From Surface to Subsurface: Diversity, Composition, and Abundance of Sessile and Endolithic Bacterial, Archaeal, and Eukaryotic Communities in Sand, Clay and Rock Substrates in the Laurentians (Quebec, Canada)
Abstract
1. Introduction
2. Materials and Methods
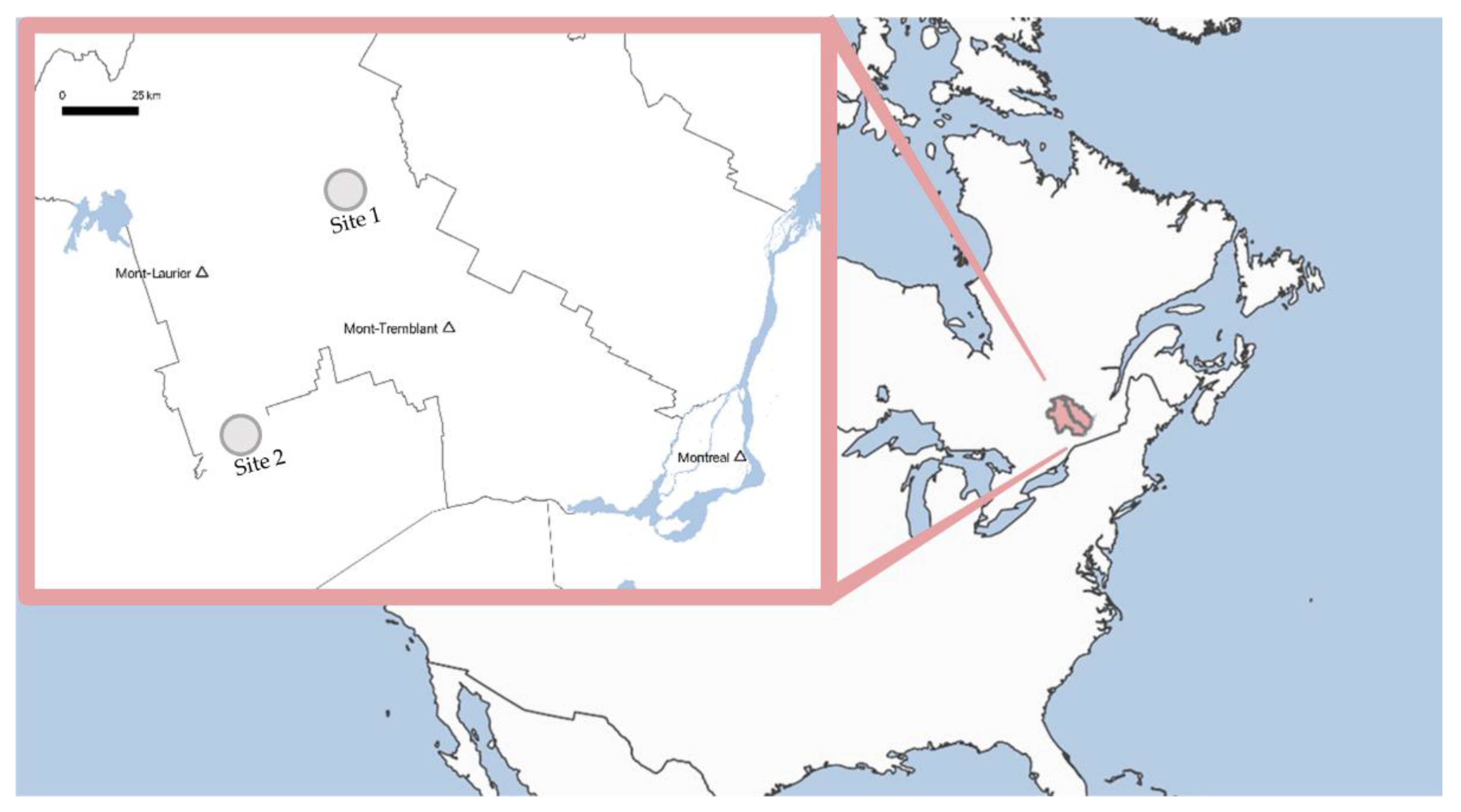
2.1. Study Sites
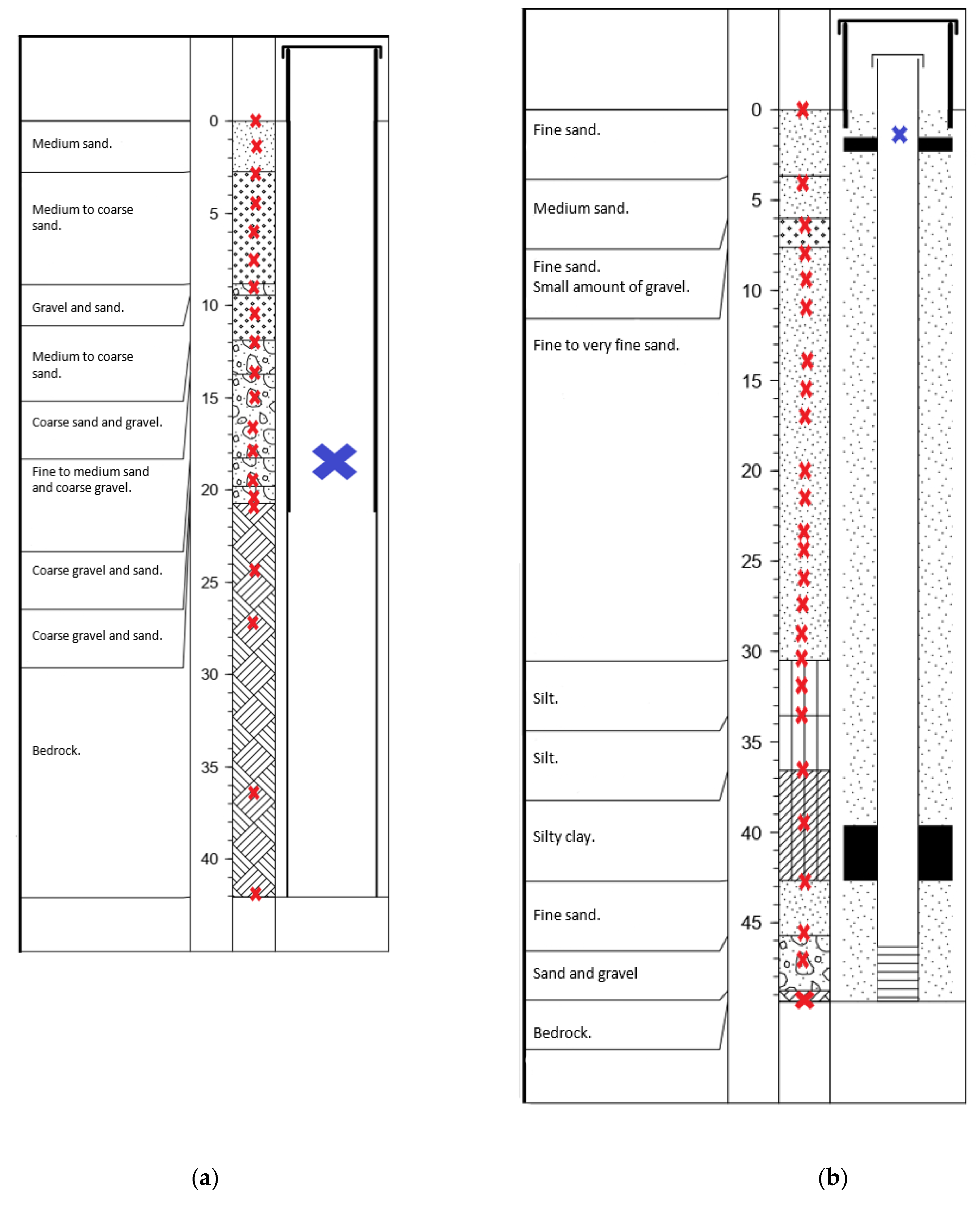
2.2. Sampling
2.3. Chemical Characteristics of Geological Material and Water Samples
2.4. DNA Extraction
2.5. PCR Amplification and Illumina Sequencing
2.6. Digital PCR
2.7. Sequence Processing
2.8. Statistical Analyses
3. Results
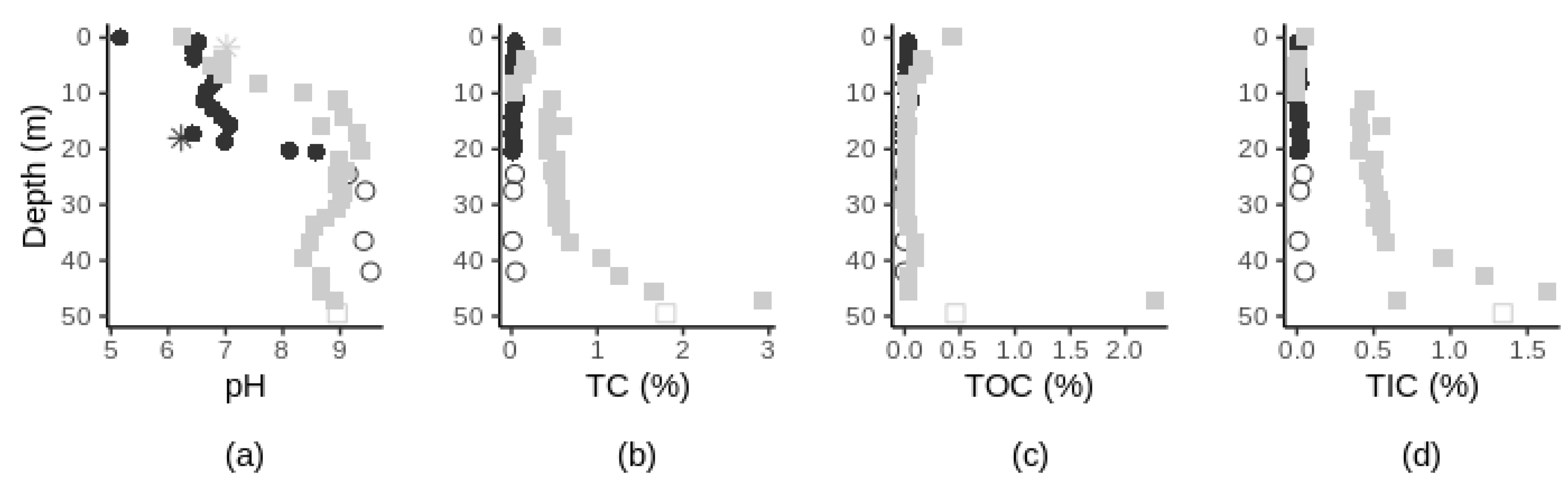
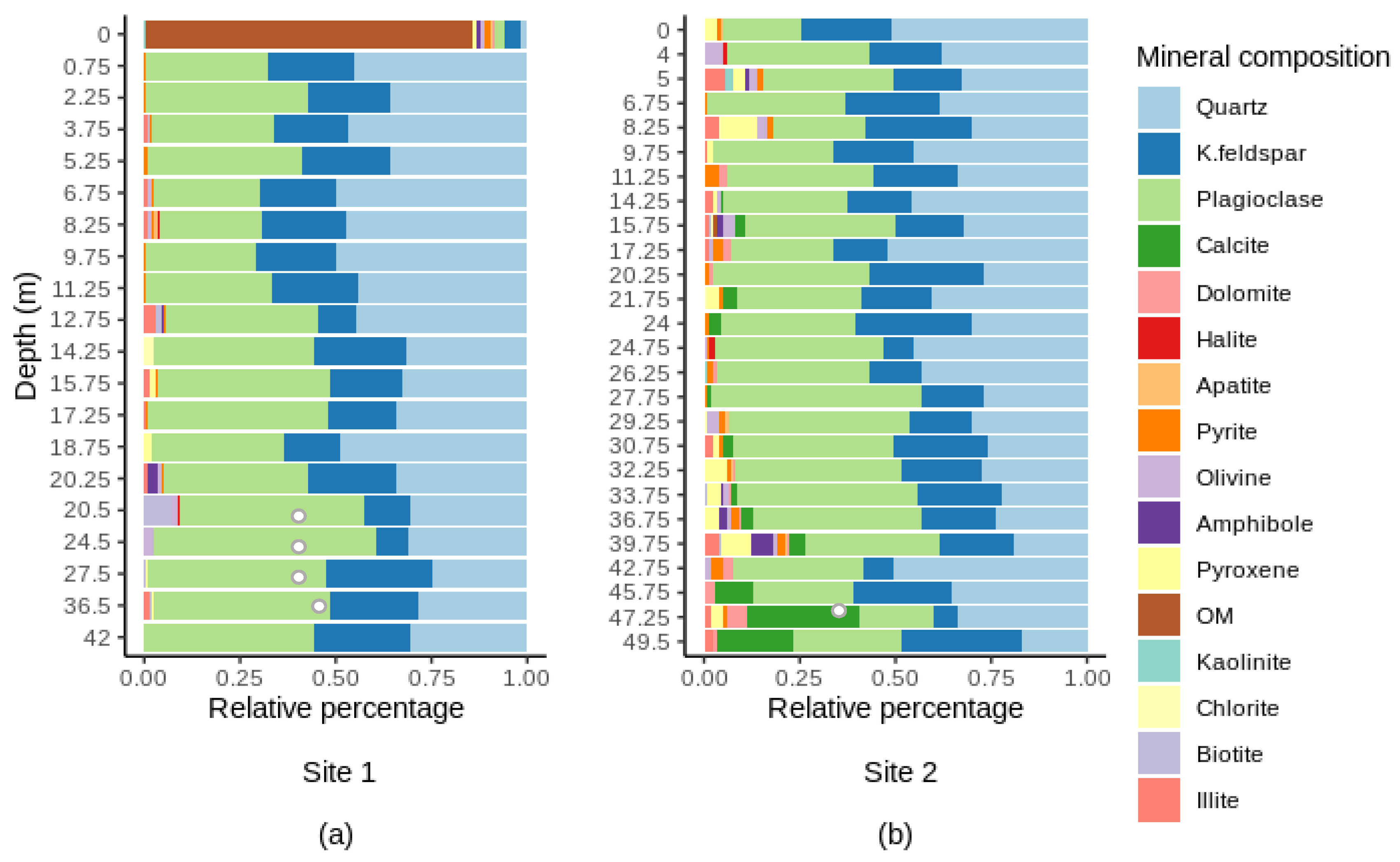
3.1. Variation of Chemical Characteristics with Depth
3.2. Taxonomic α-Diversity: Variations with Depth and Correlation with Chemical Characteristics of Geological Material
3.3. Phylogenetic α-Diversity and Correlation with Chemical Characteristics
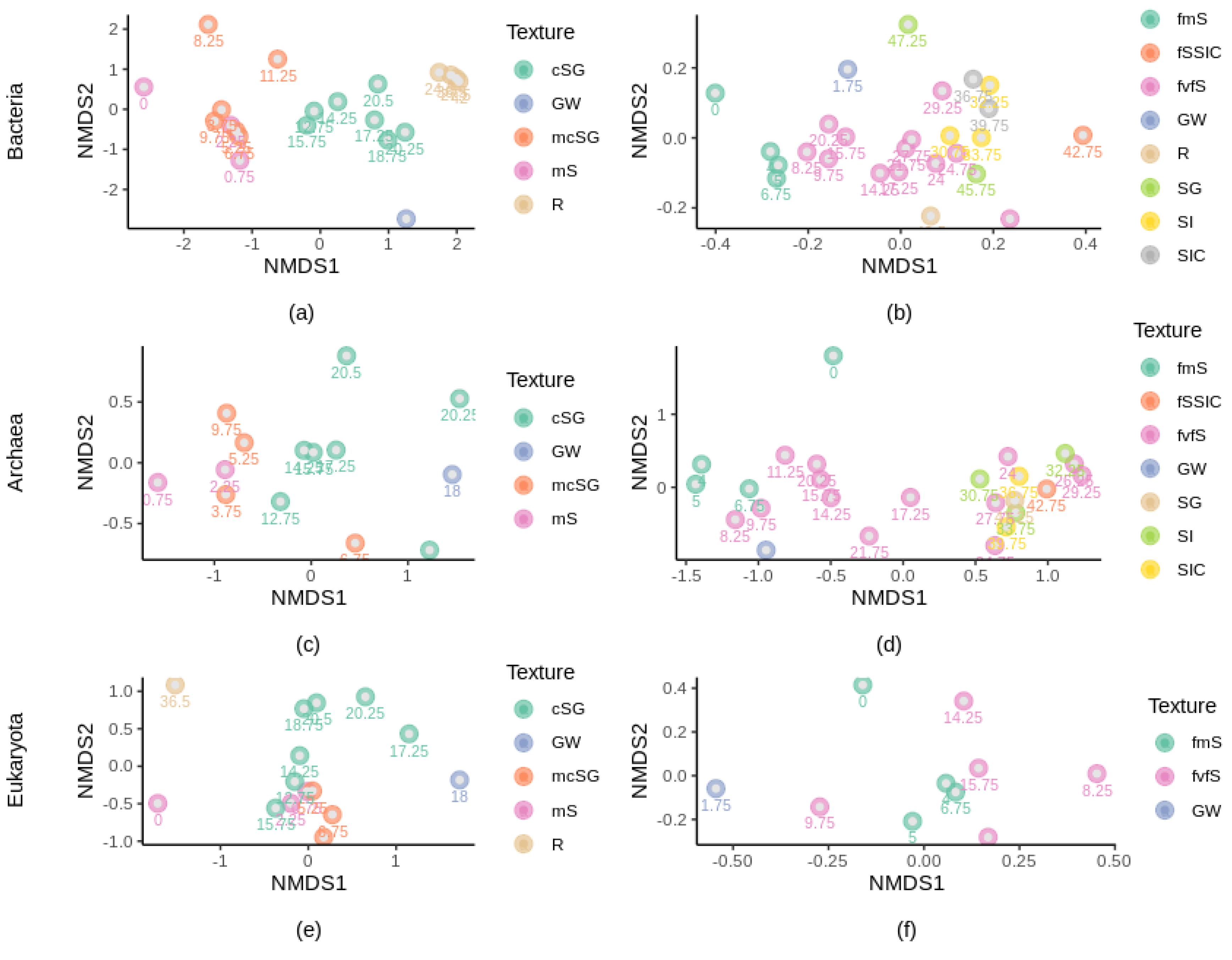
3.4. β-Diversity and Correlation with Depth and Chemical Characteristics
3.5. Absolute Bacterial Abundance
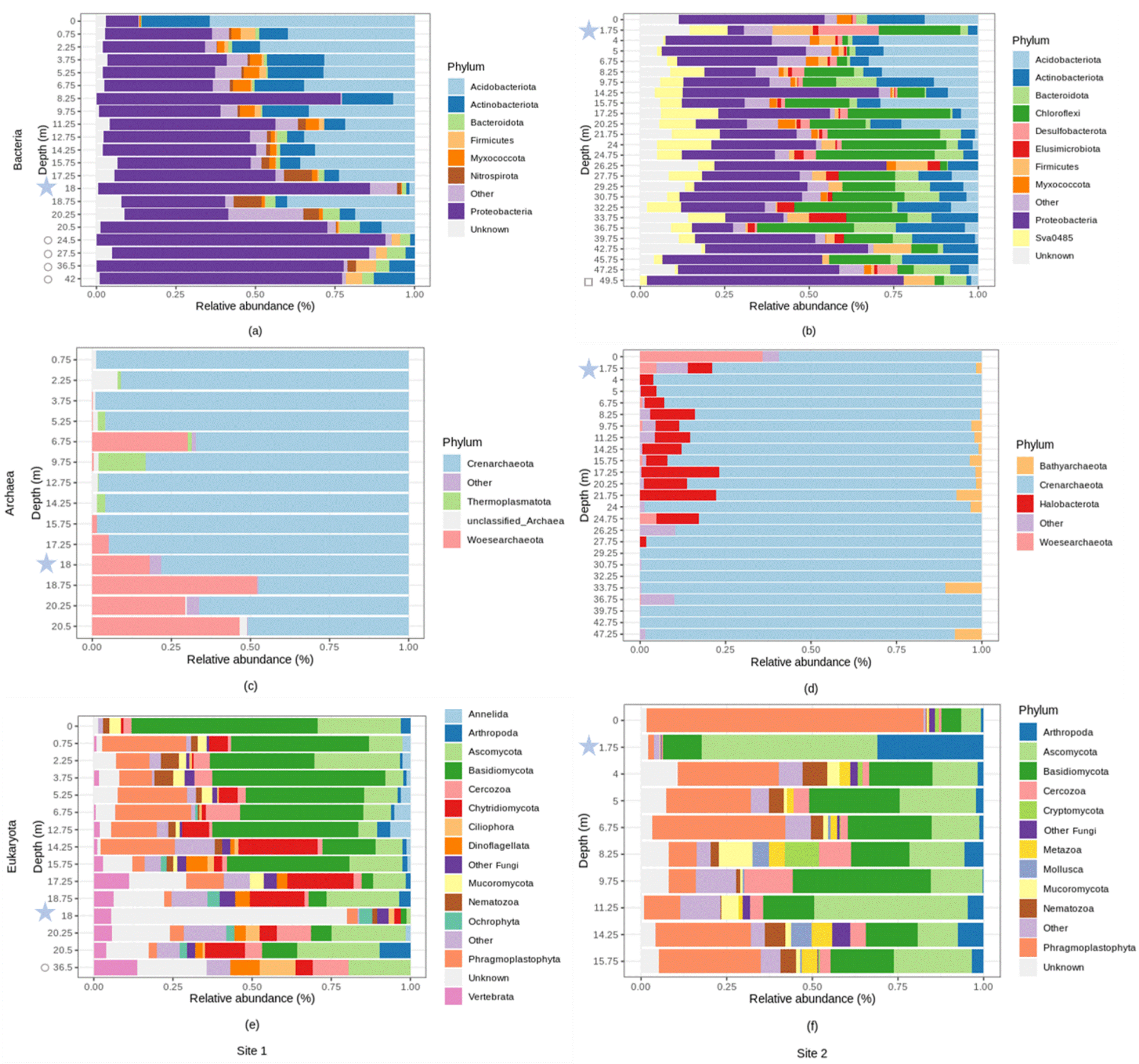
3.6. Variation in the Relative Abundance of Dominant Bacteria, Archaea, and Eukaryote Microorganisms at the Phylum and Genus Levels, with Depth
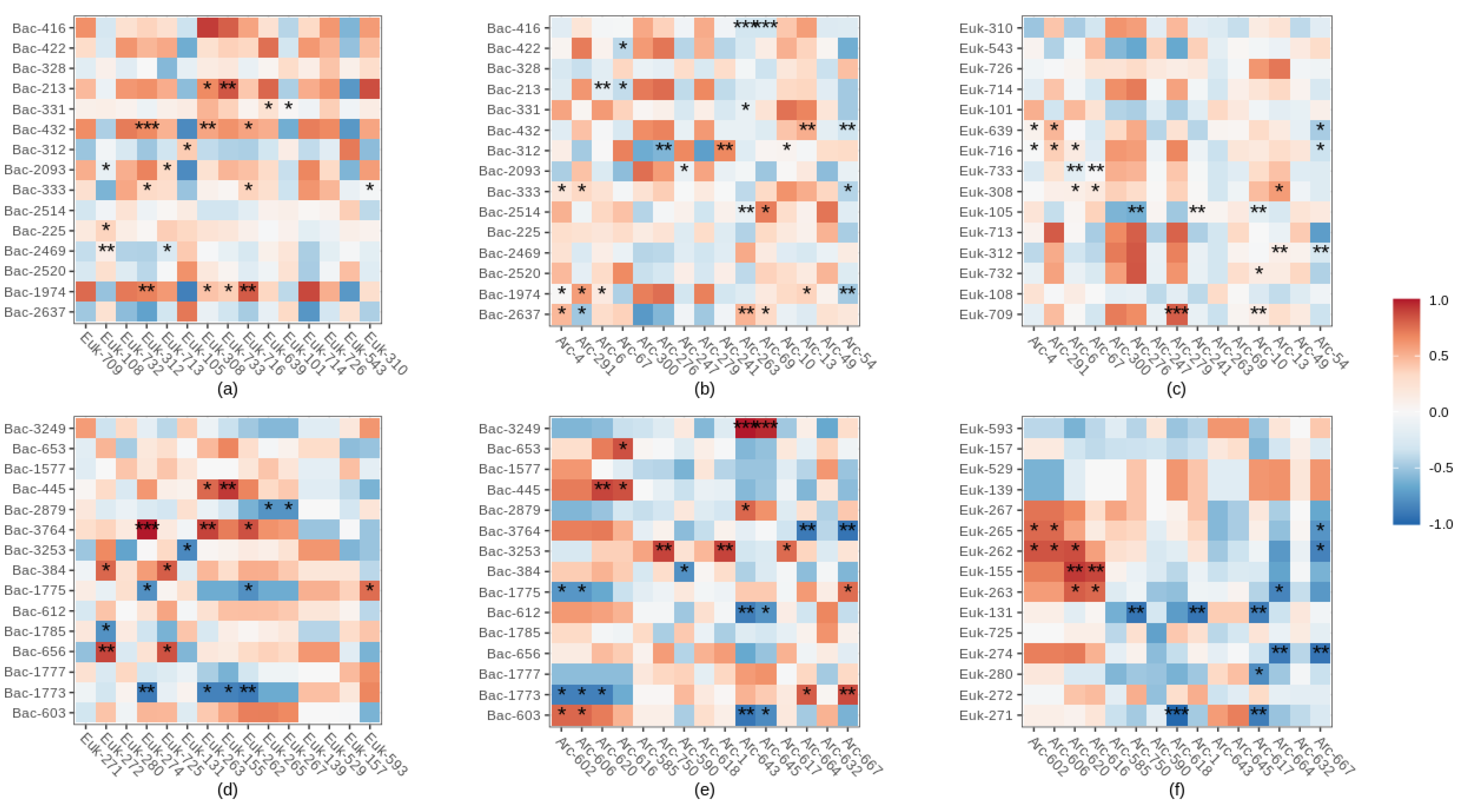
3.7. Interactions among Bacterial, Eukaryotic and Archaeal major ASVs
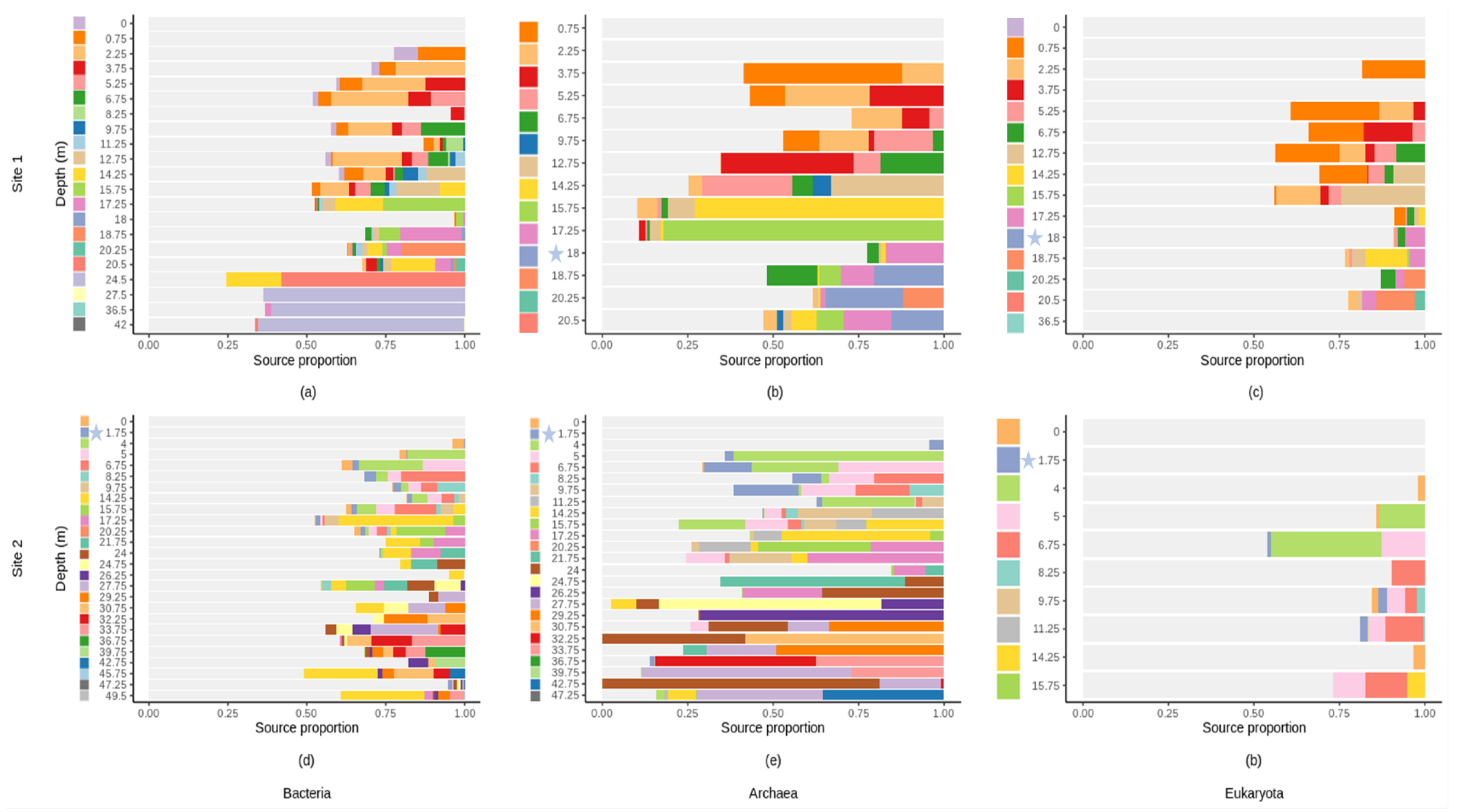
3.8. Microbial Source Tracking
4. Discussion
4.1. The Effect of Depth on Endolithic and Sessile Communities
4.2. Effect of Abiotic Characteristics on Endolithic and Sessile Communities
4.3. Potential Biotic Interactions between Bacterial, Eukaryotic and Archaeal Microorganisms
4.4. Vertical Fluid Fluxes as Sources of Microbial Communities in the Shallow Terrestrial Subsurface
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chu, H.; Sun, H.; Tripathi, B.M.; Adams, J.M.; Huang, R.; Zhang, Y.; Shi, Y. Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environ. Microbiol. 2016, 18, 1523–1533. [Google Scholar] [CrossRef]
- Hartmann, M.; Lee, S.; Hallam, S.J.; Mohn, W.W. Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands. Environ. Microbiol. 2009, 11, 3045–3062. [Google Scholar] [CrossRef]
