Abstract
The present study aimed to show the benefits of novel lactic acid bacteria (LAB) strains isolated from the caeca of healthy chickens. These novel strains, identified as Limosilactobacillus reuteri and Ligilactobacillus salivarius, displayed high levels of lactic acid production, capability of biofilm formation, high aggregation and adhesion scores, and significant survival rates under conditions mimicking the chicken gastrointestinal tract (GIT). In addition, these novel Lactobacillaceae isolates were neither hemolytic nor cytotoxic. In vivo trials were able to establish their ability to reduce necrotic enteritis. Notably, a significant weight gain was registered, on day 10 of treatment, in the group of chickens fed with a mixture of L. reuteri ICVB416 and L. salivarius ICVB430 strains, as compared with the control group. This group has also shown a reduced number of lesions in the gut compared with other infected chicken groups. This study provides in vitro and in vivo evidence supporting the benefits of these novel Lactobacillaceae isolates for their use in poultry livestock as protective cultures to control the bacterial necrotic enteritis (NE) Clostridium perfringens.
1. Introduction
The use of antibiotics as growth promoters in industrial livestock farming remains a controversial issue. Countries from the European Union (Regulation 1831/2003/EC), as well as Mexico, New Zealand and South Korea, have adopted since 2006 laws banning the use of antibiotics as a preventive measure in livestock farming [1]. The reasons for such a decision were mainly associated with the development of antimicrobial resistance (AMR), a serious global health concern.
Currently, around 700,000 people die each year in the world because of AMR-associated diseases [2]. In the USA, at least 2.8 million people suffer an antibiotic resistant infection, of which 35,000 die [3], and the EU counterpart is estimated at 33,000 annually [4]. The AMR crisis needs immediate actions according to different official sources. If left unaddressed, AMR could force 24 million people into extreme poverty by 2030, and will cause 10 million deaths per year around the world with a cumulative cost of US $100 trillion by 2050 [2,5,6].
Therefore, international organizations like WHO, FAO and OIE, but also scientists, and governments are encouraging the development of alternative approaches for replacing aging antibiotics and tackling the AMR phenomenon. In light of this, a holistic approach called One Health was proposed with the aim of rationalizing the usage of antibiotics in humans and animals, considering environmental constraints and the needs of the food industry [7]. Clostridium perfringens type G is responsible for a severe infection in chickens known as bacterial necrotic enteritis (NE). This detrimental infection was shown to be associated with the production of toxins, mainly NetB [8,9], which acts by osmotic cell lysis on enterocytes, causing their death [9,10]. It should be noted that NetB toxin is likely the main virulence factor associated with the avian NE. Clostridium strains are classified into A to G biotypes, based on the toxin produced. Thus, strains producing NetB and α-toxin are included in the newly created biotype G [9]. The α-toxin is a zinc metalloenzyme endowed with hemolytic and dermonecrotic activities [11]. This toxin is responsible for extensive plasma membrane degradation accompanied by a release of lactate dehydrogenase that characterizes necrosis [10]. It is also worth noting that NE is induced by other factors, including coccidiosis, mycotoxins, immunosuppressive agents, nutritional factors and anthropogenic factors of farm management [12]. The bacterial NE infection is responsible for major economic damages in the poultry industry [13].
To tackle AMR, different therapeutic options were suggested, including the use of prebiotics, probiotics from fungal and bacterial sources, bacteriophages, antimicrobial peptides (AMPs), antibodies, vaccines, organic acids and enzymes [14,15].
Probiotics are defined by FAO/WHO as living microorganisms able to provide benefits to the host health when adequately and sufficiently administered [16]. Lactic acid bacteria (LAB) are part of the normal chicken gut microbiota, and play a key role in the maintenance of gut homeostasis, as well as in the control of pathogens through the production of different antimicrobials including bacteriocins, ethanol and CO2 [17]. In addition to these antimicrobials, LAB may help to inhibit the invasion of pathogens through complex competitive exclusion mechanisms [18,19,20]. Some of them can modulate the host immune system by affecting the innate and adaptive immune responses, and induce the production of cytokines [21]. Notably, LAB can increase the bioavailability of nutrients such as fatty acids and vitamins, improve the digestibility of proteins, fiber and phosphorus, which are then mostly absorbed by the host [22,23,24]. LAB strains also have capabilities that can stimulate the production of mucus by the intestinal cells [25,26,27]. Some LAB species, such as Limosilactobacillus reuteri and Ligilactobacillus salivarius, are commonly found in the chicken gut microbiota. These species are reported as adequate probiotic candidates to improve zootechnical performance and poultry health [28].
In this study, we characterized in vitro the potential of newly isolated strains viz L. reuteri ICV416, L. salivarius ICV421 and L. salivarius ICV430, and then established in vivo their capability to control NE, which is a severe infection caused by Clostridium perfringens.
2. Material and Methods
2.1. Isolation of Lactic Acid Bacteria from Chicken Caeca and Their Mass Spectrometry Identification
The bacterial strains were isolated from the caeca of three different chickens (Ross 308) raised in a local farm located in the North of France. LAMVAB media, which corresponds to the de Man Rogosa and Sharpe medium supplemented with L-cysteine-HCl (0.25 g/L) (Sigma-Aldrich, St Louis, MO, USA) and vancomycin (20 mg/L) [29], was used for the isolation of the LAB strains. Therefore, isolates grown on this medium, displaying a Gram-positive staining, devoid of catalase and oxidase activities, were presumably considered as LAB. These isolates were then identified by MALDI-TOF mass spectrometry (Autoflex speed, Bruker Daltonics, Bremen, Germany) performed as described by Zidour et al. [30]. The RAPD-PCR approach using M13 primer [5′-GAGGGTGGCGGTTCT-3′] [31] was performed, enabling the discarding of the redundant LAB strains.
2.2. Anti-Clostridium perfringens Activity
The 49 LAB strains isolated in this work were tested against C. perfringens DSM 756 as well as against six other Clostridium strains isolated from sick chickens, including C. perfringens ICVB081: a strain harboring the netB gene [32]. Two methods were used to assess the anti-C. perfringens activity: the slab test using the whole bacteria and their metabolites according to the protocol described by Dec et al. [33]; and the wells test using the neutralized supernatant according to Bendali et al. [34]. These protocols are detailed thereafter.
Clostridium strains grown under anaerobic conditions were plated onto brain heart infusion (BHI) agar medium (1% (w/v) agar) by swabbing.
LAB strains were cultured in 5 mL of MRS broth at 37 °C for 20 h. The resulting culture was centrifuged (10,000× g, 10 min, 4 °C), and the culture supernatant was separated from the pellets.
For the slab test, the culture was adjusted to an OD600 of 0.4 with phosphate-buffered saline (PBS). Then, 200 μL were spread on a zone of ~4 cm diameter on Petri plates containing MRS agar. After incubation for 20 h at 37 °C, a 6 mm diameter agar column with all of its surface grown with LAB (slab) was cut and placed on top of the agar BHI already inoculated with C. perfringens.
The wells test was performed according to Bendali et al. [34]. The culture was centrifuged (10,000× g, 10 min, 4 °C), and the cell-free culture supernatant was recovered. The pH was adjusted to 6.30–6.60 using 3 M sodium hydroxide (NaOH) and then filtered through a polyethersulfone membrane of 0.45 μm porosity (Corning, Corning, NY, USA). A volume of 40 μL of the neutralized supernatant was deposited in the wells formed purposefully on the Petri plates containing the requested growth medium.
For both assays, the plates were incubated at 4 °C for 2 h in order to allow diffusion of the putative antibacterial substances. After incubation at 37 °C for 20 h under anaerobic conditions, the plates were inspected for development of zones of inhibition, and in case of a positive result, the radials were measured. Enterococcus faecalis 14 producing a leaderless class IIb enterocin 14 (Ent DD14), known for its activity against C. perfringens [35], was used as a positive control.
