Analyses of the Root-Knot Nematode (Meloidogyne graminicola) Transcriptome during Host Infection Highlight Specific Gene Expression Profiling in Resistant Rice Plants
Abstract
1. Introduction
2. Results
2.1. M. graminicola Transcriptome
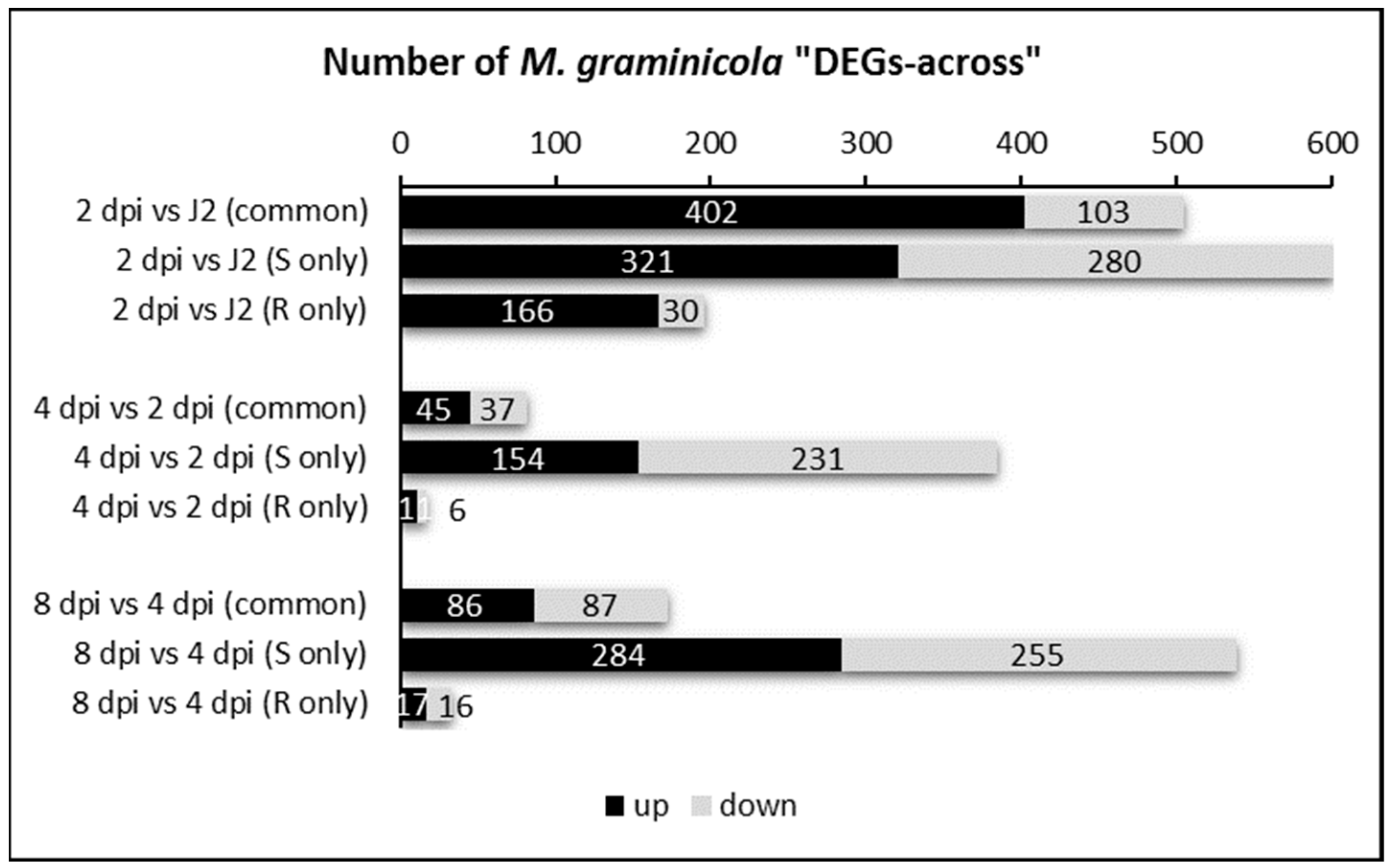
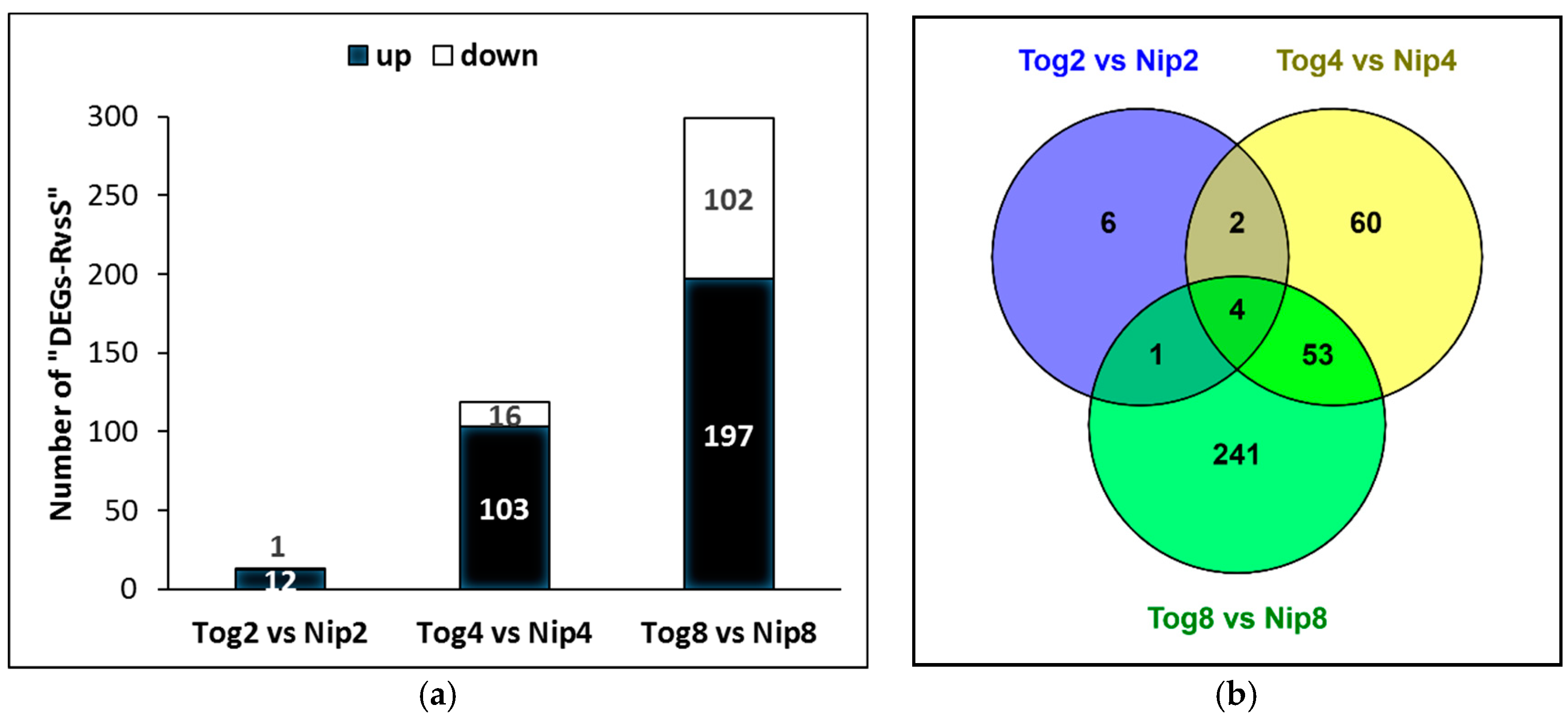
2.2. Differential Gene Expression Analysis from M. graminicola Transcriptomes in Susceptible and Resistant Rice Plants
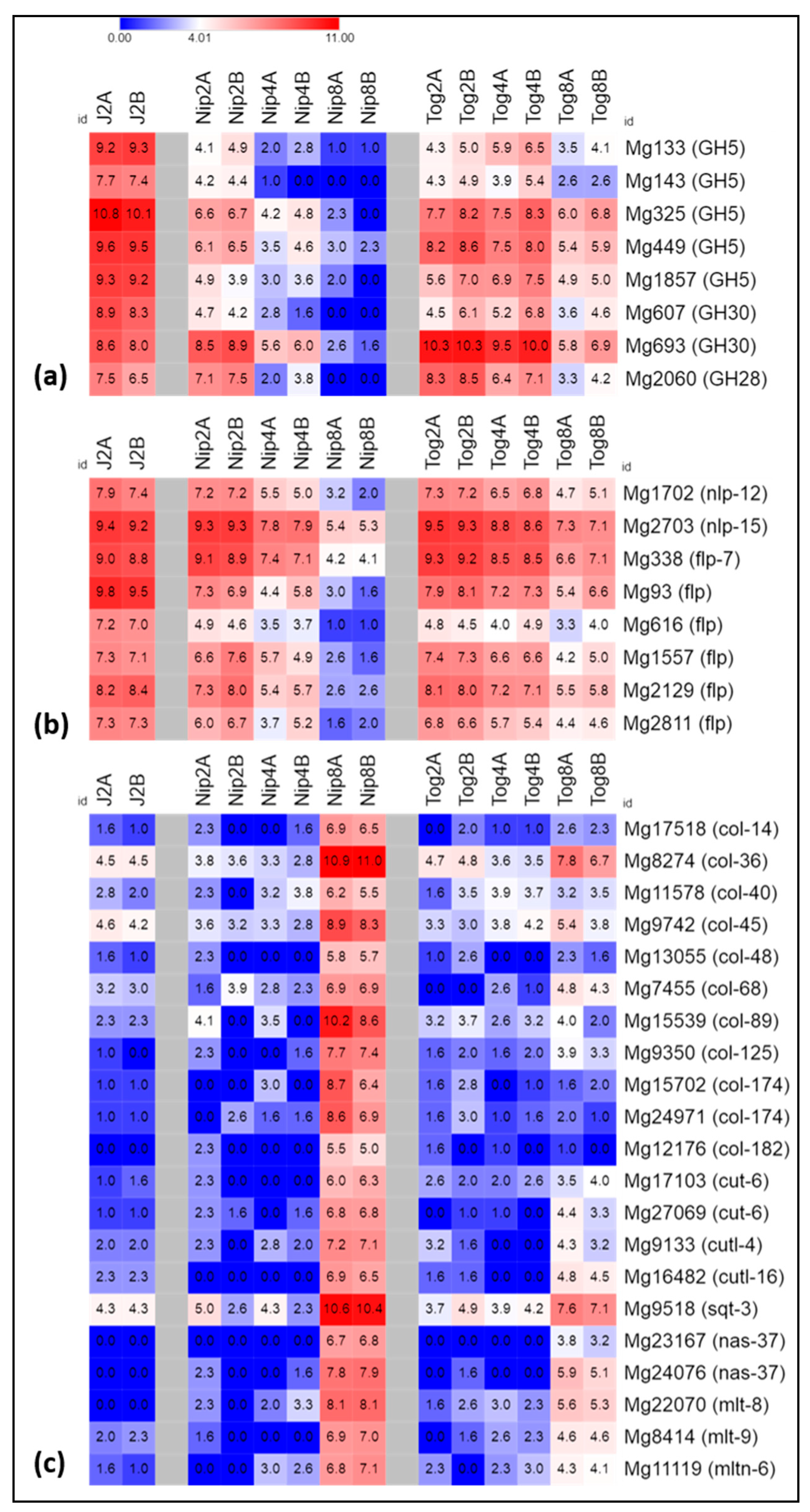
2.3. Expression of Genes Related to Host Invasion
2.4. Expression of Genes Encoding Neuropeptides
2.5. Expression of Genes Related to Nematode Development and Metabolism
2.6. Expression of Genes Related to Stress or Detoxification
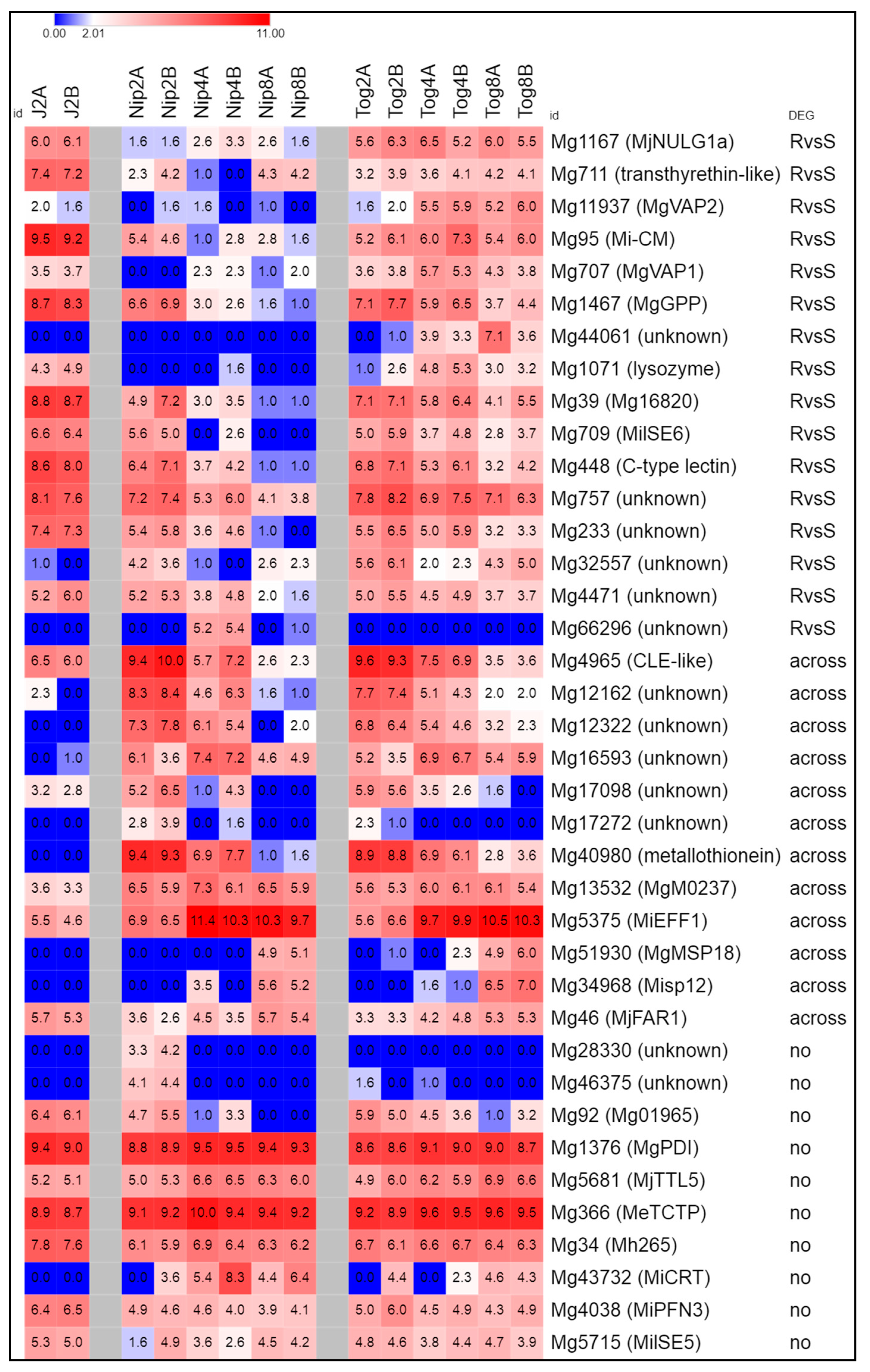
2.7. M. graminicola Genes Encoding Putative Effectors
2.8. Validation of a Set of DEGs by RT-qPCR
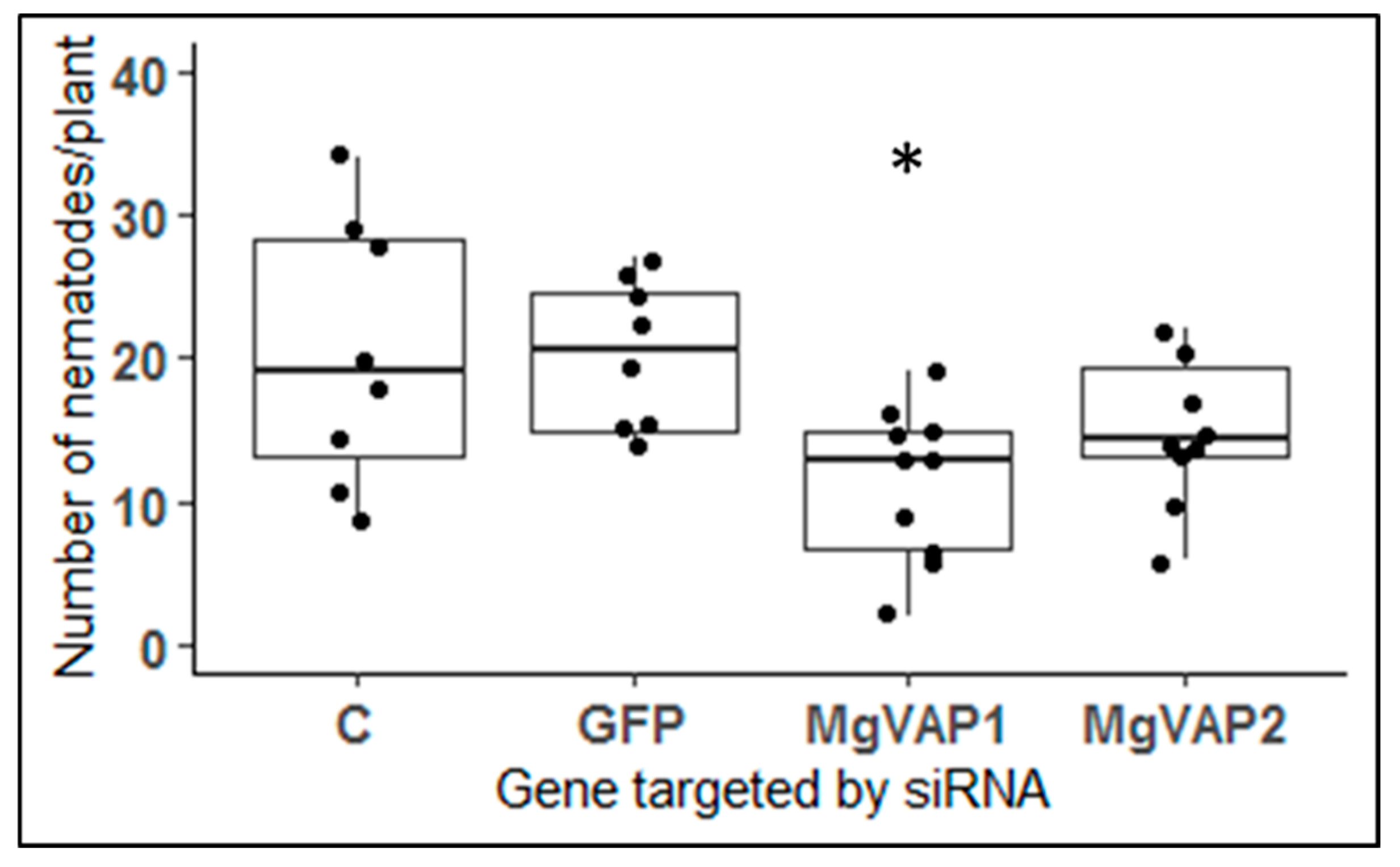
2.9. Two Up-Regulated Genes in Rice Resistant Plants Encode Putative VAP Proteins
3. Discussion
3.1. Desperately Seeking to Settle
3.2. Facing Host Defenses
3.3. M. graminicola Express Their Core Set of Effectors
3.4. A Role for VAP Genes during Infection of Resistant Plants?
4. Materials and Methods
4.1. Nematode Population
4.2. Nematode Inoculation Assays on Rice Plants
4.3. cDNA Libraries and RNA-Seq Data
4.4. Functional Annotation of Genes
4.5. Differential Expression Analyses
4.6. Cloning of M. graminicola Candidate Genes and Sequences Analyses
4.7. Validation of DEGs by RT-qPCR
4.8. In-Vitro Silencing of Mg-VAP Genes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kyndt, T.; Fernandez, D.; Gheysen, G. Plant-parasitic nematode infections in rice: Molecular and cellular insights. Annu. Rev. Phytopathol. 2014, 52, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Mantelin, S.; Bellafiore, S.; Kyndt, T. Meloidogyne graminicola: A major threat to rice agriculture. Mol. Plant Pathol. 2017, 18, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, E.; Cotroneo, A.; Carisio, L.; Troccoli, A.; Grosso, S.; Boero, C.; Capriglia, F.; De Luca, F. Detection and molecular characterization of the rice root-knot nematode Meloidogyne graminicola in Italy. Eur. J. Plant Pathol. 2017, 149, 467–476. [Google Scholar] [CrossRef]
- Nguyen, V.P.; Bellafiore, S.; Petitot, A.S.; Haidar, R.; Bak, A.; Abed, A.; Gantet, P.; Mezallira, I.; de Almeida-Engler, J.; Fernandez, D. Meloidogyne incognita—Rice (Oryza sativa) interaction: A new model system to study plant-root knot nematode interactions in monocotyledons. Rice 2014, 7, 23. [Google Scholar] [CrossRef]