- Lazar, C.S.; Lehmann, R.; Stoll, W.; Rosenberger, J.; Totsche, K.U.; Küsel, K. The endolithic bacterial diversity of shallow bedrock ecosystems. Sci. Total Environ. 2019, 679, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Flynn, T.M.; Sanford, R.A.; Ryu, H.; Bethke, C.M.; Levine, A.D.; Ashbolt, N.J.; Santo Domingo, J.W. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol. 2013, 13, 146. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, M.J.; Jung, J.Y.; Hwang, C.Y.; Kim, M.; Ro, H.M.; Chun, J.; Lee, Y.K. Vertical distribution of bacterial community is associated with the degree of soil organic matter decomposition in the active layer of moist acidic tundra. J. Microbiol. 2016, 54, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging deeper to find unique microbial communities: The strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol. Biochem. 2012, 50, 58–65. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- LaMontagne, M.G.; Schimel, J.P.; Holden, P.A. Comparison of subsurface and surface soil bacterial communities in California grassland as assessed by terminal restriction fragment length polymorphisms of PCR-amplified 16S rRNA genes. Microbial Ecol. 2003, 46, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Agnelli, A.; Ascher, J.; Corti, G.; Ceccherini, M.T.; Nannipieri, P.; Pietramellara, G. Distribution of microbial communities in a forest soil profile investigated by microbial biomass, soil respiration and DGGE of total and extracellular DNA. Soil Biol. Biochem. 2004, 36, 859–868. [Google Scholar] [CrossRef]
- Goberna, M.; Insam, H.; Klammer, S.; Pascual, J.A.; Sanchez, J. Microbial community structure at different depths in disturbed and undisturbed semiarid Mediterranean forest soils. Microbial Ecol. 2005, 50, 315–326. [Google Scholar] [CrossRef]
- Will, C.; Thürmer, A.; Wollherr, A.; Nacke, H.; Herold, N.; Schrumpf, M.; Gutknecht, J.; Wubet, T.; Buscot, F.; Daniel, R. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2010, 76, 6751–6759. [Google Scholar] [CrossRef]
- Hansel, C.M.; Fendorf, S.; Jardine, P.M.; Francis, C.A. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 2008, 74, 1620–1633. [Google Scholar] [CrossRef]