2.3. Quantification of Lactic Acid Production
The quantity of lactic acid produced by these newly isolated LAB strains was determined by high performance liquid chromatography (HPLC) assay after a standard culture in MRS medium for 20 h at 37 °C. An optical density (OD) reading at 600 nm and pH measurement were performed and followed by centrifugation (10,000× g, 10 min, 4 °C) to recover the cell-free culture supernatants, which were filtered through Millipore filters (0.2 μm) (Burlington, MA, USA). Lactic acid was quantified by the HPLC Spectra System P1000 XR (Thermo Fisher, Waltham, MA, USA). The column used was a Fast Juice Column (50 mm × 7.8 mm, Phenomenex, Torrance, CA, USA) with isocratic elution with H3PO4 (0.05 % w/w), at a flow rate of 0.8 mL/min and a temperature of 55 °C.
2.4. Biofilm Formation
Bacterial cultures, 18 h in age, recovered at an OD600nm of 5 and were diluted at 1:50 using fresh MRS broth in 96-well polystyrene plates. Once inoculated, these plates were anaerobically incubated at 37 °C for 24 h. The protocol described by Jones and Versalovic [36] was used to quantify the formed biofilms. To be brief about the culture time, the plates were washed twice to remove the non-adherent bacteria from the bottom of the wells. A solution of 0.1% (w/v) crystal violet (Sigma-Aldrich) was used to stain the bacteria forming biofilms. After 15 min of incubation at 37 °C and centrifugation (200 rpm), the plates were washed again with distilled water. Finally, 96% (v/v) of ethanol was added to remove the leftover crystal violet. The absorbance was recorded at 600 nm using a Safas MP96 microplate reader (Safas, Monaco, France) in order to quantify the biofilm formation.
2.5. Resistance to Gastrointestinal Conditions in Chicken
Conditions mimicking the different stages of the digestion in the crop, gizzard and intestine of chicken were studied. For the crop, the following conditions were applied: pH of 4.5 for 45 min, followed by a decrease of pH to 3.5 for 90 min in the presence of pepsin (3 mg/mL) (Sigma-Aldrich) and 2 g of glass beads (2 mm in diameter), shaken at 350 rpm to simulate grinding in the gizzard. Finally, an increase of pH to 6.4 and an addition of pancreatin at 1 mg/mL (Sigma-Aldrich) and bile oxgall at 0.35% (Sigma-Aldrich) were applied for 3 h to mimic the passage through the intestine [37,38]. All of these steps were performed at 41.3 °C, which corresponds to the physiological temperature of the chicken [39]. Lacticaseibacillus rhamnosus ATCC 7469 and E. faecalis 14 were used as controls [35,40]. Strain cultures, 20 h in age and grown at 37 °C, were centrifuged (5000× g, 15 min, 4 °C), and the resulting pellets were recovered, washed with 0.9 % (w/v) of sodium chloride (NaCl) and resuspended in different solutions corresponding to the conditions described above. Samples were taken and bacterial cells were counted at T = 0 after each step/compartment and cultivation on MRS agar plates. The number of bacteria and the strain viability were determined by flow cytometry (Attune NxT, Thermo Fisher, Waltham, MA, USA). The dyes used to assess LAB viability were syto-24 (Invitrogen, Waltham, MA, USA) and propidium iodide (Molecular Probes, Eugene, OR, USA) at final concentrations of 10 nM and 200 nM, respectively. Flow cytometer channels were set up as follows: FSC 100, SSC 300. After data acquisition, a common gate was determined to follow the changes in bacteria viability in all of the gastrointestinal steps.
2.6. Adhesion to Intestinal Cells
Adhesion of C. perfringens and newly isolated LAB strains on Caco-2 intestinal epithelial cells was determined in vitro. To this end, Caco-2 cells were seeded in 24-well plates, at a loading of 4 × 104 cells per well, and incubated for 7 days at 37 °C in a 95% humidity with 5% CO2 in Dulbecco’s modified eagle medium (DMEM) (Lonza, Bâle, Switzerland) supplemented with 25 mM glucose, 5 mM L-glutamine, 10 % (v/v) fetal calf serum (FBS) and 100 U/mL penicillin and streptomycin.
For the bacterial adhesion test, cultures of LAB and C. perfringens strains were prepared in MRS and BHI broth, respectively. The bacterial cells were recovered by centrifugation (8000× g, 10 min, 4 °C), washed twice with PBS buffer and finally resuspended in a non-complemented DMEM medium. Then, a volume of 500 µL of these suspensions was added to each well containing Caco-2 cells at a ratio of 1:10 (Caco-2 cell: bacterial cells). After incubation at 37 °C for 2 h, Caco-2 cell monolayers were washed twice with PBS at 37 °C, treated with 200 µL trypsin/EDTA (Gibco, Waltham, MA, USA) and incubated for 10 min at 37 °C to detach the cells. The percentage of adhesion was determined via the method described by Candela et al. [41] using real-time PCR.
2.7. qPCR Assay
An aliquot of 20 μL of each sample was transferred to a 0.2 mL tube and incubated for 10 min at room temperature with 3.8 μL of trypsin inhibitor solution (Sigma-Aldrich). The real-time quantitative PCR (QuantStudio®3, Applied Biosystems, Waltham, MA, USA) was performed with the fluorophore SYBR Green I (Molecular Probes) in order to quantify the fluorescent signal. The primers used here were CPerf165F [5′-CGCATAACGTTGAAAGATGG-3′] and CPerf269R [5′-CCTTGGTAGGCCGTTACCC-3′] for C. perfringens [42], and LactoG1F [5′-TGGAAACAGRTGCTAATACCG-3′] and LactoG1R [5′-GTCCATTGTGGAAGATTCCC-3′] for Lactobacillaceae.
Amplification was performed in a final volume of 20 μL containing 2 μL of the cell suspension, 0.4 μM of each primer and supplemented with SYBR® Select Master Mix (Applied Biosystems). The program applied was the following: pre-incubation step at 94°C for 10 min, 40 cycles of amplification (95 °C for 15 s, 60 °C for 10 s, 72 °C for 30 s) and finally 72 °C for 10 min. For absolute quantification, dilutions of each tested bacterium in PBS with concentrations of 103 to 106 CFU/mL were used as standards.
2.8. Biosafety Aspects of Newly Selected LAB Strains
2.8.1. Hemolytic Activity
LAB strains were pre-cultured in MRS broth at 37 °C for 24 h. After isolation on MRS agar, three colonies were randomly selected and plated onto a Columbia agar + horse blood (Oxoid, Basingstoke, UK) medium. The plates were incubated anaerobically at 37 °C and were inspected for any hemolysis after 24, 48 and 72 h. The presence of a clear area converges for a complete hemolysis (Β-hemolysis), whilst the greenish hemolysis (α-hemolysis) is known as an incomplete hemolysis. The absence of any hemolysis (γ-hemolysis) is usually observed for Lactobacillaeae [43]. Here, Staphylococcus aureus ATCC 25923 (Β-hemolytic) was used as a positive control strain [44].
2.8.2. Antibiotic Resistance
The bacterial cultures were swabbed uniformly onto the surface of the agar medium. After drying, antibiotic discs were applied, and the plates were incubated at 37 °C for a period of 24–48 h. After this period of incubation, the plates were carefully examined and the radials of the zones of inhibition were recorded. Sensibility and resistance traits of these novel LAB strains were determined according to the recommendations of the EUCAST manual [45]. The genome of the selected LAB strains was analyzed for the presence of antibiotic resistance genetic determinants. DNA of each strain was extracted and sequenced using the protocol described by Al Seraih et al., [46]. Genome annotation was performed using RAST (http://rast.nmpdr.org, accessed on 15 May 2018).
2.8.3. Cytotoxicity
HT29 cells were grown at 37 °C in a humid atmosphere with 5% CO2 in DMEM medium (Lonza) supplemented with 25 mM glucose, 5 mM L-glutamine, 10% (v/v) fetal calf serum (FBS), 100 U/mL penicillin and streptomycin. The test was performed with 30,000 HT29 cells.
The selected LAB strains were grown for 36 h at 37 °C in MRS broth. Cultures were centrifuged (10 min, 10,000× g, 4 °C) and resuspended in DMEM without antibiotics or FBS. Contact was made for 24 h at 37 °C and 5% CO2. Bacteria were tested at 105 CFU/well, or 107 CFU/well. Heat-inactivated bacteria (95 °C for 5 min) were used as a control to ascertain that no cytotoxic components were present on the bacterial surface.