- Goverse, A.; Smant, G. The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 2014, 52, 243–265. [Google Scholar] [CrossRef]
- Mejias, J.; Truong, N.M.; Abad, P.; Favery, B.; Quentin, M. Plant proteins and processes targeted by parasitic nematode effectors. Front. Plant Sci. 2019, 10, 970. [Google Scholar] [CrossRef]
- Haegeman, A.; Mantelin, S.; Jones, J.T.; Gheysen, G. Functional roles of effectors of plant-parasitic nematodes. Gene 2012, 492, 19–31. [Google Scholar] [CrossRef]
- Vieira, P.; Gleason, C. Plant-parasitic nematode effectors—Insights into their diversity and new tools for their identification. Curr. Opin. Plant Biol. 2019, 50, 37–43. [Google Scholar] [CrossRef]
- Haegeman, A.; Bauters, L.; Kyndt, T.; Rahman, M.M.; Gheysen, G. Identification of candidate effector genes in the transcriptome of the rice root knot nematode Meloidogyne graminicola. Mol. Plant Pathol. 2013, 14, 379–390. [Google Scholar] [CrossRef]
- Petitot, A.S.; Dereeper, A.; Agbessi, M.; Da Silva, C.; Guy, J.; Ardisson, M.; Fernandez, D. Dual RNA-seq reveals Meloidogyne graminicola transcriptome and candidate effectors during the interaction with rice plants. Mol. Plant Pathol. 2016, 17, 860–874. [Google Scholar] [CrossRef]
- Chen, J.; Lin, B.; Huang, Q.; Hu, L.; Zhuo, K.; Liao, J. A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog. 2017, 13, e1006301. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hu, L.; Sun, L.; Lin, B.; Huang, K.; Zhuo, K.; Liao, J. A novel Meloidogyne graminicola effector, MgMO237, interacts with multiple host defence-related proteins to manipulate plant basal immunity and promote parasitism. Mol. Plant Pathol. 2018, 19, 1942–1955. [Google Scholar] [CrossRef] [PubMed]
- Naalden, D.; Haegeman, A.; de Almeida-Engler, J.; Birhane Eshetu, F.; Bauters, L.; Gheysen, G. The Meloidogyne graminicola effector Mg16820 is secreted in the apoplast and cytoplasm to suppress plant host defense responses. Mol. Plant Pathol. 2018, 19, 2416–2430. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, K.; Naalden, D.; Nowak, S.; Xuan Huy, N.; Bauters, L.; Gheysen, G. A Meloidogyne graminicola C-type lectin, Mg01965, is secreted into the host apoplast to suppress plant defence and promote parasitism. Mol. Plant Pathol. 