- Amy, P.S.; Haldeman, D.L.; Ringelberg, D.; Hall, D.H.; Russell, C. Comparison of identification systems for classification of bacteria isolated from water and endolithic habitats within the deep subsurface. Appl. Environ. Microbiol. 1992, 58, 3367–3373. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Chen, X.; Hou, Y.; Zhu, B. Changes in microbial biomass, community composition and diversity, and functioning with soil depth in two alpine ecosystems on the Tibetan plateau. Plant Soil. 2021, 459, 137–153. [Google Scholar] [CrossRef]
- Shenhav, L.; Thompson, M.; Joseph, T.A.; Briscoe, L.; Furman, O.; Bogumil, D.; Mizrahi, I.; Pe’er, I.; Halperin, E. FEAST: Fast expectation-maximization for microbial source tracking. Nat. Methods 2019, 16, 627–632. [Google Scholar] [CrossRef]
- Government of Canada. Canadian Climate Normals 1981–2010 Station Data. Environ. Resour. 2021, Climate ID 7033939.
- Government of Canada. Canadian Climate Normals 1981–2010 Station Data. Environ. Resour. 2021, Climate ID 7038975.
- Ritchey, E.L.; McGrath, J.M.; Gehring, D. Determining soil texture by feel. Agric. Nat. Resour. Publ. 2015, 139. [Google Scholar]
- Eckert, D.; Sims, J.T. Recommended soil pH and lime requirement tests. In Recommended Soil Testing Procedures for the Northeastern United States; Northeast Regional Bulletin: Dover, DE, USA, 1995; Volume 493, pp. 11–16. [Google Scholar]
- Butler, B.; Hillier, S. powdR: An R package for quantitative mineralogy using full pattern summation of X-ray powder diffraction data. Comput. Geosci. 2021, 147, 104662. [Google Scholar] [CrossRef]
- Lazar, C.S.; Stoll, W.; Lehmann, R.; Herrmann, M.; Schwab, V.F.; Akob, D.M.; Küsel, K. Archaeal diversity and CO2 fixers in carbonate-/siliciclastic-rock groundwater ecosystems. Archaea 2017, 2017, 2136287. [Google Scholar] [CrossRef] [PubMed]
- Alain, K.; Callac, N.; Ciobanu, M.C.; Reynaud, Y.; Duthoit, F.; Jebbar, M. DNA extractions from deep subseafloor sediments: Novel cryogenic-mill-based procedure and comparison to existing protocols. J. Microbiol. Methods. 2011, 87, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Direito, S.O.; Marees, A.; Röling, W.F. Sensitive life detection strategies for low-biomass environments: Optimizing extraction of nucleic acids adsorbing to terrestrial and Mars analogue minerals. FEMS Microbiol. Ecol. 2012, 81, 111–123. [Google Scholar] [CrossRef]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef]
- DeLong, E.F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 1992, 89, 5685–5689. [Google Scholar] [CrossRef]