To measure the cytotoxic effect, after contact, the cells were washed twice with PBS in order to remove bacteria. DMEM medium, which is supplemented with gentamicin (50 μg/mL) and 5 % of the CCK-8 “Cell Counting Kit-8” reagent (Dojindo Molecular Technology, Rockville MD, USA), was added to each well. The OD at 480 nm was measured after 2 h. Notably, these tests were performed in triplicate.
2.9. Coccidiostat Analysis
Sensitivity to coccidiostat was determined by using the minimum inhibitory concentration (MIC) based on the method of CLSI M45 [47]. Strains were grown overnight at 37 °C in LAB susceptibility test medium (LSM) under anaerobic conditions. Coccidiostats solutions (monensin sodium salt, narasin, salinomycin, maduramicin ammonium, lasalocid A sodium salt, decoquinate, diclazuril, halofuginone hydrobromide, robenidine hydrochloride, narasin:nicarbazin) were freshly prepared on the day of testing in DMSO at concentrations suggested by the EFSA [48]. Notably, DMSO was used at 5% (v/v), at which concentration DMSO has no effect on cell viability (data not shown). A volume of 10 µL of bacterial suspension (5 × 106 cells/mL) was inoculated in the wells with coccidiostat dilutions that were prepared in 100 µL of LSM medium. The microplates were incubated and the OD600 was read at 48 h. S. aureus ATCC29213 grown in BHI for 24 h in aerobic conditions was used as a control [49,50].
2.10. In Vivo Evaluation of the Three Newly Isolated Lactobacillaceae Strains
A trial was conducted to determine the efficacy of these newly isolated LAB strains and evaluate their potential application as preventive agents to control NE in broiler chickens from a cross of the Cobb 500 (female) and Hubbard M99 (male) lines. This study was performed using L. reuteri ICV416, L. salivarius ICV421 and L. salivarius ICV430 strains regarding their promising probiotic features assessed in vitro. Notably, in vivo experiments were conducted for 17 days on 240 chickens (30 animals split in 6 cages per treatment group). The trial started on the day of hatching and set up as a near complete factorial design. During trials, LAB strains were administered alone (L. reuteri ICVB416, L. salivarius ICVB421 or L. salivarius ICVB430), in pairwise combination (L. reuteri ICVB416 and L. salivarius ICVB421; L. reuteri ICVB 416 and L. salivarius ICVB 430) and all three together, knowing that no potential cross-killing effect exists (data not shown). These strains were administered by oral gavage once daily, on days 1 and 2, and then again from day 10 to day 13.
2.10.1. Chicken Environment and Diet
The chickens were raised in cages with an average of 432 cm2 per chicken. The cages were placed at several levels in an air-conditioned room, maintained at room temperature throughout the study and cleared for testing with a level 2 biohazard. The conditions were selected according to the Guide for the Care and Use of Agricultural Animals in Research and Education (https://www.aaalac.org/about/ag_guide_3rd_ed.pdf, accessed on 3 June 2018). After the approval of the Institutional Animal Care and Use Committee (IACUC), under the number 2018–009 at the United States Department of Agriculture—Agricultural Research Service (College Station, TX, USA), the test was started. Lighting was provided 24 h a day for the duration of the study. The birds were given water and ad-libitum food. The diet consisted of the mash feed underlie of corn and soy for a starter until day 9. Then, from day 10, the grower diet included wheat. The diet’s compositions are listed in Tables S1 and S2 (Supplementary Materials).
2.10.2. Vaccination
Except for the non-challenged group, the chickens were vaccinated on day 1 with coccidiosis vaccine Advent (Huvepharma, Peachtree, GA USA) at a high dose (9×) to challenge birds. Advent vaccine contains low pathogenic, highly immunogenic and coccidiostat-sensitive strains of Eimeria maxima, E. tenella and E. acervulina. On day 9, the chickens were vaccinated intraocularly against avian infectious bursal disease with Intervet, Bursal Vac-G603.
2.10.3. C. perfringens Infection Protocol
C. perfringens–containing medium was provided by the ARS, Southern Plains Agricultural Research Center, USDA. The isolation and preparation of C. perfringens were as described by McReynolds et al. [51]. The C. perfringens pathogen was a combination of four formerly type A field isolates from three different regions (Georgia, Texas and Virginia). The wild type C. perfringens strains were grown overnight at 37 °C in thioglycollate broth as previously described [52]. After this, it was administered at a concentration of 107 CFU/mL with a volume of 3 mL by oral gavage dose in a sterile thioglycollate, using a 20-gauge dosing needle at 14–16 days of age.
2.10.4. Administration of LAB Strains
LAB strains were grown overnight at 37 °C in MRS. Cell pellets were recovered and resuspended in PBS, and 250–500 µL of obtained LAB strain suspension, at 107 CFU/mL, were administered orally to the birds on days 1 and 2. Notably, when the birds were 10–13 days old, the dosage was increased to 1 mL, but still at a concentration of 107 CFU/mL. The control groups (NCp and Cp) were treated in a similar way and received the same volumes of sterile PBS solution. The birds used for the negative control were treated first to reduce the risk of cross-contamination.
2.10.5. Measurements of Feed Uptake and Weight of Birds
The study ended on day 17. Bird performance was measured on days 0, 10, 14 and 17 of the experimental periods by recording bird weight (in g) and feed uptake for each cage. This made it possible calculate the “Feed Conversion Ratio”, which is defined as the feed consumed in kg per kg of body weight gain of the animal. This is an important measure to evaluate the economic performance of the farm [53]. On day 10, one chicken from each pen unit was necropsied, and sampling was done. At the end of the trial, all of the remaining birds were euthanized by CO2 asphyxiation. NE lesions were scored in the intestines and analyzed following the recommendations of Prescott et al. [54].
2.11. Statistical Analysis
In vitro studies were performed in triplicate. In vivo assay had five pen replicates for each treatment. Statistical comparisons between the different results obtained were made by analysis of variance ANOVA using Statgraphics® Centurion XVI software (Statpoint Technologies, Warrenton, VA, USA).
3. Results
3.1. Diversity of LAB Strain in Chicken Ceca
The 70 isolates obtained from the caeca of chickens using LAMVAB medium were identified by MALDI-TOF-MS and characterized by RAPD-PCR to discard duplicates and redundant LAB strains. This procedure made it possible to identify 28 L. salivarius strains, 14 L. reuteri, 1 Lactobacillus gallinarum, 1 Lactobacillus gasseri, 1 Lactobacillus johnsonii, 1 Limosilactobacillus antri, two Streptococcus lutetiensis and 2 Streptococcus alactolyticus.
3.2. Newly ISOLATED LAB Strains Displayed Strong Anti-Clostridium perfringens Activity
Antibacterial activity of these newly isolated LAB strains was tested against C. perfringens DSM756 as well as six other Clostridium strains isolated from sick chickens. L. salivarius ICVB421 and ICVB430 strains presented the upmost zones of inhibition, with diameters of 2.07 and 1.63 cm, respectively. L. reuteri ICVB416, L. rhamnosus ATCC 7469 and E. faecalis 14 exhibited less activity against the aforementioned target bacteria, with diameters of inhibition of 1.13, 1.07 and 0.93 cm, respectively (Tables S3 and S4). In this assessment, we noted the peculiar sensitivity of C. perfringens ICVB081 to L. salivarius strains (Figure 1).
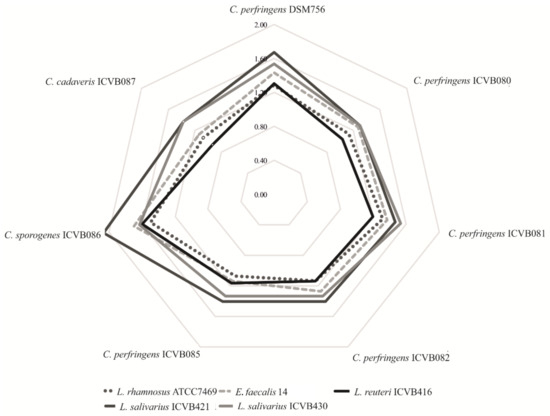
Figure 1.
Anti-Clostridium activity depicted as a spider chart. Zones of inhibition were assessed by measuring the radius of the halo in cm. Target strains are in the borders and lines show the activity of probiotic candidates.
