Factoring in the Complexity of the Cystic Fibrosis Lung to Understand Aspergillus fumigatus and Pseudomonas aeruginosa Interactions
Abstract
1. Cystic Fibrosis
2. Clinical Manifestations of Cystic Fibrosis
3. Dysbiosis of the Pulmonary Microbiome in Cystic Fibrosis
4. Pseudomonas aeruginosa Is the Predominant Respiratory Bacterial Pathogen in Cystic Fibrosis
5. Aspergillus fumigatus Is the Predominant Respiratory Mould Pathogen in Cystic Fibrosis
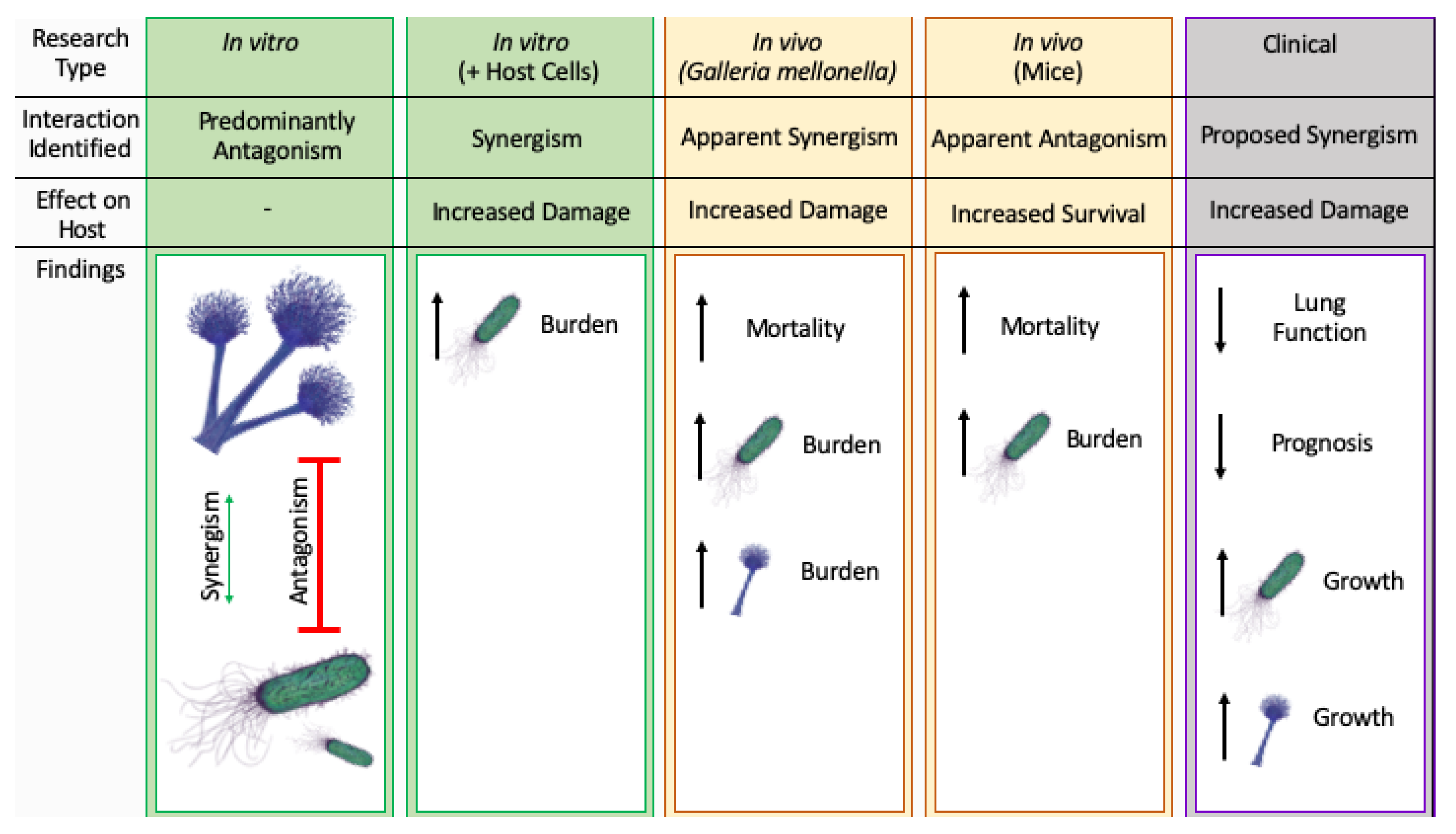
6. Combined Infection with Aspergillus fumigatus and Pseudomonas aeruginosa in Cystic Fibrosis
7. Future Directions for Clarifying A. fumigatus-P. aeruginosa Interactions in CF
Author Contributions
Funding
Conflicts of Interest
References
- Davies, J.C.; Ebdon, A.M.; Orchard, C. Recent advances in the management of cystic fibrosis. Arch. Dis. Child. 2014, 99, 1033–1036. [Google Scholar] [CrossRef]
- Cystic Fibrosis Trust. UK Cystic Fibrosis Registry Annual Data Report 2018. Available online: https://www.cysticfibrosis.org.uk (accessed on 31 July 2019).
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Andersen, D.H. Cystic fibrosis of the pancreas and its relation to celiac disease. Am. J. Dis. Child. 1938, 56, 344. [Google Scholar] [CrossRef]
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- Cohen-Cymberknoh, M.; Shoseyov, D.; Kerem, E. Managing cystic fibrosis. Am. J. Respir. Crit. Care Med. 2011, 183, 1463–1471. [Google Scholar] [CrossRef]
- Chmiel, J.F.; Konstan, M.W.; Elborn, J.S. Antibiotic and anti-inflammatory therapies for cystic fibrosis. Cold Spring Harb. Perspect. Med. 2013, 3, a009779. [Google Scholar] [CrossRef] [PubMed]
- Cantin, A.; Hartl, M.; Konstan, M.W.; Chmiel, J.F. Inflammation in cystic fibrosis lung disease: Pathogenesis and therapy. J. Cyst. Fibros. 2015, 14, 419–430. [Google Scholar] [CrossRef]
- Davis, P.B. Cystic Fibrosis Since 1938. Am. J. Respir. Crit. Care Med. 2006, 173, 475–482. [Google Scholar] [CrossRef]
- National Centre for Biotechnology Information Gene. Cftr Cf Transmembrane Conductance Regulator [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 31 July 2019).