- Gast, R.J.; Dennett, M.R.; Caron, D.A. Characterization of protistan assemblages in the Ross Sea, Antarctica, by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2004, 70, 2028–2037. [Google Scholar] [CrossRef]
- Van de Peer, Y.; De Rijk, P.; Wuyts, J.; Winkelmans, T.; De Wachter, R. The European small subunit ribosomal RNA database. Nucleic Acids Res. 2000, 28, 175–176. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Pelplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Castelle, C.J.; Probst, A.J.; Zhou, Z.; Pan, J.; Liu, Y.; Banfield, J.F.; Gu, J.D. Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome 2018, 6, 102. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Pan, J.; Wang, F.; Gu, J.D.; Li, M. Bathyarchaeota: Globally distributed metabolic generalists in anoxic environments. FEMS Microbiol. Rev. 2018, 42, 639–655. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Steven, H.M.H.; Szoecs, E.; et al. Package ‘Vegan’: Community Ecology Package. R Found. Stat. Comput. 2013, 2.5–7.2, 1–295. [Google Scholar]
- Pereira, M.B.; Wallroth, M.; Jonsson, V.; Kristiansson, E. Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genom. 2018, 19, 274. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Webb, C.O.; Ackerly, D.D.; Kembel, S.W. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 2008, 24, 18. [Google Scholar] [CrossRef]
- Mazel, F.; Davies, T.J.; Gallien, L.; Renaud, J.; Groussin, M.; Münkemüller, T.; Thuiller, W. Influence of tree shape and evolutionary time-scale on phylogenetic diversity metrics. Ecography 2016, 39, 913–920. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Kembel, S.W.; Eisen, J.A.; Pollard, K.S.; Green, J.L. The phylogenetic diversity of metagenomes. PLoS ONE 2011, 6, e23214. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Kim, M.; Kim, Y.; Byun, E.; Yang, J.W.; Ahn, J.; Lee, Y.K. Variations in bacterial and archaeal communities along depth profiles of Alaskan soil cores. Sci. Rep. 2018, 8, 504. [Google Scholar] [CrossRef]
- Baldrian, P.; Kolařík, M.; Štursová, M.; Kopecký, J.; Valášková, V.; Větrovský, T.; Žifčáková, L.; Šnajdr, J.; Rídl, J.; Vlček, C.; et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012, 6, 248–258. [Google Scholar] [CrossRef]