After neutralizing the pH of the culture-supernatant, the newly isolated Lactobacillaceae strains did not display any inhibitory activity, in contrast to the neutralized culture-supernatant of E. faecalis 14 used as the positive control (data not shown). These data suggest that antibacterial activity of the newly isolated LAB-strains was attributable to organic acids such as lactic acid, the amount of which ranged from 11.3 to 13.02 g/L for L. salivarius strains and 6.91 g/L for the L. reuteri strain (Table S5).
3.3. Biofilm Formation
These newly isolated Lactobacillaceae strains have various degrees of adhesion to polystyrene. The most efficient strains in terms of biofilm formation were L. salivarius ICVB430 and ICVB421, with absorbencies of 4.00 and 2.17 at 600 nm, whereas L. reuteri strain ICVB416 showed absorbance of 1.64 (Table S5).
3.4. Resistance to Chicken Gastrointestinal Conditions
Assessment of conditions mimicking the GIT environment made it possible to select strains with a high probability of reaching the intestine. After counting colonies on MRS agar, L. reuteri ICVB416 presented a significantly higher level of survival rate to the GIT conditions (73.47%) than other strains, which have survival rates of approximately 32% (Table S5). Nevertheless, when the living cells were quantified using the flow cytometry method, the values augmented as viable but non-culturable (VBNC) cells were included in the cells counts. Interestingly, the newly selected LAB strains possessed better survival rates than the reference strains used, L. rhamnosus ATCC7469 and E. faecalis 14 (Figure 2).
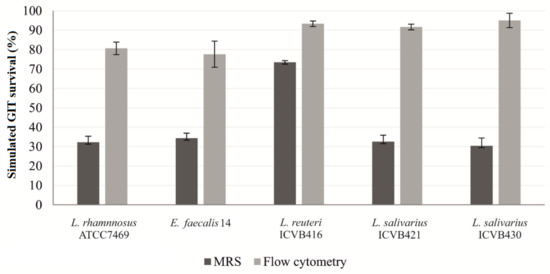
Figure 2.
Survival rates of LAB strains under conditions mimicking chicken gastrointestinal environment (%) using classical counting in MRS and flow cytometry. The cytometry assay was performed after labelling the cells with syto-24 and propidium iodide.
3.5. Adhesion to Intestinal Cells
A real-time qPCR method was used to determine the percentage of adhesion of LAB strains to eukaryotic Caco-2 cells. The percentage of adhesion was calculated in relation to the number of bacteria inoculated. L. salivarius ICVB421 has the highest percentage of adhesion (8.02%), more than the pathogenic C. perfringens DSM756 strain, which displayed a percentage of 5.85%. It should be noted that, compared to L. salivarius strains, L. reuteri ICVB416 was weakly adherent with a percentage of adhesion estimated at 1.06%,. Similarly, the reference strain L. rhamnosus ATCC7469 displayed a very low level of adhesion (0.19%), as shown in Figure 3.
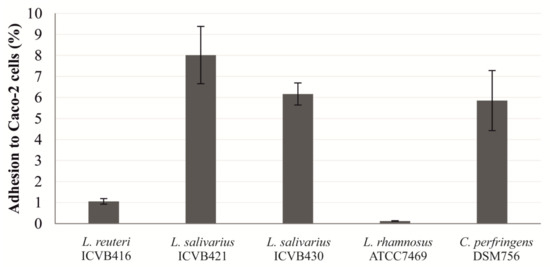
Figure 3.
Adhesion percentage of Lactobacillaceae strains to eukaryotic Caco-2 cells as determined by quantitative PCR (qPCR).
3.6. Safety Assessment of Lactobacillaceae Strains
None of the Lactobacillaceae strains tested here showed hemolytic activity after 24, 48 and 72 h of incubation. Nevertheless, L. salivarius ICVB430 and L. salivarius ICVB421 exhibited a very limited cytotoxic effect on the eukaryotic HT-29 cell line, with more than 70% viability, but only when they were tested at a concentration of 107 CFU/well. When the three Lactobacillaceae strains were tested together, further cytotoxicity was observed decreasing the HT-29 cell viability to less than 40%. When tested at a concentration of 105 CFU/well, no cytotoxic effect on HT-29 cells was registered (Figure 4). Regarding the antibiotic resistance, L. reuteri and L. salivarius strains exhibited resistance to penicillin G and vancomycin according to critical points established by EUCAST. Notably, resistance to trimethoprim-sulfamethoxazole was observed for L. salivarius ICVB430 (Table 1).
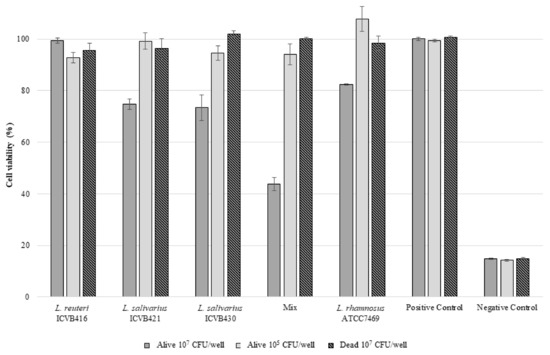
Figure 4.
Cell viability (%) of the eukaryotic cell line HT-29 after 24 h contact with LAB. ICVB416: L. reuteri ICVB416; ICVB421: L. salivarius ICVB421; ICVB430: L. salivarius ICVB430. MIX: Mixture in equal volumes of three precedent strains. ATCC7469: L. rhamnosus (reference strain). Positive control: Medium without bacteria. Negative control: Triton 0.1%.

Table 1.
Antibiograms of Lactobacillaceae strains tested in vivo. The sizes of inhibition zones are indicated in mm between brackets.
3.7. Coccidiostat Analysis
The Lactobacillaceae strains tested were resistant to diclazuril and sensitive to the rest of the coccidiostat tested. The MIC range was lower than 1 ppm when these Lactobacillaceae strains were tested against narasin, salinomycin, maduramicin ammonium and lasalocid A sodium (Table 2).

Table 2.
Minimal inhibitory concentration (MIC) of coccidiostat usually used in poultry production. Values are expressed in ppm.
3.8. In Vivo Trials
Zootechnical Performance during the Start-Up Phase (0–14 Days)
The administration of Lactobacillaceae strains alone or in mixture in the first two days of the bird’s life did not significantly influence the weight or feed intake of the chickens (Table 3). On day 10, the group treated with the combination of L. reuteri ICVB416 and L. salivarius ICVB430 presented a significantly higher weight than the control. From days 10 to 14, no significant weight gain or increase in food consumption were observed in the Lactobacillaceae-treated groups. Different effects were found from the evaluations of the lesion scores. Interestingly, the group treated concomitantly with L. reuteri ICVB416 and L. salivarius ICVB430 showed lower lesion scores in comparison to other infected chicken groups (Figure 5 and Figure 6).

Table 3.
Effects on chicken weight, feed consumption and FCR (Feed Conversion Ratio) after Lactobacillaceae supplementation. FCR is the ratio of measuring the efficiency with which the bodies of livestock convert animal feed into the desired output.
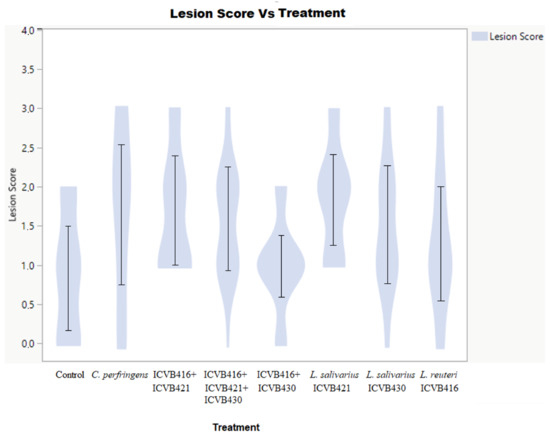
Figure 5.
Lesion score layout. It shows the distribution of lesions depending on the treatment. It shows proportionally the number of animals that presented each type of lesion score depending on the treatment.
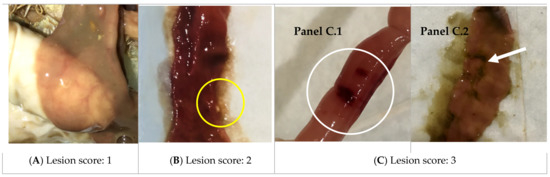
Figure 6.