- Viart, V.; Bergougnoux, A.; Bonini, J.; Varilh, J.; Chiron, R.; Tabary, O.; Molinari, N.; Claustres, M.; Taulan-Cadars, M. Transcription factors and miRNAs that regulate fetal to adult CFTR expression change are new targets for cystic fibrosis. Eur. Respir. J. 2014, 45, 116–128. [Google Scholar] [CrossRef]
- Hwang, T.C.; Sheppard, D.N. Gating of the CFTR Cl− channel by ATP-driven nucleotide-binding domain dimerisation. J. Physiol. 2009, 587, 2151–2161. [Google Scholar] [CrossRef]
- Ji, H.L.; Chalfant, M.L.; Jovov, B.; Lockhart, J.P.; Parker, S.B.; Fuller, C.; Stanton, B.A.; Benos, D.J. The cytosolic termini of the beta- and gamma-ENaC subunits are involved in the functional interactions between cystic fibrosis transmembrane conductance regulator and epithelial sodium channel. J. Biol. Chem. 2000, 275, 27947–27956. [Google Scholar] [PubMed]
- Nagel, G.; Szellas, T.; Riordan, J.R.; Friedrich, T.; Hartung, K. Non-specific activation of the epithelial sodium channel by the CFTR chloride channel. EMBO Rep. 2001, 2, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Leroy, C.; Privé, A.; Bourret, J.C.; Berthiaume, Y.; Ferraro, P.; Brochiero, E. Regulation of ENaC and CFTR expression with K+ channel modulators and effect on fluid absorption across alveolar epithelial cells. Am. J. Physiol. Cell. Mol. Physiol. 2006, 291, L1207–L1219. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Leng, Q.; Egan, M.E.; Caplan, M.J.; Boulpaep, E.L.; Giebisch, G.H.; Hebert, S.C. CFTR is required for PKA-regulated ATP sensitivity of Kir1.1 potassium channels in mouse kidney. J. Clin. Investig. 2006, 116, 797–807. [Google Scholar] [CrossRef]
- Wang, J.; Haanes, K.A.; Novak, I. Purinergic regulation of CFTR and Ca2+-activated Cl− channels and K+ channels in human pancreatic duct epithelium. Am. J. Physiol. Physiol. 2013, 304, C673–C684. [Google Scholar] [CrossRef]
- Borowitz, U. CFTR, bicarbonate, and the pathophysiology of cystic fibrosis. Pediatr. Pulmonol. 2015, 50, S24–S30. [Google Scholar] [CrossRef]
- Gabriel, S.E.; Clarke, L.L.; Boucher, R.C.; Stutts, M.J. CFTR and outward rectifying chloride channels are distinct proteins with a regulatory relationship. Nature 1993, 363, 263–266. [Google Scholar] [CrossRef]
- Fischer, H.; Illek, B.; Sachs, L.; Finkbeiner, W.E.; Widdicombe, J.H. CFTR and calcium-activated chloride channels in primary cultures of human airway gland cells of serous or mucous phenotype. Am. J. Physiol. Cell. Mol. Physiol. 2010, 299, L585–L594. [Google Scholar] [CrossRef]
- Mall, M.A.; Galietta, L. Targeting ion channels in cystic fibrosis. J. Cyst. Fibros. 2015, 14, 561–570. [Google Scholar] [CrossRef]
- Cystic Fibrosis Genetic Analysis Consortium. Cystic Fibrosis Mutation Database. Available online: www.genet.sickkids.on.ca/cftr (accessed on 5 July 2019).
- Castellani, C.; Cuppens, H.; Macek, M., Jr.; Cassiman, J.J.; Kerem, E.; Durie, P.; Tullis, E.; Assael, B.M.; Bombieri, C.; Brown, A.; et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J. Cyst. Fibros. 2008, 7, 179–196. [Google Scholar] [CrossRef]
- Vernon, R.M.; Chong, P.A.; Lin, H.; Yang, Z.; Zhou, Q.; Aleksandrov, A.A.; Dawson, J.E.; Riordan, J.R.; Brouillette, C.G.; Thibodeau, P.H.; et al. Stabilization of a nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator yields insight into disease-causing mutations. J. Biol. Chem. 2017, 292, 14147–14164. [Google Scholar] [CrossRef]
- Boyle, M.P.; De Boeck, K. A new era in the treatment of cystic fibrosis: Correction of the underlying CFTR defect. Lancet Respir. Med. 2013, 1, 158–163. [Google Scholar] [CrossRef]
- Bono-Neri, F.; Romano, C.; Isedeh, A. Cystic fibrosis: Advancing along the continuum. J. Pediatr. Health Care 2019, 33, 242–254. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, B.P.; Freedman, S.D. Cystic fibrosis. Lancet 2009, 373, 1891–1904. [Google Scholar] [CrossRef]
- Lao, O.; Andrés, A.M.; Mateu, E.; Bertranpetit, J.; Calafell, F.; Andr, A.M. Spatial patterns of cystic fibrosis mutation spectra in European populations. Eur. J. Hum. Genet. 2003, 11, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Lewis, H.A.; Zhao, X.; Wang, C.; Sauder, J.M.; Rooney, I.; Noland, B.W.; Lorimer, D.; Kearins, M.C.; Conners, K.; Condon, B.; et al. Impact of the ΔF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J. Biol. Chem. 2004, 280, 1346–1353. [Google Scholar] [CrossRef]
- Protasevich, I.; Yang, Z.; Wang, C.; Atwell, S.; Zhao, X.; Emtage, S.; Wetmore, D.; Hunt, J.F.; Brouillette, C.G. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010, 19, 1917–1931. [Google Scholar] [CrossRef]
- Du, K.; Lukacs, G.L. Cooperative assembly and misfolding of CFTR domains in vivo. Mol. Biol. Cell 2009, 20, 1903–1915. [Google Scholar] [CrossRef]
- Fernandez, E.F.; De Santi, C.; De Rose, V.; Greene, C.M. CFTR dysfunction in cystic fibrosis and chronic obstructive pulmonary disease. Expert Rev. Respir. Med. 2018, 12, 483–492. [Google Scholar] [CrossRef]
- Malhotra, S.; Hayes, D.; Wozniak, D.J. Cystic fibrosis and Pseudomonas aeruginosa: The Host-microbe interface. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef]