- Ko, D.; Yoo, G.; Yun, S.T.; Jun, S.C.; Chung, H. Bacterial and fungal community composition across the soil depth profiles in a fallow field. J. Ecol. Environ. 2017, 41, 34. [Google Scholar] [CrossRef]
- Feng, H.; Guo, J.; Wang, W.; Song, X.; Yu, S. Soil depth determines the composition and diversity of bacterial and archaeal communities in a poplar plantation. Forests 2019, 10, 550. [Google Scholar] [CrossRef]
- Cao, P.; Zhang, L.M.; Shen, J.P.; Zheng, Y.M.; Di, H.J.; He, J.Z. Distribution and diversity of archaeal communities in selected Chinese soils. FEMS Microbiol. Ecol. 2012, 80, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Akob, D.M.; Küsel, K. Where microorganisms meet rocks in the Earth’s Critical Zone. Biogeosciences 2011, 8, 3531–3543. [Google Scholar] [CrossRef]
- Caruso, T.; Schaefer, I.; Monson, F.; Keith, A.M. Oribatid mites show how climate and latitudinal gradients in organic matter can drive large-scale biodiversity patterns of soil communities. J. Biogeogr. 2019, 46, 611–620. [Google Scholar] [CrossRef]
- Tucker, C.M.; Cadotte, M.W.; Carvalho, S.B.; Davies, T.J.; Ferrier, S.; Fritz, S.A.; Grenyer, R.; Helmus, M.R.; Jin, L.S.; Mooers, A.O.; et al. A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 2017, 92, 698–715. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Wegner, C.E.; Taubert, M.; Geesink, P.; Lehmann, K.; Yan, L.; Lehmann, R.; Totsche, K.U.; Küsel, K. Predominance of Cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 2019, 10, 1407. [Google Scholar] [CrossRef] [PubMed]
- Anacker, B.L.; Harrison, S.P. Historical and ecological controls on phylogenetic diversity in Californian plant communities. Am. Nat. 2012, 180, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Abu-Ashour, J.; Joy, D.M.; Lee, H.; Whiteley, H.R.; Zelin, S. Transport of microorganisms through soil. Water Air Soil Pollut. 1994, 75, 141–158. [Google Scholar] [CrossRef]
- Hamarashid, N.H.; Othman, M.A.; Hussain, M.A.H. Effects of soil texture on chemical compositions, microbial populations and carbon mineralization in soil. Egypt J. Exp. Biol. 2010, 6, 59–64. [Google Scholar]