Necrotic enteritis lesions. (A) Score 1: intestine is thin and friable; (B) Score 2: the picture shows the thin, friable intestine with a small patch of necrosis (yellow circle). There is also redness in the intestinal tract, and some gray mucus is characteristic; (C) Score 3: the intestine shows a larger lesion visible through the outer wall of the intestinal tract. There are larger patches of necrosis (white circle and arrow), which are the green areas stained by bile. Panel C.1 shows the closed tube and panel C.2 shows the open tube and exposed inner side above.
4. Discussion
Microbial infections such as NE can lead to critical damage and a significant number of deaths when no treatments are applied [55,56,57]. NE is known to afflict the GIT of poultry and cause major economic losses around the world, which can reach up to 6 billion US $ per year [58]. Administration of antibiotics such as penicillin G, amoxicillin, ampicillin, bacitracin, neomycin and tylosin have been suggested as means of preventing NE [59], but strains of C. perfringens with resistant phenotypes have been reported [60,61], which delineate the potential of this etiologic agent to defy aging antibiotics. Further measures such as vaccination, bacteriophages, usage of AMPs, prebiotics and probiotics have been insistently proposed to control the bacterial NE infection [62,63].
Lactobacillaceae strains isolated in this work include L. reuteri and L. salivarius, which are particularly active against C. perfringens through their production of lactic acid as assessed in vitro. Although this inhibitory activity is exerted in a strain-dependent manner, the data obtained here are in agreement with those formerly reported [64,65,66]. Lactic acid causes damages in the cell membrane, leading to a cascade of deleterious effects such as inhibition of enzymatic activities, alteration of DNA structure and cell death [67,68].
Besides, LAB strain can form biofilms which can be a strategy to control growth of pathogenic bacteria. The formation of biofilms is noticeably important for selecting and designing probiotic candidate strains; even this criterion remains versatile within the LAB group [36]. It is worth noting that the biofilm formation was reported for LAB strains as a means of controlling pathogens such as Listeria and Salmonella [69,70]. The adhesion of LAB to intestinal cells is considered an element in selecting and characterizing strains, which are candidates for probiotic claims. LAB-strains with such adhesion ability can stand as a barrier against enteropathogens, offering advantages to the host by increasing their time of transit in the gut, helping them compete for nutrients, discarding pathogens from binding sites and increasing the host’s immunity [71,72,73]. In this study, we show that strains of L. salivarius are more adherent to Caco-2 cells than those of L. reuteri. The scores of adhesion obtained for these newly isolated LAB strains are globally in strong agreement with those reported in the literature [64,74]. The ability of bacteria to defy and survive under the GIT conditions can be simulated in vitro, and the resulting data are crucial for validating their suitability for probiotic applications. Interestingly, L. reuteri showed a better potential capability to survive the passage through the simulated chicken GIT than other LAB strains tested under similar conditions. This peculiar advantage can be linked to its ability to precipitate the bile salts via the action of bile salt hydrolase [75,76,77].
The method of assessing GIT survival by agar enumeration is deemed tedious and does not take into account the VBNC [78]. The survival of LAB strains was confirmed by the flow cytometry method, which includes both viable bacteria and VBNC. As expected, the survival percentages obtained by this method were significantly elevated, compared to data obtained after their growth and enumeration on the MRS media. These observations were remarkably obtained and validated for all strains, and scores were comprised between 80 to 90%.
It is worth noting that all of the selected Lactobacillaceae strains were devoid of hemolytic activity, and assessment of their cytotoxicity on eukaryotic cells revealed a very limited effect of L. salivarius strains on HT-29 cells at a concentration of 107 CFU/well, or in conjunction with other selected Lactobacillaceae strains. Nevertheless, at 105 CFU/well, this cytotoxic effect was abolished, underlining the importance of cell concentrations, as has been previously reported by Er et al. [79]. The three selected Lactobacillacae strains exhibited resistance to some antibiotics, but such a feature is not deemed restrictive for their application [80].
Genome analyses of the LAB strains isolated here enabled us to locate gene coding for resistance to antibiotics on chromosomes and permitted us to identify the coding for efflux pumps, which are known to use the energy of ion or substrate-product gradients to expel cytotoxic compounds [81]. On the other hand, resistance to vancomycin is believed to naturally occur in Limosilactobacillus and Ligilactobacillus species [82], and the genome analyses carried out in this study did not show any genetic mobile element. Taken together, these in vitro tests made it possible to design the most promising candidates for probiotic applications.
Our focus was concentrated on the use of these newly isolated strains as means to control the bacterial NE, which continues to emerge in the poultry livestock as a harmful bacterial infection with large economic consequences. To confirm and strengthen all of the data obtained in vitro, additional trials were undertaken to provide evidence of in vivo efficacy. To that end, coccidiostats, which are frequently administered for their action against the Eimeria parasite, were used. Nonetheless, the effects of coccidiostats were tested on newly isolated Lactobacillaceae, and the data obtained for monensin sodium, diclazuril, salinomycin, lasalocid A and robenidine were in strong agreement with previously reported data [83,84]. These newly isolated Lactobacillaceae strains were sensitive to narasin, with values of 0.016 to 0.125 ppm; below are those reported for L. fermentum strains [85].
The administration of Lactobacillaceae strains, alone or in conjunction during the first two days of the chicken’s life, did not influence their weights or feed consumption. Only the group treated with L. reuteri ICVB416 + L. salivarius ICVB430 showed a weight gain, compared to the control on day 10. It is reported that probiotics are most apparent in the first days of life, when the initial microbiota is in development [86]. The effectiveness of probiotics is noticeable after a change in the diet, following any stress disturbance or antibiotic uptake [87]. Moreover, administration of high concentrations of L. salivarius (109 CFU/g) impact the microbiota by translating them into an increase in body weight [88]. Other studies have suggested that no significant effects are registered on animal performance when treated with Lactobacillaceae probiotics [89,90].
The results obtained after infection with C. perfringens revealed the advantages of the combination of L. reuteri ICVB416 and L. salivarius ICVB430, since a significant effect on the weight gain and lesion score of chickens was registered. The synergistic effect between two, or even more, probiotic strains has already been reported. A previous study by Mappley et al. [91] mentioned the benefits of the combination of L. reuteri LM1 and L. salivarius LM2 against avian intestinal spirochetosis [91]. In line with this, Carter et al. [92] reported a protective effect when L. salivarius 59 and E. faecium PXN33 were used in combination to reduce Salmonella loads in the chicken. Different studies pointed out the protective role of Lactobacillaceae strains against C. perfringens [93,94,95]. Notable, Lactobacillaceae probiotics have been proposed as preventive treatments for the bacterial NE. This is the case for L. johnsonii FI9785, which was shown to reduce the extent of colonization and persistence of C. perfringens in 20-day-old chicks when it was administered at 109 CFU [96]. On the other hand, L. fermentum 1.2029 brought about a reduction of lesions due to NE in chickens by modulating the immune response of the intestinal mucosa [97].
5. Conclusions
The combination of L. reuteri ICVB416 and L. salivarius ICVB430 has a good in vitro probiotic capacity, which is translated into an increased performance and preventative treatment against the bacterial NE in chickens. Additional experiments are requested to determine when and how these newly isolated Lactobacillus strains are expected to be administered during the chicken’s life.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10010152/s1, Table S1: Feed formula, Table S2: Nutrient composition, Table S3: Anti-Clostridium perfringens DSM756 activities of lactic acid bacteria, Table S4: Anti-Clostridium perfringens ANSES Clin1 activities of lactic acid bacteria, Table S5: Characterization of LAB from the study.
Author Contributions
Conceptualization, D.D., E.A. and N.V.-S.; Methodology, N.V.-S., Y.B., R.R., C.P., C.B. and F.G.; Software, N.V.-S.; Validation, D.D., E.A., Y.B. and F.G.; Formal Analysis, D.D.; Investigation, N.V.-S. and Y.B.; Resources, D.D., E.A. and R.R.; Data Curation, N.V.-S.; Writing—Original Draft Preparation, N.V.-S. and Y.B.; Writing—Review & Editing, D.D. and Y.B.; Visualization, D.D.; Supervision, D.D., E.A. and F.G.; Project Administration, D.D. and E.A.; Funding Acquisition, D.D. and E.A. All authors have read and agreed to the published version of the manuscript.