2019, 20, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.L.; Wang, Z.H.; Maria, M.; Qu, N.; Zheng, J.W. Meloidogyne graminicola protein disulfide isomerase may be a nematode effector and is involved in protection against oxidative damage. Sci. Rep. 2019, 9, 11949. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Mao, Z.; Zhai, M.; Zeng, F.; Yang, Y.; Xie, B. Transcriptome profiling of Cucumis metuliferus infected by Meloidogyne incognita provides new insights into putative defense regulatory network in Cucurbitaceae. Sci. Rep. 2017, 7, 3544. [Google Scholar] [CrossRef] [PubMed]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef]
- Dimkpa, S.O.; Lahari, Z.; Shrestha, R.; Douglas, A.; Gheysen, G.; Price, A.H. A genome-wide association study of a global rice panel reveals resistance in Oryza sativa to root-knot nematodes. J. Exp. Bot. 2016, 67, 1191–1200. [Google Scholar] [CrossRef]
- Mhatre, P.H.; Singh, P.; Malik, S.; Kaur, S.; Singh, A.K.; Mohan, S.; Sirohi, A. Histopathological changes and evaluation of resistance in Asian rice (Oryza sativa L.) against rice root-knot nematode, Meloidogyne graminicola Golden and Birch. Indian J. Genet. 2015, 75, 41–48. [Google Scholar] [CrossRef]
- Phan, N.T.; De Waele, D.; Lorieux, M.; Xiong, L.; Bellafiore, S. A Hypersensitivity-like Response to Meloidogyne graminicola in rice (Oryza sativa). Phytopathology 2018, 108, 521–528. [Google Scholar] [CrossRef]
- Soriano, I.; Schmit, V.; Brar, D.; Prot, J.C.; Reversat, G. Resistance to rice root-knot nematode Meloidogyne graminicola identified in Oryza longistaminata and O. glaberrima. Nematology 1999, 1, 395–398. [Google Scholar] [CrossRef]
- Petitot, A.S.; Kyndt, T.; Haidar, R.; Dereeper, A.; Collin, M.; de Almeida Engler, J.; Gheysen, G.; Fernandez, D. Transcriptomic and histological responses of the African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Ann. Bot. 2017, 119, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Mattos, V.S.; Leite, R.R.; Cares, J.E.; Gomes, A.C.M.M.; Moita, A.W.; Lobo, V.L.S.; Carneiro, R.M.D.G. Oryza glumaepatula, a new source of resistance to Meloidogyne graminicola and histological characterization of its defense mechanisms. Phytopathology 2019, 109, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Somvanshi, V.S.; Tathode, M.; Shukla, R.N.; Rao, U. Nematode genome announcement: A draft genome for rice root-knot nematode, Meloidogyne graminicola. J. Nematol. 2018, 50, 111–116. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Perfus-Barbeoch, L.; Aury, J.M.; Da Rocha, M.; Gouzy, J.; Sallet, E.; Martin-Jimenez, C.; Bailly-Bechet, M.; Castagnone-Sereno, P.; Flot, J.F.; et al. Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS Genet. 2017, 13, e1006777. [Google Scholar] [CrossRef]