- Sergeev, V.; Chou, F.Y.; Lam, G.Y.; Hamilton, C.M.; Wilcox, P.G.; Quon, B.S. The extrapulmonary effects of cystic fibrosis transmembrane conductance regulator modulators in cystic fibrosis. Ann. Am. Thorac. Soc. 2020, 17, 147–154. [Google Scholar] [CrossRef]
- Moran, A.; Dunitz, J.; Nathan, B.; Saeed, A.; Holme, B.; Thomas, W. Cystic fibrosis–Related diabetes: Current trends in prevalence, incidence, and mortality. Diabetes Care 2009, 32, 1626–1631. [Google Scholar] [CrossRef]
- Harris, W.T.; Kirk, K.L. CFTR and cystic fibrosis. In Ion Channels and Transporters of Epithelia in Health and Disease; Springer Science and Business Media LLC: Berlin, Germany, 2015; pp. 519–552. [Google Scholar]
- Graeber, S.Y.; Zhou-Suckow, Z.; Schatterny, J.; Hirtz, S.; Boucher, R.C.; Mall, M.A. Hypertonic saline is effective in the prevention and treatment of mucus obstruction, but not airway inflammation, in mice with chronic obstructive lung disease. Am. J. Respir. Cell Mol. Biol. 2013, 49, 410–417. [Google Scholar] [CrossRef]
- Worlitzsch, D.; Tarran, R.; Ulrich, M.; Schwab, U.; Cekici, A.; Meyer, K.C.; Birrer, P.; Bellon, G.; Berger, J.; Weiss, T.; et al. Effects of reduced mucus oxygen concentration in airway pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 2002, 3, 317–325. [Google Scholar] [CrossRef]
- Pezzulo, A.A.; Tang, X.X.; Hoegger, M.J.; Abou Alaiwa, M.H.; Ramachandran, S.; Moninger, T.O.; Karp, P.H.; Wohlford-Lenane, C.L.; Haagsman, H.P.; van Eijk, M.; et al. Reduced airway surface ph impairs bacterial killing in the porcine cystic fibrosis lung. Nature 2012, 487, 109–113. [Google Scholar] [CrossRef]
- Berkebile, A.R.; McCray, P.B. Effects of airway surface liquid pH on host defense in cystic fibrosis. Int. J. Biochem. Cell Biol. 2014, 52, 124–129. [Google Scholar] [CrossRef]
- Song, Y.; Salinas, D.; Nielson, D.; Verkman, A.S. Hyperacidity of secreted fluid from submucosal glands in early cystic fibrosis. Am. J. Physiol. Physiol. 2006, 290, C741–C749. [Google Scholar] [CrossRef]
- Stoltz, D.; Meyerholz, D.K.; Welsh, M.J. Origins of cystic fibrosis lung disease. N. Engl. J. Med. 2015, 372, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ostedgaard, L.; Alaiwa, M.H.A.; Lu, L.; Fischer, A.J.; Stoltz, D.A. Mucociliary transport in healthy and cystic fibrosis pig airways. Ann. Am. Thorac. Soc. 2018, 15, S171–S176. [Google Scholar] [CrossRef]
- Gifford, A.M.; Chalmers, J.D. The role of neutrophils in cystic fibrosis. Curr. Opin. Hematol. 2014, 21, 16–22. [Google Scholar] [CrossRef]
- Guan, X.; Hou, Y.; Sun, F.; Yang, Z.; Li, C. Dysregulated chemokine signaling in cystic fibrosis lung disease: A Potential therapeutic target. Curr. Drug Targets 2016, 17, 1535–1544. [Google Scholar] [CrossRef]
- Castellani, S.; D’Oria, S.; Diana, A.; Polizzi, A.M.; Di Gioia, S.; Mariggiò, M.A.; Guerra, L.; Favia, M.; Vinella, A.; Leonetti, G.; et al. G-CSF and GM-CSF modify neutrophil functions at concentrations found in cystic fibrosis. Sci. Rep. 2019, 9, 12937. [Google Scholar] [CrossRef]
- Colombo, C.; Costantini, D.; Rocchi, A.; Cariani, L.; Garlaschi, M.L.; Tirelli, S.; Calori, G.; Copreni, E.; Conese, M. Cytokine levels in sputum of cystic fibrosis patients before and after antibiotic therapy. Pediatr. Pulmonol. 2005, 40, 15–21. [Google Scholar] [CrossRef]
- Zaman, M.M.; Gelrud, A.; Junaidi, O.; Regan, M.M.; Warny, M.; Shea, J.C.; Kelly, C.; O’Sullivan, B.P.; Freedman, S.D. Interleukin 8 secretion from monocytes of subjects heterozygous for the deltaf508 cystic fibrosis transmembrane conductance regulator gene mutation is altered. Clin. Diagn. Lab. Immunol. 2004, 11, 819–824. [Google Scholar] [CrossRef]
- Freedman, S.D.; Blanco, P.G.; Zaman, M.M.; Shea, J.C.; Ollero, M.; Hopper, I.K.; Weed, D.A.; Gelrud, A.; Regan, M.M.; Laposata, M.; et al. Association of cystic fibrosis with abnormalities in fatty acid metabolism. N. Engl. J. Med. 2004, 350, 560–569. [Google Scholar] [CrossRef]
- Karp, C.L.; Flick, L.M.; Park, K.W.; Softic, S.; Greer, T.M.; Keledjian, R.; Yang, R.; Uddin, J.; Guggino, W.B.; Atabani, S.F.; et al. Defective lipoxin-mediated anti-inflammatory activity in the cystic fibrosis airway. Nat. Immunol. 2004, 5, 388–392. [Google Scholar] [CrossRef]
- Cohen-Cymberknoh, M.; Kerem, E.; Ferkol, T.; Elizur, A. Airway inflammation in cystic fibrosis: Molecular mechanisms and clinical implications. Thorax 2013, 68, 1157–1162. [Google Scholar] [CrossRef]
- Hartl, D.; Gaggar, A.; Bruscia, E.; Hector, A.; Marcos, V.; Jung, A.; Greene, C.M.; McElvaney, G.; Mall, M.; Doring, G. Innate immunity in cystic fibrosis lung disease. J. Cyst. Fibros. 2012, 11, 363–382. [Google Scholar] [CrossRef]
- Elborn, S.; Vallieres, E. Cystic fibrosis gene mutations: Evaluation and assessment of disease severity. Adv. Genom. Genet. 2014, 4, 161–172. [Google Scholar] [CrossRef]
- Dickson, R.P.; Huffnagle, G.B. The lung microbiome: New principles for respiratory bacteriology in health and disease. PLOS Pathog. 2015, 11, e1004923. [Google Scholar] [CrossRef]