Funding
NVS was supported by a PhD grant awarded by Phileo Lesaffre and ANRT.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care and Use Committee (IACUC) with number for the in vivo trial conducted at the USDA is 2018-009.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw data will be provided upon request.
Acknowledgments
This work was support by Phileo, from la Société Lessafre, through ANRT Ph.D. grant to Nuria Vieco. Part of this work was supported by Alibiotech CPER/FEDER 2015–2020 grant from la région des Haut-de-France (http://www.alibiotech.eu/, accessed on 10 December 2021). We would also like to thank the French Agency for Food, Environmental and Occupational Health & Safety (ANSES), which kindly provided the Clostridium strains used in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Laxminarayan, R.; van Boeckel, T.; Teillant, A. The Economic Costs of Withdrawing Antimicrobial Growth Promoters from the Livestock Sector; OECD Food, Agriculture and Fisheries Papers; OECD: Paris, France, 2015; Volume 78. [Google Scholar]
- WHO New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 16 June 2021).
- CDC Antibiotic Resistance Threatens Everyone. Available online: https://www.cdc.gov/drugresistance/index.html (accessed on 10 June 2021).
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Rev. Antimicrob. Resist. 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 16 June 2021).
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Antibiotic Resistance: Moving from Individual Health Norms to Social Norms in One Health and Global Health. Front. Microbiol. 2020, 11, 1914. [Google Scholar] [CrossRef]
- Keyburn, A.L.; Boyce, J.D.; Vaz, P.; Bannam, T.L.; Ford, M.E.; Parker, D.; Di Rubbo, A.; Rood, J.I.; Moore, R.J. NetB, a New Toxin That Is Associated with Avian Necrotic Enteritis Caused by Clostridium perfringens. PLOS Pathog. 2008, 4, e26. [Google Scholar] [CrossRef] [Green Version]
- Rood, J.I.; Adams, V.; Lacey, J.; Lyras, D.; McClane, B.A.; Melville, S.B.; Moore, R.J.; Popoff, M.R.; Sarker, M.R.; Songer, J.G.; et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 2018, 53, 5–10. [Google Scholar] [CrossRef]
- Navarro, M.A.; McClane, B.A.; Uzal, F.A. Mechanisms of Action and Cell Death Associated with Clostridium perfringens Toxins. Toxins 2018, 10, 212. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, J.; Nagahama, M.; Oda, M. Clostridium perfringens Alpha-Toxin: Characterization and Mode of Action. J. Biochem. 2004, 136, 569–574. [Google Scholar] [CrossRef] [Green Version]
- Moore, R.J. Necrotic enteritis predisposing factors in broiler chickens. Avian Pathol. 2016, 45, 275–281. [Google Scholar] [CrossRef] [Green Version]
- M’Sadeq, S.A.; Wu, S.; Swick, R.; Choct, M. Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide. Anim. Nutr. 2015, 1, 1–11. [Google Scholar] [CrossRef]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.W.; Harper, D.; et al. Alternatives to antibiotics—A pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Seal, B.S.; Lillehoj, H.S.; Donovan, D.M.; Gay, C.G. Alternatives to antibiotics: A symposium on the challenges and solutions for animal production. Anim. Heal. Res. Rev. 2013, 14, 78–87. [Google Scholar] [CrossRef] [Green Version]
- FAO/WHO Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Guidelines for the Evaluation of Probiotics in Food. Available online: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (accessed on 30 April 2020).
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [Green Version]
- Callaway, T.R.; Edrington, T.S.; Anderson, R.C.; Harvey, R.B.; Genovese, K.J.; Kennedy, C.N.; Venn, D.W.; Nisbet, D.J. Probiotics, prebiotics and competitive exclusion for prophylaxis against bacterial disease. Anim. Health Res. Rev. 2008, 9, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Klose, V.; Mohnl, M.; Plail, R.; Schatzmayr, G.; Loibner, A.-P. Development of a competitive exclusion product for poultry meeting the regulatory requirements for registration in the European Union. Mol. Nutr. Food Res. 2006, 50, 563–571. [Google Scholar] [CrossRef]
- Siedler, S.; Rau, M.H.; Bidstrup, S.; Vento, J.M.; Aunsbjerg, S.D.; Bosma, E.F.; McNair, L.; Beisel, C.L.; Neves, A.R. Competitive Exclusion is a major bioprotective mechanism of Lactobacilli against fungal spoilage in fermented milk products. Appl. Environ. Microbiol. 2020, 86, e02312-19. [Google Scholar] [CrossRef] [Green Version]
- Kiczorowska, B.; Samolińska, W.; Al-Yasiry, A.R.M.; Kiczorowski, P.; Winiarska-Mieczan, A. The natural feed additives as immunostimulants in monogastric animal nutrition—A review. Ann. Anim. Sci. 2017, 17, 605–625. [Google Scholar] [CrossRef] [Green Version]
- Leblanc, J.; Laiño, J.E.; Del Valle, M.J.; Vannini, V.; Van Sinderen, D.; Taranto, M.; De Valdez, G.F.; De Giori, G.S.; Sesma, F. B-Group vitamin production by lactic acid bacteria—Current knowledge and potential applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef]
- Liao, S.F.; Nyachoti, M. Using probiotics to improve swine gut health and nutrient utilization. Anim. Nutr. 2017, 3, 331–343. [Google Scholar] [CrossRef]
- Sharifuzzaman, S.; Austin, B. Probiotics for disease control in aquaculture. Diagn. Control Dis. Fish Shellfish 2017, 189–222. [Google Scholar] [CrossRef]
- Ljungh, A.; Wadström, T. Lactic Acid Bacteria as Probiotics. Curr. Issues Intest. Microbiol. 2006, 7, 73–89. [Google Scholar]
- Park, Y.H.; Hamidon, F.; Rajangan, C.; Soh, K.P.; Gan, C.Y.; Lim, T.S.; Abdullah, W.N.W.; Liong, M.T. Application of probiotics for the production of safe and high-quality poultry meat. Korean J. Food Sci. Anim. Resour. 2016, 36, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Vieco-Saiz, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Khalique, A.; Zeng, D.; Shoaib, M.; Wang, H.; Qing, X.; Rajput, D.S.; Pan, K.; Ni, X. Probiotics mitigating subclinical necrotic enteritis (SNE) as potential alternatives to antibiotics in poultry. AMB Express 2020, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hartemink, R.; Domenech, V.; Rombouts, F. LAMVAB—A new selective medium for the isolation of lactobacilli from faeces. J. Microbiol. Methods 1997, 29, 77–84. [Google Scholar] [CrossRef]
- Zidour, M.; Chevalier, M.; Belguesmia, Y.; Cudennec, B.; Grard, T.; Drider, D.; Souissi, S.; Flahaut, C. Isolation and characterization of bacteria colonizing acartia tonsa copepod eggs and displaying antagonist effects against Vibrio anguillarum, Vibrio alginolyticus and other pathogenic strains. Front. Microbiol. 2017, 8, 1919. [Google Scholar] [CrossRef]
- Stenlid, J.; Karlsson, J.-O.; Högberg, N. Intraspecific genetic variation in Heterobasidion annosum revealed by amplification of minisatellite DNA. Mycol. Res. 1994, 98, 57–63. [Google Scholar] [CrossRef]
- Vieco-Saiz, N.; Belguesmia, Y.; Vachée, A.; Le Maréchal, C.; Salvat, G.; Drider, D. Antibiotic resistance, genome analysis and further safe traits of Clostridium perfringens ICVB082; A strain capable of producing an inhibitory compound directed only against a closely related pathogenic strain. Anaerobe 2020, 62, 102177. [Google Scholar] [CrossRef]