- Abad, P.; Gouzy, J.; Aury, J.M.; Castagnone-Sereno, P.; Danchin, E.G.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef]
- Opperman, C.H.; Bird, D.M.; Williamson, V.M.; Rokhsar, D.S.; Burke, M.; Cohn, J.; Cromer, J.; Diener, S.; Gajan, J.; Graham, S.; et al. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. USA 2008, 105, 14802–14807. [Google Scholar] [CrossRef]
- Grove, C.; Cain, S.; Chen, W.J.; Davis, P.; Harris, T.; Howe, K.L.; Kishore, R.; Lee, R.; Paulini, M.; Raciti, D.; et al. Using WormBase: A genome biology resource for Caenorhabditis elegans and related nematodes. Methods Mol. Biol. 2018, 1757, 399–470. Available online: http://www.wormbase.org (accessed on 19 December 2019). [CrossRef]
- Li, C.; Kim, K. Neuropeptides. In WormBook; The C. elegans Research Community; 2008; Available online: http://www.wormbook.org (accessed on 5 May 2020). [CrossRef]
- Triantaphyllou, A.; Hirschmann, H. Post-infection development of Meloidogyne incognita Chitwood, 1949. Ann. Inst. Phytopathol. Benaki 1960, 3, 3–11. [Google Scholar]
- Page, A.P.; Stepek, G.; Winter, A.D.; Pertab, D. Enzymology of the nematode cuticle: A potential drug target? Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 133–141. [Google Scholar] [CrossRef]
- Zhuo, K.; Chen, J.; Lin, B.; Wang, J.; Sun, F.; Hu, L.; Liao, J. A novel Meloidogyne enterolobii effector MeTCTP promotes parasitism by suppressing programmed cell death in host plants. Mol. Plant Pathol. 2017, 18, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Zhuo, K.; Wu, P.; Cui, R.; Zhang, L.H.; Liao, J. A novel effector protein, MJ-NULG1a, targeted to giant cell nuclei plays a role in Meloidogyne javanica parasitism. Mol. Plant-Microbe Interact. 2013, 26, 55–66. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, G.; Dong, R.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Two chorismate mutase genes from the root-knot nematode Meloidogyne incognita. Mol. Plant Pathol. 2005, 6, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Mao, Z.; Zhang, X.; Ling, J.; Lin, R.; Zhang, X.; Liu, R.; Wang, Y.; Yang, Y.; Cheng, X.; et al. The novel secreted Meloidogyne incognita effector MiISE6 targets the host nucleus and facilitates parasitism in Arabidopsis. Front. Plant Sci. 2018, 9, 252. [Google Scholar] [CrossRef] [PubMed]