- Surette, M.G. The cystic fibrosis lung microbiome. Ann. Am. Thorac. Soc. 2014, 11, S61–S65. [Google Scholar] [CrossRef] [PubMed]
- Marsland, B.J.; Gollwitzer, E.S. Host–microorganism interactions in lung diseases. Nat. Rev. Immunol. 2014, 14, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Krüger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.D.; Niemiec, M.J. Fungal-bacterial interactions in health and disease. Pathogens 2019, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Lynch, S.V. The emerging relationship between the airway microbiota and chronic respiratory disease: Clinical implications. Expert Rev. Respir. Med. 2011, 5, 809–821. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, Y.J.; Lipuma, J.J. The microbiome in cystic fibrosis. Clin. Chest Med. 2016, 37, 59–67. [Google Scholar] [CrossRef]
- Bevivino, A.; Bacci, G.; Drevinek, P.; Nelson, M.T.; Hoffman, L.; Mengoni, A. Deciphering the ecology of cystic fibrosis bacterial communities: Towards systems-level integration. Trends Mol. Med. 2019, 25, 1110–1122. [Google Scholar] [CrossRef]
- Wylie, K.M.; Weinstock, G.M.; Storch, G.A. Emerging view of the human virome. Transl. Res. 2012, 160, 283–290. [Google Scholar] [CrossRef]
- Billard, L.; Le Berre, R.; Pilorgé, L.; Payan, C.; Héry-Arnaud, G.; Vallet, S. Viruses in cystic fibrosis patients’ airways. Crit. Rev. Microbiol. 2017, 43, 690–708. [Google Scholar] [CrossRef]
- Kiedrowski, M.R.; Bomberger, J.M. Viral-bacterial co-infections in the cystic fibrosis respiratory tract. Front. Immunol. 2018, 9, 3067. [Google Scholar] [CrossRef]
- Rosenfeld, M.; Ramsey, B.W.; Gibson, R.L. Pseudomonas acquisition in young patients with cystic fibrosis: Pathophysiology, diagnosis, and management. Curr. Opin. Pulm. Med. 2003, 9, 492–497. [Google Scholar] [CrossRef]
- Cystic Fibrosis Foundation. US 2018 Patient Registry Annual Data Report. Available online: https://www.cff.org (accessed on 24 July 2020).
- Stutman, H.R.; Lieberman, J.M.; Nussbaum, E.; Marks, M.I. Antibiotic prophylaxis in infants and young children with cystic fibrosis: A randomized controlled trial. J. Pediatr. 2002, 140, 299–305. [Google Scholar] [CrossRef]
- Ratjen, F.; Comes, G.; Paul, K.; Posselt, H.; Wagner, T.; Harms, K. Effect of continuous antistaphylococcal therapy on the rate of P. aeruginosa acquisition in patients with cystic fibrosis. Pediatr. Pulmonol. 2001, 31, 13–16. [Google Scholar] [CrossRef]
- Treggiari, M.M.; Rosenfeld, M.; Retsch-Bogart, G.; Gibson, R.; Ramsey, B. Approach to eradication of initial Pseudomonas aeruginosa infection in children with cystic fibrosis. Pediatr. Pulmonol. 2007, 42, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Stuart, B.; Lin, J.H.; Mogayzel, J.P.J. Early eradication of Pseudomonas aeruginosa in patients with cystic fibrosis. Paediatr. Respir. Rev. 2010, 11, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Hewer, S.C.L.; Smyth, A.R. Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis. Cochrane Database Syst. Rev. 2017, 2017, CD004197. [Google Scholar] [CrossRef]
- Zhao, J.; Schloss, P.D.; Kalikin, L.M.; Carmody, L.A.; Foster, B.K.; Petrosino, J.F.; Cavalcoli, J.D.; VanDevanter, D.R.; Murray, S.; Li, J.Z.; et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 2012, 109, 5809–5814. [Google Scholar] [CrossRef]
- Martiniano, S.L.; Nick, J.A.; Daley, C.L. Nontuberculous mycobacterial infections in cystic fibrosis. Clin. Chest Med. 2016, 37, 83–96. [Google Scholar] [CrossRef]
- Lamoureux, C.; Guilloux, C.-A.; Beauruelle, C.; Jolivet-Gougeon, A.; Héry-Arnaud, G. Anaerobes in cystic fibrosis patients’ airways. Crit. Rev. Microbiol. 2019, 45, 103–117. [Google Scholar] [CrossRef]
- Jones, A.M. Anaerobic bacteria in cystic fibrosis: Pathogens or harmless commensals? Thorax 2011, 66, 558–559. [Google Scholar] [CrossRef]
- Armstead, J.; Morris, J.; Denning, D.W. Multi-country estimate of different manifestations of aspergillosis in cystic fibrosis. PLoS ONE 2014, 9, e98502. [Google Scholar] [CrossRef]
- Weaver, D.; Gago, S.; Bromley, M.; Bowyer, P. The human lung mycobiome in chronic respiratory disease: Limitations of methods and our current understanding. Curr. Fungal Infect. Rep. 2019, 13, 109–119. [Google Scholar] [CrossRef]
- Engel, T.G.; Slabbers, L.; De Jong, C.; Melchers, W.J.; Hagen, F.; Verweij, P.E.; Merkus, P.; Meis, J.F. Prevalence and diversity of filamentous fungi in the airways of cystic fibrosis patients–A Dutch, multicentre study. J. Cyst. Fibros. 2019, 18, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Ranjendran, R.; Ramage, G. Pathogenesis of Fungal Infections in Cystic Fibrosis. Curr. Fungal Infect. Rep. 2016, 10, 163–169. [Google Scholar] [CrossRef] [PubMed]
- King, J.; Brunel, S.F.; Warris, A. Aspergillus infections in cystic fibrosis. J. Infect. 2016, 72, S50–S55. [Google Scholar] [CrossRef] [PubMed]