- Dec, M.; Puchalski, A.; Urban-Chmiel, R.; Wernicki, A. Screening of Lactobacillus strains of domestic goose origin against bacterial poultry pathogens for use as probiotics. Poult. Sci. 2014, 93, 2464–2472. [Google Scholar] [CrossRef]
- Bendali, F.; Hébraud, M.; Sadoun, D. Anti-bacterial and anti-adherence activities of a probiotic strain of Lactobacillus paracasei subsp. paracasei against Listeria monocytogenes. Int. J. Appl. Microbiol. Biotechnol. Res. 2014, 2, 52. [Google Scholar]
- Caly, D.L.; Chevalier, M.; Flahaut, C.; Cudennec, B.; Al Atya, A.K.; Chataigné, G.; D’Inca, R.; Auclair, E.; Drider, D. The safe enterocin DD14 is a leaderless two-peptide bacteriocin with anti-Clostridium perfringens activity. Int. J. Antimicrob. Agents 2017, 49, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Versalovic, J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009, 9, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Haro, M.; Taggart, M.; Green, A.J.; Mateo, R. Avian digestive tract simulation to study the effect of grit geochemistry and food on pb shot bioaccessibility. Environ. Sci. Technol. 2009, 43, 9480–9486. [Google Scholar] [CrossRef] [PubMed]
- Musikasang, H.; Tani, A.; H-Kittikun, A.; Maneerat, S. Probiotic potential of lactic acid bacteria isolated from chicken gastrointestinal digestive tract. World J. Microbiol. Biotechnol. 2009, 25, 1337–1345. [Google Scholar] [CrossRef]
- Troxell, B.; Petri, N.; Daron, C.; Pereira, R.; Mendoza, M.; Hassan, H.M.; Koci, M.D. Poultry body temperature contributes to invasion control through reduced expression of Salmonella pathogenicity island 1 genes in Salmonella enterica Serovars Typhimurium and Enteritidis. Appl. Environ. Microbiol. 2015, 81, 8192–8201. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-Q.; Zhu, Y.-H.; Zhang, H.-F.; Yue, Y.; Cai, Z.-X.; Lü, Q.-P.; Zhang, L.; Weng, X.-G.; Zhang, F.-J.; Zhou, N.; et al. Risks Associated with high-dose Lactobacillus rhamnosus in an Escherichia coli model of piglet diarrhoea: Intestinal microbiota and immune imbalances. PLoS ONE 2012, 7, e40666. [Google Scholar] [CrossRef]
- Candela, M.; Perna, F.; Carnevali, P.; Vitali, B.; Ciati, R.; Gionchetti, P.; Rizzello, F.; Campieri, M.; Brigidi, P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008, 125, 286–292. [Google Scholar] [CrossRef]
- Wu, S.-B.; Rodgers, N.; Choct, M. Real-time pcr assay for Clostridium perfringens in broiler chickens in a challenge model of necrotic enteritis. Appl. Environ. Microbiol. 2011, 77, 1135–1139. [Google Scholar] [CrossRef] [Green Version]
- Pisano, M.B.; Viale, S.; Conti, S.; Fadda, M.E.; Deplano, M.; Melis, M.P.; Deiana, M.; Cosentino, S. Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from sardinian dairy products. BioMed Res. Int. 2014, 2014, 286390. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zheng, Y.; Gao, H.; Xu, P.; Wang, M.; Li, A.; Miao, M.; Xie, X.; Deng, Y.; Zhou, H.; et al. Identification and characterization of Staphylococcus aureus strains with an incomplete hemolytic phenotype. Front. Cell. Infect. Microbiol. 2016, 6, 146. [Google Scholar] [CrossRef] [Green Version]
- Matuschek, E.; Brown, D.F.J.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef] [Green Version]
- Al Seraih, A.; Belguesmia, Y.; Cudennec, B.; Baah, J.; Drider, D. In silico and experimental data claiming safety aspects and beneficial attributes of the bacteriocinogenic strain Enterococcus faecalis B3A-B3B. Probiotics Antimicrob. Proteins 2017, 10, 456–465. [Google Scholar] [CrossRef]
- CLSI M45Ed3: Test Infrequently Isolated/Fastidious Bacteria. Available online: https://clsi.org/standards/products/microbiology/documents/m45/ (accessed on 24 August 2021).
- EFSA Guidance on the Characterisation of Microorganisms Used as Feed Additives or as Production Organisms. Autorité Européenne de Sécurité des Aliments. Available online: https://www.efsa.europa.eu/fr/efsajournal/pub/5206 (accessed on 24 August 2021).
- Chan, W.Y.; Hickey, E.E.; Khazandi, M.; Page, S.W.; Trott, D.J.; Hill, P.B. In vitro antimicrobial activity of narasin against common clinical isolates associated with canine otitis externa. Vet. Dermatol. 2018, 29, 149-e57. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Scientific Opinion on safety and efficacy of Cygro® 10G (maduramicin ammonium α) for chickens for fattening. EFSA J. 2011, 9, 1952. [Google Scholar] [CrossRef]
- McReynolds, J.L.; Byrd, J.A.; Anderson, R.C.; Moore, R.W.; Edrington, T.S.; Genovese, K.J.; Poole, T.L.; Kubena, L.F.; Nisbet, D.J. Evaluation of immunosuppressants and dietary mechanisms in an experimental disease model for necrotic enteritis. Poult. Sci. 2004, 83, 1948–1952. [Google Scholar] [CrossRef]
- Dromigny, E.; Bourrion, F.; Rugraf, Y.; Bolton, F.J.; Leden, N. New media for detection and counting of clostridia in foods. Lett. Appl. Microbiol. 1997, 24, 19–22. [Google Scholar] [CrossRef]
- Wenk, C.; Pfirter, H.; Bickel, H. Energetic aspects of feed conversion in growing pigs. Livest. Prod. Sci. 1980, 7, 483–495. [Google Scholar] [CrossRef]
- Prescott, J.F.; Sivendra, R.; Barnum, D.A. The use of bacitracin in the prevention and treatment of experimentally-induced necrotic enteritis in the chicken. Can. Vet. J. 1978, 19, 181–183. [Google Scholar]
- Cooper, K.K.; Songer, J.G. Necrotic enteritis in chickens: A paradigm of enteric infection by Clostridium perfringens type A. Anaerobe 2009, 15, 55–60. [Google Scholar] [CrossRef]
- Misiakos, E.P.; Bagias, G.; Patapis, P.; Sotiropoulos, D.; Kanavidis, P.; Machairas, A. Current concepts in the management of necrotizing fasciitis. Front. Surg. 2014, 1, 36. [Google Scholar] [CrossRef] [Green Version]
- Shiroff, A.M.; Herlitz, G.N.; Gracias, V.H. Necrotizing soft tissue infections. J. Intensiv. Care Med. 2012, 29, 138–144. [Google Scholar] [CrossRef]
- Wade, B.; Keyburn, A.L. The true cost of necrotic enteritis. Poultry World 2015, 31, 16–17. [Google Scholar]
- Landoni, M.; Albarellos, G. The use of antimicrobial agents in broiler chickens. Vet. J. 2015, 205, 21–27. [Google Scholar] [CrossRef]
- Mwangi, S.; Timmons, J.; Fitz-Coy, S.; Parveen, S. Characterization of Clostridium perfringens recovered from broiler chicken affected by necrotic enteritis. Poult. Sci. 2019, 98, 128–135. [Google Scholar] [CrossRef]
- Osman, K.; ElHariri, M. Antibiotic resistance of Clostridium perfringens isolates from broiler chickens in Egypt. Rev. Sci. Tech. de l’OIE 2013, 32, 841–850. [Google Scholar] [CrossRef]
- Adhikari, P.; Kiess, A.; Jha, R. An approach to alternative strategies to control avian coccidiosis and necrotic enteritis. J. Appl. Poult. Res. 2020, 29, 515–534. [Google Scholar] [CrossRef]
- Golić, N.; Veljović, K.; Popović, N.; Djokić, J.; Strahinić, I.; Mrvaljević, I.; Terzić-Vidojević, A. In vitro and in vivo antagonistic activity of new probiotic culture against Clostridium difficile and Clostridium perfringens. BMC Microbiol. 2017, 17, 108. [Google Scholar] [CrossRef] [Green Version]
- Ait-Seddik, H.; Bendali, F.; Cudennec, B.; Drider, D. Anti-pathogenic and probiotic attributes of Lactobacillus salivarius and Lactobacillus plantarum strains isolated from feces of Algerian infants and adults. Res. Microbiol. 2017, 168, 244–254. [Google Scholar] [CrossRef]