- Jaouannet, M.; Perfus-Barbeoch, L.; Deleury, E.; Magliano, M.; Engler, G.; Vieira, P.; Danchin, E.G.; Da Rocha, M.; Coquillard, P.; Abad, P.; et al. A root-knot nematode-secreted protein is injected into giant cells and targeted to the nuclei. New Phytol. 2012, 194, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Grossi-de-Sa, M.; Petitot, A.S.; Xavier, D.A.; Sá, M.E.L.; Mezzalira, I.; Beneventi, M.A.; Martins, N.F.; Baimey, H.K.; Albuquerque, E.V.S.; Grossi-de-Sa, M.F.; et al. Rice susceptibility to root-knot nematodes is enhanced by the Meloidogyne incognita MSP18 effector gene. Planta 2019, 250, 1215–1227. [Google Scholar] [CrossRef]
- Xie, J.; Li, S.; Mo, C.; Wang, G.; Xiao, X.; Xiao, Y. A novel Meloidogyne incognita effector Misp12 suppresses plant defense response at latter stages of nematode parasitism. Front. Plant Sci. 2016, 7, 964. [Google Scholar] [CrossRef]
- Iberkleid, I.; Vieira, P.; de Almeida Engler, J.; Firester, K.; Spiegel, Y.; Horowitz, S.B. Fatty acid-and retinol-binding protein, Mj-FAR-1 induces tomato host susceptibility to root-knot nematodes. PLoS ONE 2013, 8, e64586. [Google Scholar] [CrossRef]
- Lin, B.; Zhuo, K.; Chen, S.; Hu, L.; Sun, L.; Wang, X.; Zhang, L.H.; Liao, J. A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 2016, 209, 1159–1173. [Google Scholar] [CrossRef]
- Gleason, C.; Polzin, F.; Habash, S.S.; Zhang, L.; Utermark, J.; Grundler, F.M.; Elashry, A. Identification of two Meloidogyne hapla genes and an investigation of their roles in the plant-nematode interaction. Mol. Plant-Microbe Interact. 2017, 30, 101–112. [Google Scholar] [CrossRef]
- Jaouannet, M.; Magliano, M.; Arguel, M.J.; Gourgues, M.; Evangelisti, E.; Abad, P.; Rosso, M.N. The root-knot nematode calreticulin Mi-CRT is a key effector in plant defense suppression. Mol. Plant-Microbe Interact. 2013, 26, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Leelarasamee, N.; Zhang, L.; Gleason, C. The root-knot nematode effector MiPFN3 disrupts plant actin filaments and promotes parasitism. PLoS Pathog. 2018, 14, e1006947. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Mao, Z.; Zhang, X.; Zhang, X.; Wang, Y.; Ling, J.; Lin, R.; Li, D.; Kang, X.; Sun, W.; et al. A Meloidogyne incognita effector MiISE5 suppresses programmed cell death to promote parasitism in host plant. Sci. Rep. 2018, 8, 7256. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Torres, J.L.; Wilbers, R.H.; Gawronski, P.; Boshoven, J.C.; Finkers-Tomczak, A.; Cordewener, J.H.; America, A.H.; Overmars, H.A.; Van′t Klooster, J.W.; Baranowski, L.; et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA 2012, 109, 10119–10124. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Torres, J.L.; Wilbers, R.H.; Warmerdam, S.; Finkers-Tomczak, A.; Diaz-Granados, A.; van Schaik, C.C.; Helder, J.; Bakker, J.; Goverse, A.; Schots, A.; et al. Apoplastic Venom Allergen-like Proteins of cyst nematodes modulate the activation of basal plant innate immunity by cell surface receptors. PLoS Pathog. 2014, 10, e1004569. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Shields, J.; Allen, R.; Hussey, R.S. Molecular cloning and characterisation of a venom allergen AG5-like cDNA from Meloidogyne incognita. Int. J. Parasitol. 2000, 30, 77–81. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Hu, Y.; Fu, P.; Xu, J. Molecular cloning and analysis of a new venom allergen-like protein gene from the root-knot nematode Meloidogyne incognita. Exp. Parasitol. 2007, 117, 133–140. [Google Scholar] [CrossRef]
- Danchin, E.G.; Rosso, M.N.; Vieira, P.; de Almeida-Engler, J.; Coutinho, P.M.; Henrissat, B.; Abad, P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Nat. Acad. Sci. USA 2010, 107, 17651–17656. [Google Scholar] [CrossRef]
- Vieira, P.; Mowery, J.; Eisenback, J.D.; Shao, J.; Nemchinov, L.G. Cellular and transcriptional responses of resistant and susceptible cultivars of alfalfa to the root lesion nematode, Pratylenchus penetrans. Front. Plant Sci. 2019, 10, 971. [Google Scholar] [CrossRef]
- Gillet, F.X.; Bournaud, C.; de Souza Júnior, J.D.A.; Grossi-de-Sa, M.F. Plant-parasitic nematodes: Towards understanding molecular players in stress responses. Ann. Bot. 2017, 119, 775–789. [Google Scholar] [CrossRef]
- Williamson, V.M.; Hussey, R.S. Nematode pathogenesis and resistance in plants. Plant Cell 1996, 8, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Urwin, P.E.; Lilley, C.J.; Atkinson, H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant-Microbe Interact. 2002, 15, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Wilbers, R.H.P.; Schneiter, R.; Holterman, M.H.M.; Drurey, C.; Smant, G.; Asojo, O.A.; Maizels, R.M.; Lozano-Torres, J.L. Secreted venom allergen-like proteins of helminths: Conserved modulators of host responses in animals and plants. PLoS Pathog. 2018, 14, e1007300. [Google Scholar] [CrossRef]