- Lipuma, J.J. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 2010, 23, 299–323. [Google Scholar] [CrossRef]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Genet. 2012, 10, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.L.; Gibson, R.L.; McNamara, S.; Yim, D.; Emerson, J.; Rosenfeld, M.; Hiatt, P.; McCoy, K.; Castile, R.; Smith, A.L.; et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 2001, 183, 444–452. [Google Scholar] [CrossRef]
- Reece, E.; Segurado, R.; Jackson, A.D.; McClean, S.; Renwick, J.; Greally, P. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: An Irish registry analysis. BMC Pulm. Med. 2017, 17, 70. [Google Scholar] [CrossRef]
- Gonçalves-De-Albuquerque, C.F.; Silva, A.R.; Burth, P.; Rocco, P.R.M.; Castro-Faria, M.V.; Castro-Faria-Neto, H.C. Possible mechanisms of Pseudomonas aeruginosa-associated lung disease. Int. J. Med. Microbiol. 2016, 306, 20–28. [Google Scholar] [CrossRef]
- Diaz, M.H.; Shaver, C.M.; King, J.D.; Musunuri, S.; Kazzaz, J.A.; Hauser, A.R. Pseudomonas aeruginosa induces localized immunosuppression during pneumonia. Infect. Immun. 2008, 76, 4414–4421. [Google Scholar] [CrossRef]
- Manago, A.; Becker, K.A.; Carpinteiro, A.; Wilker, B.; Soddemann, M.; Seitz, A.P.; Edwards, M.J.; Grassmé, H.; Szabò, I.; Gulbins, E. Pseudomonas aeruginosa pyocyanin induces neutrophil death via mitochondrial reactive oxygen species and mitochondrial acid sphingomyelinase. Antioxid. Redox Sign. 2015, 22, 1097–1110. [Google Scholar] [CrossRef]
- Laarman, A.J.; Bardoel, B.W.; Ruyken, M.; Fernie, J.; Milder, F.J.; Van Strijp, J.A.; Rooijakkers, S.H.M. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J. Immunol. 2011, 188, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Hogardt, M.; Heesemann, J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr. Top Microbiol. Immunol. 2013, 358, 91–118. [Google Scholar] [PubMed]
- Mayer-Hamblett, N.; Rosenfeld, M.; Gibson, R.L.; Ramsey, B.W.; Kulasekara, H.D.; Retsch-Bogart, G.Z.; Morgan, W.; Wolter, D.J.; Pope, C.E.; Houston, L.S.; et al. Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomes. Am. J. Respir. Crit. Care Med. 2014, 190, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Cigana, C.; Lore’, N.I.; Riva, C.; De Fino, I.; Spagnuolo, L.; Sipione, B.; Rossi, G.; Nonis, A.; Cabrini, G.; Bragonzi, A. Tracking the immunopathological response to Pseudomonas aeruginosa during respiratory infections. Sci. Rep. 2016, 6, 21465. [Google Scholar] [CrossRef]
- Hogardt, M.; Heesemann, J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 2010, 300, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Winstanley, C.; O’Brien, S.; Brockhurst, M.A. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016, 24, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Vandeplassche, E.; Sass, A.M.; Lemarcq, A.; Dandekar, A.A.; Coenye, T.; Crabbé, A. In vitro evolution of Pseudomonas aeruginosa AA2 biofilms in the presence of cystic fibrosis lung microbiome members. Sci. Rep. 2019, 9, 12814–12859. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.Ø.; Fiandaca, M.J.; Pedersen, J.; Hansen, C.R.; Andersen, C.B.; Pressler, T.; Givskov, M.; Høiby, N. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 2009, 44, 547–558. [Google Scholar] [CrossRef]
- Olivares, E.; Badel-Berchoux, S.; Provot, C.; Prévost, G.; Bernardi, T.; Jehl, F. Clinical impact of antibiotics for the treatment of Pseudomonas aeruginosa biofilm infections. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- McDaniel, C.T.; Panmanee, W.; Hassett, D.J. An overview of infections in cystic fibrosis airways and the role of environmental conditions on Pseudomonas aeruginosa biofilm formation and viability. In Cystic Fibrosis in the Light of New Research; IntechOpen: London, UK, 2015. [Google Scholar]
- Li, Z.; Kosorok, M.R.; Farrell, P.M.; Laxova, A.; West, S.E.H.; Green, C.G.; Collins, J.; Rock, M.J.; Splaingard, M.L. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 2005, 293, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef] [PubMed]
- Young, R.L.; Malcolm, K.C.; Kret, J.E.; Caceres, S.M.; Poch, K.R.; Nichols, D.P.; Taylor-Cousar, J.L.; Saavedra, M.T.; Randell, S.H.; Vasil, M.L.; et al. Neutrophil extracellular trap (NET)-mediated killing of Pseudomonas aeruginosa: Evidence of acquired resistance within the CF airway, independent of CFTR. PLoS ONE 2011, 6, e23637. [Google Scholar] [CrossRef] [PubMed]
- Pier, G.B.; Coleman, F.; Grout, M.; Franklin, M.; Ohman, D.E. Role of alginate o acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 2001, 69, 1895–1901. [Google Scholar] [CrossRef]
- Malhotra, S.; Limoli, D.H.; English, A.E.; Parsek, M.R.; Wozniak, D.J. Mixed communities of mucoid and nonmucoid Pseudomonas aeruginosa exhibit enhanced resistance to host antimicrobials. mBio 2018, 9, e00275-18. [Google Scholar] [CrossRef]
- Goltermann, L.; Tolker-Nielsen, T. Importance of the exopolysaccharide matrix in antimicrobial tolerance of Pseudomonas aeruginosa aggregates. Antimicrob. Agents Chemother. 2017, 61, e02696-16. [Google Scholar] [CrossRef]
- Van De Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latgé, J.P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Genet. 2017, 15, 661–674. [Google Scholar] [CrossRef]