- Hor, Y.Y.; Liong, M.T. Use of extracellular extracts of lactic acid bacteria and bifidobacteria for the inhibition of dermatological pathogen Staphylococcus aureus. Dermatol. Sin. 2014, 32, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Vodnar, D.C.; Paucean, A.; Dulf, F.V.; Socaciu, C. HPLC Characterization of lactic acid formation and FTIR fingerprint of probiotic bacteria during fermentation processes. Not. Bot. Horti Agrobot. 2010, 38, 109–113. [Google Scholar] [CrossRef]
- Stanojević-Nikolić, S.; Dimić, G.; Mojović, L.; Pejin, J.; Djukić-Vuković, A.; Kocić-Tanackov, S. Antimicrobial activity of lactic acid against pathogen and spoilage microorganisms. J. Food Process. Preserv. 2015, 40, 990–998. [Google Scholar] [CrossRef]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.T.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; Franco, B.D.G.D.M. Use of potential probiotic lactic acid bacteria (lab) biofilms for the control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157:H7 biofilms formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibarreche, M.P.; Castellano, P.; Vignolo, G. Evaluation of anti-Listeria meat borne Lactobacillus for biofilm formation on selected abiotic surfaces. Meat Sci. 2014, 96, 295–303. [Google Scholar] [CrossRef]
- Mare, L.; Wolfaardt, G.; Dicks, L. Adhesion of Lactobacillus plantarum 423 and Lactobacillus salivarius 241 to the intestinal tract of piglets, as recorded with fluorescent in situ hybridization (FISH), and production of plantaricin 423 by cells colonized to the ileum. J. Appl. Microbiol. 2006, 100, 838–845. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef] [Green Version]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Messaoudi, S.; Kergourlay, G.; Dalgalarrondo, M.; Choiset, Y.; Ferchichi, M.; Prevost, H.; Pilet, M.-F.; Chobert, J.-M.; Manai, M.; Dousset, X. Purification and characterization of a new bacteriocin active against Campylobacter produced by Lactobacillus salivarius SMXD51. Food Microbiol. 2012, 32, 129–134. [Google Scholar] [CrossRef]
- Barache, N.; Ladjouzi, R.; Belguesmia, Y.; Bendali, F.; Drider, D. Abundance of Lactobacillus plantarum strains with beneficial attributes in blackberries (Rubus sp.), fresh figs (Ficus carica), and prickly pears (Opuntia ficus-indica) grown and harvested in Algeria. Probiotics Antimicrob. Proteins 2020, 12, 1514–1523. [Google Scholar] [CrossRef]
- De Boever, P.; Wouters, R.; Verschaeve, L.; Berckmans, P.; Schoeters, G.; Verstraete, W. Protective effect of the bile salt hydrolase-active Lactobacillus reuteri against bile salt cytotoxicity. Appl. Microbiol. Biotechnol. 2000, 53, 709–714. [Google Scholar] [CrossRef]
- Martoni, C.J.; Labbé, A.; Ganopolsky, J.G.; Prakash, S.; Jones, M.L. Changes in bile acids, FGF-19 and sterol absorption in response to bile salt hydrolase active L. reuteri NCIMB 30242. Gut Microbes 2015, 6, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.M.T.; Pyle, B.H.; Camper, A.K. Specific and rapid enumeration of viable but nonculturable and viable-culturable gram-negative bacteria by using flow cytometry. Appl. Environ. Microbiol. 2010, 76, 5088–5096. [Google Scholar] [CrossRef] [Green Version]
- Er, S.; KOPARAL, A.T.; Kivanç, M. Cytotoxic effects of various lactic acid bacteria on Caco-2 cells. Turk. J. Boil. 2015, 39, 23–30. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sánchez, B.; Reyes-Gavilán, C.G.D.L.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-Z.; Nikaido, H. Efflux-mediated drug resistance in bacteria: An update. Drugs 2009, 69, 1555–1623. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Aquilina, G.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; De Knecht, J.; Dierick, N.A.; Gralak, M.A.; Gropp, J.; Halle, I.; Kroker, R.; et al. Scientific Opinion on Safety and Efficacy of Clinacox® 0.5% (Diclazuril) for Turkeys for Fattening. Available online: https://www.efsa.europa.eu/fr/efsajournal/pub/2115 (accessed on 16 June 2021).
- Dec, M.; Puchalski, A.; Stępień-Pyśniak, D.; Marek, A.; Urban-Chmiel, R. Susceptibility of chicken Lactobacillus bacteria to coccidiostats. J. Vet. Med Sci. 2020, 82, 333–336. [Google Scholar] [CrossRef] [Green Version]
- Marounek, M.; Rada, V. Susceptibility of poultry Lactobacilli to ionophore antibiotics. J. Vet. Med. Ser. B 1995, 42, 193–196. [Google Scholar] [CrossRef]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef]
- Song, J.; Xiao, K.; Ke, Y.L.; Jiao, L.F.; Hu, C.H.; Diao, Q.Y.; Shi, B.; Zou, X.T. Effect of a probiotic mixture on intestinal microflora, morphology, and barrier integrity of broilers subjected to heat stress. Poult. Sci. 2014, 93, 581–588. [Google Scholar] [CrossRef]
- Sureshkumar, S.; Lee, H.C.; Jung, S.K.; Kim, D.; Oh, K.B.; Yang, H.; Jo, Y.J.; Lee, H.S.; Lee, S.; Byun, S.J. Inclusion of Lactobacillus salivarius strain revealed a positive effect on improving growth performance, fecal microbiota and immunological responses in chicken. Arch. Microbiol. 2020, 203, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Ashayerizadeh, A.; Dabiri, N.; Mirzadeh, K.H.; Ghorbani, M.R. Effects of dietary inclusion of several biological feed additives on growth response of broiler chickens. J. Cell Anim. Biol. 2011, 5, 61–65. [Google Scholar]
- Nakphaichit, M.; Thanomwongwattana, S.; Phraephaisarn, C.; Sakamoto, N.; Keawsompong, S.; Nakayama, J.; Nitisinprasert, S. The effect of including Lactobacillus reuteri KUB-AC5 during post-hatch feeding on the growth and ileum microbiota of broiler chickens. Poult. Sci. 2011, 90, 2753–2765. [Google Scholar] [CrossRef] [PubMed]
- Mappley, L.J.; Tchórzewska, M.A.; Cooley, W.A.; Woodward, M.J.; La Ragione, R.M. Lactobacilli antagonize the growth, motility, and adherence of Brachyspira pilosicoli: A potential intervention against avian intestinal Spirochetosis. Appl. Environ. Microbiol. 2011, 77, 5402–5411. [Google Scholar] [CrossRef] [Green Version]
- Carter, A.; Adams, M.; La Ragione, R.; Woodward, M.J. Colonisation of poultry by Salmonella Enteritidis S1400 is reduced by combined administration of Lactobacillus salivarius 59 and Enterococcus faecium PXN-33. Vet. Microbiol. 2017, 199, 100–107. [Google Scholar] [CrossRef]
- Gérard, P.; Brézillon, C.; Quéré, F.; Salmon, A.; Rabot, S. Characterization of cecal microbiota and response to an orally administered Lactobacillus probiotic strain in the broiler chicken. J. Mol. Microbiol. Biotechnol. 2007, 14, 115–122. [Google Scholar] [CrossRef]
- Kizerwetter-Swida, M.; Binek, M. Protective effect of potentially probiotic Lactobacillus strain on infection with pathogenic bacteria in chickens. Pol. J. Vet. Sci. 2009, 12, 15–20. [Google Scholar]
- Li, Z.; Wang, W.; Liu, D.; Guo, Y. Effects of Lactobacillus acidophilus on the growth performance and intestinal health of broilers challenged with Clostridium perfringens. J. Anim. Sci. Biotechnol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- La Ragione, R.; Narbad, A.; Gasson, M.; Woodward, M. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett. Appl. Microbiol. 2004, 38, 197–205. [Google Scholar] [CrossRef]
- Cao, L.; Yang, X.; Li, Z.; Sun, F.; Wu, X.; Yao, J. Reduced lesions in chickens with Clostridium perfringens-induced necrotic enteritis by Lactobacillus fermentum 1.2029. Poult. Sci. 2012, 91, 3065–3071. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).