- Fang, K.S.; Vitale, M.P.; Fehlner, P.; King, T.P. cDNA cloning and primary structure of a white-face hornet venom allergen, antigen 5. Proc. Natl. Acad. Sci. USA 1988, 85, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Maleita, C.; Egas, C.; Abrantes, I.; Curtis, R. Significant effects of RNAi silencing of the venom allergen-like protein (Mhivap-1) of the root-knot nematode Meloidogyne hispanica in the early events of infection. Plant Pathol. 2017, 66, 1329–1337. [Google Scholar] [CrossRef]
- Luo, S.; Liu, S.; Kong, L.; Peng, H.; Huang, W.; Jian, H.; Peng, D. Two venom allergen-like proteins, HaVAP1 and HaVAP2, are involved in the parasitism of Heterodera avenae. Mol. Plant Pathol. 2019, 20, 471–484. [Google Scholar] [CrossRef]
- Chaudhary, S.; Dutta, T.K.; Shivakumara, T.N.; Rao, U. RNAi of an esophageal gland specific Mi-msp-1 gene alters the early stage infection behaviour of root-knot nematode, Meloidogyne incognita. J. Gen. Plant Pathol. 2019, 85, 232–242. [Google Scholar] [CrossRef]
- Kang, J.S.; Koh, Y.H.; Moon, Y.S.; Lee, S.H. Molecular properties of a venom allergen like protein suggest a parasitic function in the pinewood nematode Bursaphelenchus xylophilus. Int. J. Parasitol. 2012, 42, 63–70. [Google Scholar] [CrossRef]
- Reversat, G.; Boyer, J.; Sannier, C.; Pando-Bahuon, A. Use of a mixture of sand and water-absorbent synthetic polymer as substrate for the xenic culturing of plant-parasitic nematodes in the laboratory. Nematology 1999, 1, 209–212. [Google Scholar] [CrossRef]
- Howe, K.L.; Bolt, B.J.; Shafie, M.; Kersey, P.; Berriman, M. WormBase ParaSite—A comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017, 215, 2–10. Available online: http://www.parasite.wormbase.org (accessed on 4 December 2019). [CrossRef]
- Monat, C.; Pera, B.; Ndjiondjop, M.N.; Sow, M.; Tranchant-Dubreuil, C.; Bastianelli, L.; Ghesquière, A.; Sabot, F. De novo assemblies of three Oryza glaberrima accessions provide first insights about Pan-genome of african rices. Genome Biol. Evol. 2017, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Holik, A.Z.; Su, S.; Jansz, N.; Chen, K.; Leong, H.S.; Blewitt, M.E.; Asselin-Labat, M.L.; Smyth, G.K.; Ritchie, M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015, 43, e97. [Google Scholar] [CrossRef]
- Venny Oliveros, J.C. An Interactive Tool for Comparing Lists with Venn′s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 27 February 2020).