- Ballard, E.; Melchers, W.J.G.; Zoll, J.; Brown, A.J.P.; Verweij, P.E.; Warris, A. In-host microevolution of Aspergillus fumigatus: A phenotypic and genotypic analysis. Fungal Genet. Biol. 2018, 113, 1–13. [Google Scholar] [CrossRef]
- Warris, A.; Verweij, P.E. Clinical implications of environmental sources for Aspergillus. Med. Mycol. 2005, 43, 59–65. [Google Scholar] [CrossRef]
- Gago, S.; Denning, D.W.; Bowyer, P. Pathophysiological aspects of Aspergillus colonization in disease. Med. Mycol. 2019, 57, S219–S227. [Google Scholar] [CrossRef]
- Kosmidis, C.; Denning, D.W. Republished: The clinical spectrum of pulmonary aspergillosis. Postgrad. Med. J. 2015, 91, 403–410. [Google Scholar] [CrossRef]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2016, 10, 296–322. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases—Estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Escobar, N.; Ordonez, S.R.; Wösten, H.A.B.; Haas, P.J.A.; De Cock, H.; Haagsman, H.P. Hide, keep quiet, and keep low: Properties that make Aspergillus fumigatus a successful lung pathogen. Front. Microbiol. 2016, 7, 438. [Google Scholar] [CrossRef]
- Wiesner, D.L.; Klein, B.S. Lung epithelium: Barrier immunity to inhaled fungi and driver of fungal-associated allergic asthma. Curr. Opin. Microbiol. 2017, 40, 8–13. [Google Scholar] [CrossRef]
- Overton, N.; Gago, S.; Bowyer, P. Immunogenetics of chronic and allergic aspergillosis. In Immunogenetics of Fungal Diseases; Springer Science and Business Media LLC: Berlin, Germany, 2017; Volume 108, pp. 153–171. [Google Scholar]
- McCormick, A.; Heesemann, L.; Wagener, J.; Marcos, V.; Hartl, D.; Loeffler, J.; Heesemann, J.; Ebel, F. NETs formed by human neutrophils inhibit growth of the pathogenic mold Aspergillus fumigatus. Microbes Infect. 2010, 12, 928–936. [Google Scholar] [CrossRef]
- Moss, R.B. Treating allergic bronchopulmonary aspergillosis: The way forward. Eur. Respir. J. 2016, 47, 385–387. [Google Scholar] [CrossRef]
- Baxter, C.; Dunn, G.; Jones, A.M.; Webb, K.; Gore, R.; Richardson, M.D.; Denning, D.W. Novel immunologic classification of aspergillosis in adult cystic fibrosis. J. Allergy Clin. Immunol. 2013, 132, 560–566.e10. [Google Scholar] [CrossRef]
- Agarwal, R.; Chakrabarti, A.; Shah, A.; Gupta, D.; Meis, J.F.G.M.; Guleria, R.; Moss, R.; Denning, D.W. ABPA complicating asthma ISHAM working group Allergic bronchopulmonary aspergillosis: Review of literature and proposal of new diagnostic and classification criteria. Clin. Exp. Allergy 2013, 43, 850–873. [Google Scholar] [CrossRef]
- Denning, D.W.; Pashley, C.H.; Hartl, D.; Wardlaw, A.J.; Godet, C.; Del Giacco, S.; Delhaes, L.; Sergejeva, S. Fungal allergy in asthma–state of the art and research needs. Clin. Transl. Allergy 2014, 4, 14. [Google Scholar] [CrossRef]
- Brandt, C.; Roehmel, J.; Rickerts, V.; Melichar, V.; Niemann, N.; Schwarz, C. Aspergillus Bronchitis in Patients with Cystic Fibrosis. Mycopathologia 2017, 183, 61–69. [Google Scholar] [CrossRef]
- Chrdle, A.; Mustakim, S.; Bright-Thomas, R.J.; Baxter, C.G.; Felton, T.; Denning, D.W. Aspergillus bronchitis without significant immunocompromise. Ann. Acad. Sci. 2012, 1272, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cheng, W.; He, X.; Liu, Y. The co-colonization prevalence of Pseudomonas aeruginosa and Aspergillus fumigatus in cystic fibrosis: A systematic review and meta-analysis. Microb. Pathog. 2018, 125, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Paugam, A.; Baixench, M.T.; Demazes-Dufeu, N.; Burgel, P.R.; Sauter, E.; Kanaan, R.; Dusser, D.; Dupouy-Camet, J.; Hubert, D. Characteristics and consequences of airway colonization by filamentous fungi in 201 adult patients with cystic fibrosis in France. Med. Mycol. 2010, 48, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Bakare, N.; Rickerts, V.; Bargon, J.; Just-Nübling, G. Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses 2003, 46, 19–23. [Google Scholar] [CrossRef]
- Granchelli, A.M.; Adler, F.R.; Keogh, R.H.; Kartsonaki, C.; Cox, D.R.; Liou, T.G. Microbial interactions in the cystic fibrosis airway. J. Clin. Microbiol. 2018, 56, JCM.00354-18. [Google Scholar] [CrossRef]
- Hector, A.; Kirn, T.; Ralhan, A.; Graepler-Mainka, U.; Berenbrinker, S.; Riethmueller, J.; Hogardt, M.; Wagner, M.; Pfleger, A.; Autenrieth, I.; et al. Microbial colonization and lung function in adolescents with cystic fibrosis. J. Cyst. Fibros. 2016, 15, 340–349. [Google Scholar] [CrossRef]
- Amin, R.; Dupuis, A.; Aaron, S.D.; Ratjen, F. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 2010, 137, 171–176. [Google Scholar] [CrossRef]
- Briard, B.; Rasoldier, V.; Bomme, P.; ElAouad, N.; Guerreiro, C.; Chassagne, P.; Muszkieta, L.; Latge, J.P.; Mulard, L.; Beauvais, A. Dirhamnolipids secreted from Pseudomonas aeruginosa modify antifungal susceptibility of Aspergillus fumigatus by inhibiting beta1,3 glucan synthase activity. ISME J. 2017, 11, 1578–1591. [Google Scholar] [CrossRef]
- Mowat, E.; Rajendran, R.; Williams, C.; McCulloch, E.; Jones, B.; Lang, S.; Ramage, G. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 2010, 313, 96–102. [Google Scholar] [CrossRef]