- Griffiths, R.I.; Whiteley, A.S.; O’Donnell, A.G.; Bailey, M.J. Influence of depth and sampling time on bacterial community structure in an upland grassland soil. FEMS Microbiol. Ecol. 2003, 43, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Li, X.; Li, X.; Zhang, H. Shifts in bacterial community composition increase with depth in three soil types from paddy fields in China. Pedobiologia 2019, 77, 150589. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009, 3, 442–453. [Google Scholar] [CrossRef]
- Govil, T.; Rathinam, N.K.; Salem, D.R.; Sani, R.K. Taxonomical diversity of extremophiles in the deep biosphere. In Microbial Diversity in the Genomic Era; Academic Press: New York, NY, USA, 2019; pp. 631–656. [Google Scholar]
- Bräuer, S.L.; Cadillo-Quiroz, H.; Ward, R.J.; Yavitt, J.B.; Zinder, S.H. Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int. J. Syst. Evol. Microbiol. 2011, 61, 45–52. [Google Scholar] [CrossRef]
- Mondav, R.; Woodcroft, B.J.; Kim, E.H.; McCalley, C.K.; Hodgkins, S.B.; Crill, P.M.; Chanton, J.; Hurst, G.B.; VerBerkmoes, N.C.; Saleska, S.R.; et al. Discovery of a novel methanogen prevalent in thawing permafrost. Nat. Commun. 2014, 5, 6312. [Google Scholar] [CrossRef]
- Haroon, M.F.; Hu, S.; Shi, Y.; Imelfort, M.; Keller, J.; Hugenholtz, P.; Tyson, G.W. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 2013, 500, 567–570. [Google Scholar] [CrossRef]
- Li, Y.; Adams, J.; Shi, Y.; Wang, H.; He, J.S.; Chu, H. Distinct Soil Microbial Communities in habitats of differing soil water balance on the Tibetan Plateau. Sci. Rep. 2017, 7, 46407. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Liang, W.; Shi, Y.; Lin, X.; Zhang, H.; Wu, X.; Xie, G.; Chain, P.; Grognan, P.; Chu, H. Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecology 2014, 95, 3190–3202. [Google Scholar] [CrossRef]
- Kang, B.; Bowatte, S.; Hou, F. Soil microbial communities and their relationships to soil properties at different depths in an alpine meadow and desert grassland in the Qilian mountain range of China. J. Arid Environ. 2021, 184, 104316. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Z.K.; Yip, D.; Morris, R.H.; Lebreux, S.J.; Cregger, M.A.; Klingeman, D.M.; Hui, D.; Hettich, R.L.; Wilhelm, S.W.; et al. One-time nitrogen fertilization shifts switchgrass soil microbiomes within a context of larger spatial and temporal variation. PLoS ONE 2019, 14, e0211310. [Google Scholar] [CrossRef] [PubMed]
