999 | 0.437 | 0.563 | 0.787 | 0.213 | 0.940 | 0.060 |
| YA | −0.539 | 0.461 | 0.495 | 0.505 | −0.510 | 0.490 | 0.212 | 0.788 | 0.930 | 0.070 | 0.903 | 0.097 |
| Proteobacteria | Acidobacteria | Actinobacteria | Planctomycetes | Total Bacteria | Diversity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | p | CC | p | CC | p | CC | p | CC | p | CC | p | |
| pH | 0.924 | 0.076 | −0.979 | 0.021 | 0.997 | 0.003 | −0.921 | 0.079 | 0.850 | 0.150 | 0.968 | 0.032 |
| At | −0.914 | 0.086 | 0.586 | 0.414 | −0.736 | 0.264 | 0.879 | 0.121 | −0.215 | 0.785 | −0.517 | 0.483 |
| ST | 0.530 | 0.470 | −0.844 | 0.156 | 0.788 | 0.212 | −0.602 | 0.398 | 0.983 | 0.017 | 0.930 | 0.070 |
| SH | −0.905 | 0.095 | 0.970 | 0.030 | −0.996 | 0.004 | 0.922 | 0.078 | −0.864 | 0.136 | −0.980 | 0.020 |
| SOM | −0.488 | 0.512 | 0.179 | 0.821 | −0.191 | 0.809 | 0.271 | 0.729 | 0.230 | 0.770 | 0.064 | 0.936 |
| TN | 0.526 | 0.474 | −0.537 | 0.463 | 0.400 | 0.600 | −0.272 | 0.728 | 0.280 | 0.720 | 0.278 | 0.722 |
| AN | 0.579 | 0.421 | −0.581 | 0.419 | 0.455 | 0.545 | −0.332 | 0.668 | 0.315 | 0.685 | 0.328 | 0.672 |
| AK | 0.424 | 0.576 | −0.489 | 0.511 | 0.321 | 0.679 | −0.162 | 0.838 | 0.276 | 0.724 | 0.228 | 0.772 |
| AP | 0.874 | 0.126 | −0.959 | 0.041 | −0.761 | 0.239 | −0.761 | 0.239 | 0.781 | 0.219 | 0.841 | 0.159 |
| PPO | 0.289 | 0.711 | −0.643 | 0.357 | 0.598 | 0.402 | −0.423 | 0.577 | 0.890 | 0.110 | 0.798 | 0.202 |
| POD | 0.834 | 0.166 | −0.652 | 0.348 | 0.651 | 0.349 | −0.654 | 0.346 | 0.280 | 0.720 | 0.449 | 0.551 |
| UE | −0.985 | 0.015 | 0.913 | 0.087 | −0.982 | 0.018 | 0.979 | 0.021 | −0.697 | 0.303 | −0.889 | 0.111 |
| DHA | 0.780 | 0.220 | −0.809 | 0.191 | 0.716 | 0.284 | −0.589 | 0.411 | 0.563 | 0.437 | 0.612 | 0.388 |
| SC | −0.860 | 0.140 | 0.866 | 0.134 | −0.804 | 0.196 | 0.699 | 0.301 | −0.612 | 0.388 | −0.696 | 0.304 |
| ALP | −0.953 | 0.047 | 0.972 | 0.028 | −0.995 | 0.005 | 0.931 | 0.069 | −0.805 | 0.195 | −0.941 | 0.060 |
| ACP | −0.861 | 0.139 | 0.663 | 0.337 | −0.843 | 0.157 | 0.963 | 0.037 | −0.468 | 0.532 | −0.739 | 0.261 |
| SYT | 0.964 | 0.036 | −0.902 | 0.098 | 0.985 | 0.015 | −0.988 | 0.012 | 0.717 | 0.283 | 0.910 | 0.090 |
| YA | 0.736 | 0.264 | −0.618 | 0.382 | 0.796 | 0.204 | −0.895 | 0.105 | 0.530 | 0.470 | 0.759 | 0.241 |
| XB | 0.993 | 0.007 | −0.916 | 0.084 | 0.971 | 0.029 | −0.959 | 0.041 | 0.676 | 0.324 | 0.864 | 0.136 |
| QB | 0.856 | 0.144 | −0.770 | 0.230 | 0.910 | 0.090 | −0.964 | 0.036 | 0.646 | 0.354 | 0.861 | 0.139 |
| JA | 0.739 | 0.261 | −0.684 | 0.316 | 0.835 | 0.165 | −0.895 | 0.105 | 0.630 | 0.370 | 0.826 | 0.174 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Jin, H.; Zhang, J.; Xu, Z.; Yang, X.; Liu, H.; Xu, X.; Min, D.; Lu, D.; Qin, B. Effects of Allelochemicals, Soil Enzyme Activities, and Environmental Factors on Rhizosphere Soil Microbial Community of Stellera chamaejasme L. along a Growth-Coverage Gradient. Microorganisms 2022, 10, 158. https://doi.org/10.3390/microorganisms10010158
Cheng J, Jin H, Zhang J, Xu Z, Yang X, Liu H, Xu X, Min D, Lu D, Qin B. Effects of Allelochemicals, Soil Enzyme Activities, and Environmental Factors on Rhizosphere Soil Microbial Community of Stellera chamaejasme L. along a Growth-Coverage Gradient. Microorganisms. 2022; 10(1):158. https://doi.org/10.3390/microorganisms10010158
Chicago/Turabian StyleCheng, Jinan, Hui Jin, Jinlin Zhang, Zhongxiang Xu, Xiaoyan Yang, Haoyue Liu, Xinxin Xu, Deng Min, Dengxue Lu, and Bo Qin. 2022. "Effects of Allelochemicals, Soil Enzyme Activities, and Environmental Factors on Rhizosphere Soil Microbial Community of Stellera chamaejasme L. along a Growth-Coverage Gradient" Microorganisms 10, no. 1: 158. https://doi.org/10.3390/microorganisms10010158
APA StyleCheng, J., Jin, H., Zhang, J., Xu, Z., Yang, X., Liu, H., Xu, X., Min, D., Lu, D., & Qin, B. (2022). Effects of Allelochemicals, Soil Enzyme Activities, and Environmental Factors on Rhizosphere Soil Microbial Community of Stellera chamaejasme L. along a Growth-Coverage Gradient. Microorganisms, 10(1), 158. https://doi.org/10.3390/microorganisms10010158