- Briard, B.; Bomme, P.; Lechner, B.E.; Mislin, G.L.; Lair, V.; Prevost, M.C.; Latge, J.P.; Haas, H.; Beauvais, A. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep. 2015, 5, 8220. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, W. Interaction between Pseudomonas aeruginosa and Aspergillus fumigatus in cystic fibrosis. Peer J. 2018, 6, e5931. [Google Scholar] [CrossRef]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.C.; Dietl, A.M.; Visca, P.; Haas, H.; et al. Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J. Bacteriol. 2018, 200, e00345-17. [Google Scholar] [CrossRef] [PubMed]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.C.; Dietl, A.M.; Visca, P.; Haas, H.; et al. Aspergillus-Pseudomonas interaction, relevant to competition in airways. Med Mycol. 2019, 57, S228–S232. [Google Scholar] [CrossRef] [PubMed]
- Briard, B.; Mislin, G.L.A.; Latge, J.P.; Beauvais, A. Interactions between Aspergillus fumigatus and pulmonary bacteria: Current state of the field, new data, and future perspective. J. Fungi 2019, 5, 48. [Google Scholar] [CrossRef]
- Penner, J.C.; Ferreira, J.A.G.; Secor, P.R.; Sweere, J.M.; Birukova, M.K.; Joubert, L.-M.; Haagensen, J.A.J.; Garcia, O.; Malkovskiy, A.V.; Kaber, G.; et al. Pf4 bacteriophage produced by Pseudomonas aeruginosa inhibits Aspergillus fumigatus metabolism via iron sequestration. Microbiology 2016, 162, 1583–1594. [Google Scholar] [CrossRef]
- Reece, E.; Doyle, S.; Greally, P.; Renwick, J.; McClean, S. Aspergillus fumigatus inhibits pseudomonas aeruginosa in co-culture: Implications of a mutually antagonistic relationship on virulence and inflammation in the CF airway. Front. Microbiol. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Briard, B.; Heddergott, C.; Latge, J.P. Volatile compounds emitted by Pseudomonas aeruginosa stimulate growth of the fungal pathogen Aspergillus fumigatus. mBio 2016, 7, 00219. [Google Scholar] [CrossRef]
- Moree, W.J.; Phelan, V.V.; Wu, C.H.; Bandeira, N.A.G.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef]
- Sass, G.; Ansari, S.R.; Dietl, A.M.; Déziel, E.; Haas, H.; Stevens, D.A. Intermicrobial interaction: Aspergillus fumigatus siderophores protect against competition by Pseudomonas aeruginosa. PLoS ONE 2019, 14, e0216085. [Google Scholar] [CrossRef]
- Nazik, H.; Sass, G.; Ansari, S.R.; Ertekin, R.; Haas, H.; Déziel, E.; Stevens, D.A. Novel intermicrobial molecular interaction: Pseudomonas aeruginosa Quinolone Signal (PQS) modulates Aspergillus fumigatus response to iron. Microbiology 2020, 166, 44–55. [Google Scholar] [CrossRef]
- Margalit, A.; Carolan, J.C.; Shehan, D.; Kavanagh, K. The Aspergillus fumigatus secretome alters the proteome of Pseudomonas aeruginosa to stimulate bacterial growth: Implications for co-infection. Mol. Cell. Proteom. 2020, 1–45. [Google Scholar] [CrossRef]
- Anand, R.; Clemons, K.V.; Stevens, D.A. Effect of anaerobiasis or hypoxia on Pseudomonas aeruginosa inhibition of Aspergillus fumigatus biofilm. Arch. Microbiol. 2017, 199, 881–890. [Google Scholar] [CrossRef]
- Anand, R.; Moss, R.B.; Sass, G.; Banaei, N.; Clemons, K.V.; Martinez, M.; Stevens, D.A. Small colony variants of Pseudomonas aeruginosa display heterogeneity in inhibiting Aspergillus fumigatus biofilm. Mycopathologia 2017, 183, 263–272. [Google Scholar] [CrossRef]
- Ferreira, J.A.G.; Penner, J.C.; Moss, R.B.; Haagensen, J.A.J.; Clemons, K.V.; Spormann, A.M.; Nazik, H.; Cohen, K.; Banaei, N.; Carolino, E.; et al. Inhibition of Aspergillus fumigatus and its biofilm by Pseudomonas aeruginosa is dependent on the source, phenotype and growth conditions of the bacterium. PLoS ONE 2015, 10, e0134692. [Google Scholar] [CrossRef]
- Scott, J.; Sueiro-Olivares, M.; Ahmed, W.; Heddergott, C.; Zhao, C.; Thomas, R.; Bromley, M.; Latgé, J.-P.; Krappmann, S.; Fowler, S.; et al. Pseudomonas aeruginosa-derived volatile sulfur compounds promote distal Aspergillus fumigatus growth and a synergistic pathogen-pathogen interaction that increases pathogenicity in co-infection. Front. Microbiol. 2019, 10, 2311. [Google Scholar] [CrossRef]
- Margalit, A.; Kavanagh, K.; Carolan, J.C. Characterization of the proteomic response of A549 cells following sequential exposure to Aspergillus fumigatus and Pseudomonas aeruginosa. J. Proteome Res. 2019, 19, 279–291. [Google Scholar] [CrossRef]
- Smith, K.; Rajendran, R.; Kerr, S.; Lappin, D.F.; Mackay, W.; Williams, C.; Ramage, G. Aspergillus fumigatus enhances elastase production in Pseudomonas aeruginosa co-cultures. Med. Mycol. 2015, 53, 645–655. [Google Scholar] [CrossRef]
- Yonezawa, M.; Sugiyama, H.; Kizawa, K.; Hori, R.; Mitsuyama, J.; Araki, H.; Shimakura, M.; Minami, S.; Watanabe, Y.; Yamaguchi, K. A new model of pulmonary superinfection with Aspergillus fumigatus and Pseudomonas aeruginosa in mice. J. Infect. Chemother. 2000, 6, 155–161. [Google Scholar] [CrossRef]