- Borgonie, G.; García-Moyano, A.; Litthauer, D.; Bert, W.; Bester, A.; van Heerden, E.; Moller, C.; Erasmus, M.; Onstott, T.C. Nematoda from the terrestrial deep subsurface of South Africa. Nature 2011, 474, 79–82. [Google Scholar] [CrossRef]
- Wan, X.; Gao, Q.; Zhao, J.; Feng, J.; van Nostrand, J.D.; Yang, Y.; Zhou, J. Biogeographic patterns of microbial association networks in paddy soil within Eastern China. Soil Biol. Biochem. 2020, 142, 107696. [Google Scholar] [CrossRef]
- Mackenzie, B.W.; Chang, K.; Zoing, M.; Jain, R.; Hoggard, M.; Biswas, K.; Douglas, R.G.; Taylor, M.W. Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Sci. Rep. 2019, 9, 17416. [Google Scholar] [CrossRef]
- Kneip, C.; Lockhart, P.; Voβ, C.; Maier, U.G. Nitrogen fixation in eukaryotes–new models for symbiosis. BMC Evol. Biol. 2007, 7, 55. [Google Scholar] [CrossRef]
- Ibelings, B.W.; De Bruin, A.; Kagami, M.; Rijkeboer, M.; Brehm, M.; Donk, E.V. Host parasite interactions between freshwater phytoplankton and chytrid fungi (chytridiomycota) 1. J. Phycol. 2004, 40, 437–453. [Google Scholar] [CrossRef]
- Kieft, T.L.; Murphy, E.M.; Haldeman, D.L.; Amy, P.S.; Bjornstad, B.N.; McDonald, E.V.; Ringelberg, D.B.; White, D.C.; Stair, J.; Griffiths, R.P.; et al. Microbial transport, survival, and succession in a sequence of buried sediments. Microb. Ecol. 1998, 36, 336–348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rinke, C.; Rubino, F.; Messer, L.F.; Youssef, N.; Parks, D.H.; Chuvochina, M.; Brown, M.; Jeffries, T.; Tyson, G.W.; Seymour, J.R.; et al. A phylogenomic and ecological analysis of the globally abundant Marine Group II archaea (Ca. Poseidoniales ord. nov.). ISME J. 2019, 13, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T.; Chao, E.E.; Thompson, C.E.; Hourihane, S.L. Oikomonas, a distinctive zooflagellate related to chrysomonads. Arch. Protistenkd. 1996, 146, 273–279. [Google Scholar] [CrossRef]
- Griebler, C.; Mindl, B.; Slezak, D.; Geiger-Kaiser, M. Distribution patterns of attached and suspended bacteria in pristine and contaminated shallow aquifers studied with an in situ sediment exposure microcosm. Aquat. Microb. Ecol. 2002, 28, 117–129. [Google Scholar] [CrossRef]
| Site | Samples | Depth (m) | Abundance (Copies g−1) |
|---|---|---|---|
| Site 1 | RR-01 | 0 | 682,273,937.5 |
| RR-02 | 0.75 | 109,185 | |
| RR-03 | 2.25 | 3,618,757.75 | |