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Arguel, M.J.; Jaouannet, M.; Magliano, M.; Abad, P.; Rosso, M.N. siRNAs trigger efficient silencing of a parasitism gene in plant parasitic root-knot nematodes. Genes 2012, 3, 391–408. [Google Scholar] [CrossRef]
- Byrd, D.W.; Kirkpatrick, T., Jr.; Barker, K.R. An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 1983, 15, 142–143. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; Available online: http://www.R-project.org (accessed on 5 June 2020).

| M. graminicola Transcript | Predicted Function | RNA Seq (R versus S) | qPCR (R versus S) | ||||
|---|---|---|---|---|---|---|---|
| 2 dpi | 4 dpi | 8 dpi | 2 dpi | 4 dpi | 8 dpi | ||
| Mg449 | Endoglucanase | 2.17 * | 3.69 * | 2.99 * | 2.70 * | 5.14 * | 5.45 * |
| Mg693 | Glucuronoxylanase | 1.57 | 3.95 * | 4.38 * | 1.88 | 3.66 * | 5.82 * |
| Mg1702 | Neuropeptide “nlp” | 0.07 | 1.40 | 2.19 * | 0.70 | 2.40 * | 4.98 * |
| Mg2129 | Neuropeptide “flp” | 0.33 | 1.58 | 3.14 * | 1.03 | 2.06 | 4.69 * |
| Mg707 | VAP | 9.26 | 3.27 * | 2.57 * | 2.42 | 4.99 * | 5.62 * |
| Mg11937 | VAP | 1.38 | 5.03 * | 5.86 * | 2.08 | 5.63 * | 6.78 * |
| Mg757 | Unknown | 0.67 | 1.57 | 2.8 * | 1.11 | 2.83 * | 4.40 * |
| Mg4965 | CLE-like | −0.26 | 0.60 | 1.11 | 1.09 | 0.96 | 1.19 |
| Mg12322 | Unknown | −0.92 | −0.72 | 1.68 | −0.81 | −0.15 | 3.67 |
| Mg13532 | Unknown | −0.79 | −0.75 | −0.44 | −0.49 | −0.27 | 1.04 |
| Mg44061 | Unknown | 5.42 | 9.55 | 12.09 * | 0.001 | 1.89 | 5.02 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petitot, A.-S.; Dereeper, A.; Da Silva, C.; Guy, J.; Fernandez, D. Analyses of the Root-Knot Nematode (Meloidogyne graminicola) Transcriptome during Host Infection Highlight Specific Gene Expression Profiling in Resistant Rice Plants. Pathogens 2020, 9, 644. https://doi.org/10.3390/pathogens9080644
Petitot A-S, Dereeper A, Da Silva C, Guy J, Fernandez D. Analyses of the Root-Knot Nematode (Meloidogyne graminicola) Transcriptome during Host Infection Highlight Specific Gene Expression Profiling in Resistant Rice Plants. Pathogens. 2020; 9(8):644. https://doi.org/10.3390/pathogens9080644
Chicago/Turabian StylePetitot, Anne-Sophie, Alexis Dereeper, Corinne Da Silva, Julie Guy, and Diana Fernandez. 2020. "Analyses of the Root-Knot Nematode (Meloidogyne graminicola) Transcriptome during Host Infection Highlight Specific Gene Expression Profiling in Resistant Rice Plants" Pathogens 9, no. 8: 644. https://doi.org/10.3390/pathogens9080644
APA StylePetitot, A.-S., Dereeper, A., Da Silva, C., Guy, J., & Fernandez, D. (2020). Analyses of the Root-Knot Nematode (Meloidogyne graminicola) Transcriptome during Host Infection Highlight Specific Gene Expression Profiling in Resistant Rice Plants. Pathogens, 9(8), 644. https://doi.org/10.3390/pathogens9080644