- Mitsuyama, J.; Kizawa, K.; Minami, S.; Watanabe, Y.; Yamaguchi, K. Evaluation of antimicrobial agents using an experimental pulmonary superinfection model with Aspergillus fumigatus and Pseudomonas aeruginosa in leukopenic mice. J. Infect. Chemother. 2003, 9, 144–150. [Google Scholar] [CrossRef]
- Shirazi, F.; Ferreira, J.A.G.; Stevens, D.A.; Clemons, K.V.; Kontoyiannis, D.P. Biofilm filtrates of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients inhibit preformed Aspergillus fumigatus biofilms via apoptosis. PLoS ONE 2016, 11, e0150155. [Google Scholar] [CrossRef]
- Döring, G.; Flume, P.; Heijerman, H.; Elborn, J.S. Treatment of lung infection in patients with cystic fibrosis: Current and future strategies. J. Cyst. Fibros. 2012, 11, 461–479. [Google Scholar] [CrossRef]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef]
- Khabnadideh, S.; Rezaei, Z.; Ghasemi, Y.; Najafabady, N.M. Antibacterial Activity of Some New Azole Compounds. Anti-Infective Agents 2012, 10, 26–33. [Google Scholar] [CrossRef]
- Day, S.; Lalitha, P.; Haug, S.; Fothergill, A.W.; Cevallos, V.; Vijayakumar, R.; Prajna, N.V.; Acharya, N.R.; McLeod, S.D.; Lietman, T.M. Activity of antibiotics against Fusarium; and Aspergillus. Br. J. Ophthalmol. 2009, 93, 116. [Google Scholar] [CrossRef]
- Azevedo, M.M.; Teixeira-Santos, R.; Silva, A.P.; Cruz, L.; Ricardo, E.; Pina-Vaz, C.; Rodrigues, A.G. The effect of antibacterial and non-antibacterial compounds alone or associated with antifugals upon fungi. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Venturini, T.; Al-Hatmi, A.M.S.; Rossato, L.; Azevedo, M.I.; Keller, J.T.; Weiblen, C.; Santurio, J.M.; Alves, S.H. Do antibacterial and antifungal combinations have better activity against clinically relevant Fusarium species? in vitro synergism. Int. J. Antimicrob. Agents 2018, 51, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Eldesouky, H.E.; Li, X.; Abutaleb, N.S.; Mohammad, H.; Seleem, M.N. Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int. J. Antimicrob. Agents 2018, 52, 754–761. [Google Scholar] [CrossRef]
- Wambaugh, A.; Denham, M.; Ayala, S.T.; Brammer, M.; Stonhill, B.A.; Brown, M. Synergistic and antagonistic drug interactions in the treatment of systemic fungal infections. eLife 2020, 9. [Google Scholar] [CrossRef]
- Fiori, A.; Van Dijck, P. Potent synergistic effect of doxycycline with fluconazole against Candida albicans is mediated by interference with iron homeostasis. Antimicrob. Agents Chemother. 2012, 56, 3785. [Google Scholar] [CrossRef]
- Galdino, A.C.M.; Viganor, L.; De Castro, A.A.; Da Cunha, E.F.F.; Mello, T.P.; Mattos, L.M.; Pereira, M.D.; Hunt, M.C.; O’Shaughnessy, M.; Howe, O.; et al. Disarming Pseudomonas aeruginosa virulence by the inhibitory action of 1,10-phenanthroline-5,6-dione-based compounds: Elastase b (lasb) as a chemotherapeutic target. Front. Microbiol. 2019, 10, 1701. [Google Scholar] [CrossRef]
- Cathcart, G.R.A.; Quinn, D.; Greer, B.; Harriott, P.; Lynas, J.F.; Gilmore, B.F.; Walker, B. Novel inhibitors of the pseudomonas aeruginosa virulence factor LasB: A potential therapeutic approach for the attenuation of virulence mechanisms in pseudomonal infection. Antimicrob. Agents Chemother. 2011, 55, 2670–2678. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Sass, G.; Swietnicki, W.; Stevens, D.A. Review of potential pseudomonas weaponry, relevant to the Pseudomonas–Aspergillus interplay, for the mycology community. J. Fungi 2020, 6, 81. [Google Scholar] [CrossRef] [PubMed]
| CFTR Mutation Class | ||||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | |
| CFTR Defect | No functional CFTR protein | CFTR trafficking defect | Defective channel regulation | Decreased channel conductance | Reduced CFTR synthesis | Decreased CFTR stability |
| Mutation Type | Nonsense; frameshift; canonical splice | Missense; amino acid deletion | Missense; amino acid change | Missense; amino acid change | Splicing defect; missense | Missense; amino acid change |
| Examples of Causative Mutation | G542X | ΔF508 | G551D | R117H | 3849 + 10kbC→T | 4326delTC |
| W1282X | N1303K | D178R | R347P | 2789 + 5G→A | G1412X | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beswick, E.; Amich, J.; Gago, S. Factoring in the Complexity of the Cystic Fibrosis Lung to Understand Aspergillus fumigatus and Pseudomonas aeruginosa Interactions. Pathogens 2020, 9, 639. https://doi.org/10.3390/pathogens9080639
Beswick E, Amich J, Gago S. Factoring in the Complexity of the Cystic Fibrosis Lung to Understand Aspergillus fumigatus and Pseudomonas aeruginosa Interactions. Pathogens. 2020; 9(8):639. https://doi.org/10.3390/pathogens9080639
Chicago/Turabian StyleBeswick, Emily, Jorge Amich, and Sara Gago. 2020. "Factoring in the Complexity of the Cystic Fibrosis Lung to Understand Aspergillus fumigatus and Pseudomonas aeruginosa Interactions" Pathogens 9, no. 8: 639. https://doi.org/10.3390/pathogens9080639
APA StyleBeswick, E., Amich, J., & Gago, S. (2020). Factoring in the Complexity of the Cystic Fibrosis Lung to Understand Aspergillus fumigatus and Pseudomonas aeruginosa Interactions. Pathogens, 9(8), 639. https://doi.org/10.3390/pathogens9080639