| RR-04 | 3.75 | 252,068 | |
| RR-05 | 5.25 | 344,150.25 | |
| RR-06 | 6.75 | 440,495.5 | |
| RR-11 | 14.25 | 203,594.5 | |
| RR-12 | 15.75 | 569,690.5 | |
| RR-14 | 18.75 | 475,723.25 | |
| RR-16 | 20.5 | 18,367,280.5 | |
| Site 2 | NDDL-01 | 0 | 539,684,562.5 |
| NDDL-02 | 4 | 222,415.5 | |
| NDDL-03 | 5 | 190,000.75 | |
| NDDL-04 | 6.75 | 273854.25 | |
| NDDL-05 | 8.25 | 83,918.75 | |
| NDDL-06 | 9.75 | 520,608 | |
| NDDL-07 | 11.25 | 365,820.5 | |
| NDDL-08 | 14.25 | 367,227 | |
| NDDL-09 | 15.75 | 432,781.5 | |
| NDDL-10 | 17.25 | 145,616.25 | |
| NDDL-11 | 20.25 | 89,283.75 | |
| NDDL-12 | 21.75 | 34,321.5 | |
| NDDL-13 | 24 | 498,270.75 | |
| NDDL-14 | 24.75 | 35,184.25 | |
| NDDL-17 | 29.25 | 60,160.5 | |
| NDDL-20 | 33.75 | 33,647.25 | |
| NDDL-21 | 36.75 | 18,212 | |
| NDDL-22 | 39.75 | 97,846 | |
| NDDL-25 | 47.25 | 20,474 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyer, J.; Zakhary, S.; Larocque, M.; Lazar, C.S. From Surface to Subsurface: Diversity, Composition, and Abundance of Sessile and Endolithic Bacterial, Archaeal, and Eukaryotic Communities in Sand, Clay and Rock Substrates in the Laurentians (Quebec, Canada). Microorganisms 2022, 10, 129. https://doi.org/10.3390/microorganisms10010129
Meyer J, Zakhary S, Larocque M, Lazar CS. From Surface to Subsurface: Diversity, Composition, and Abundance of Sessile and Endolithic Bacterial, Archaeal, and Eukaryotic Communities in Sand, Clay and Rock Substrates in the Laurentians (Quebec, Canada). Microorganisms. 2022; 10(1):129. https://doi.org/10.3390/microorganisms10010129
Chicago/Turabian StyleMeyer, Julia, Sheri Zakhary, Marie Larocque, and Cassandre S. Lazar. 2022. "From Surface to Subsurface: Diversity, Composition, and Abundance of Sessile and Endolithic Bacterial, Archaeal, and Eukaryotic Communities in Sand, Clay and Rock Substrates in the Laurentians (Quebec, Canada)" Microorganisms 10, no. 1: 129. https://doi.org/10.3390/microorganisms10010129
APA StyleMeyer, J., Zakhary, S., Larocque, M., & Lazar, C. S. (2022). From Surface to Subsurface: Diversity, Composition, and Abundance of Sessile and Endolithic Bacterial, Archaeal, and Eukaryotic Communities in Sand, Clay and Rock Substrates in the Laurentians (Quebec, Canada). Microorganisms, 10(1), 129. https://doi.org/10.3390/microorganisms10010129






