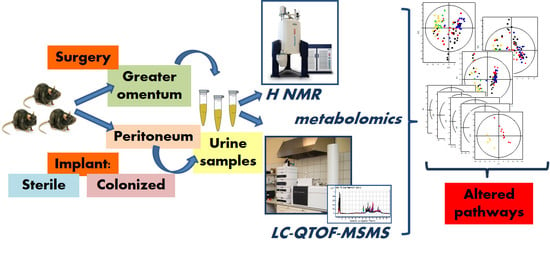
LC-QTOF-MS and 1H NMR Metabolomics Verifies Potential Use of Greater Omentum for Klebsiella pneumoniae Biofilm Eradication in Rats
Abstract
1. Introduction
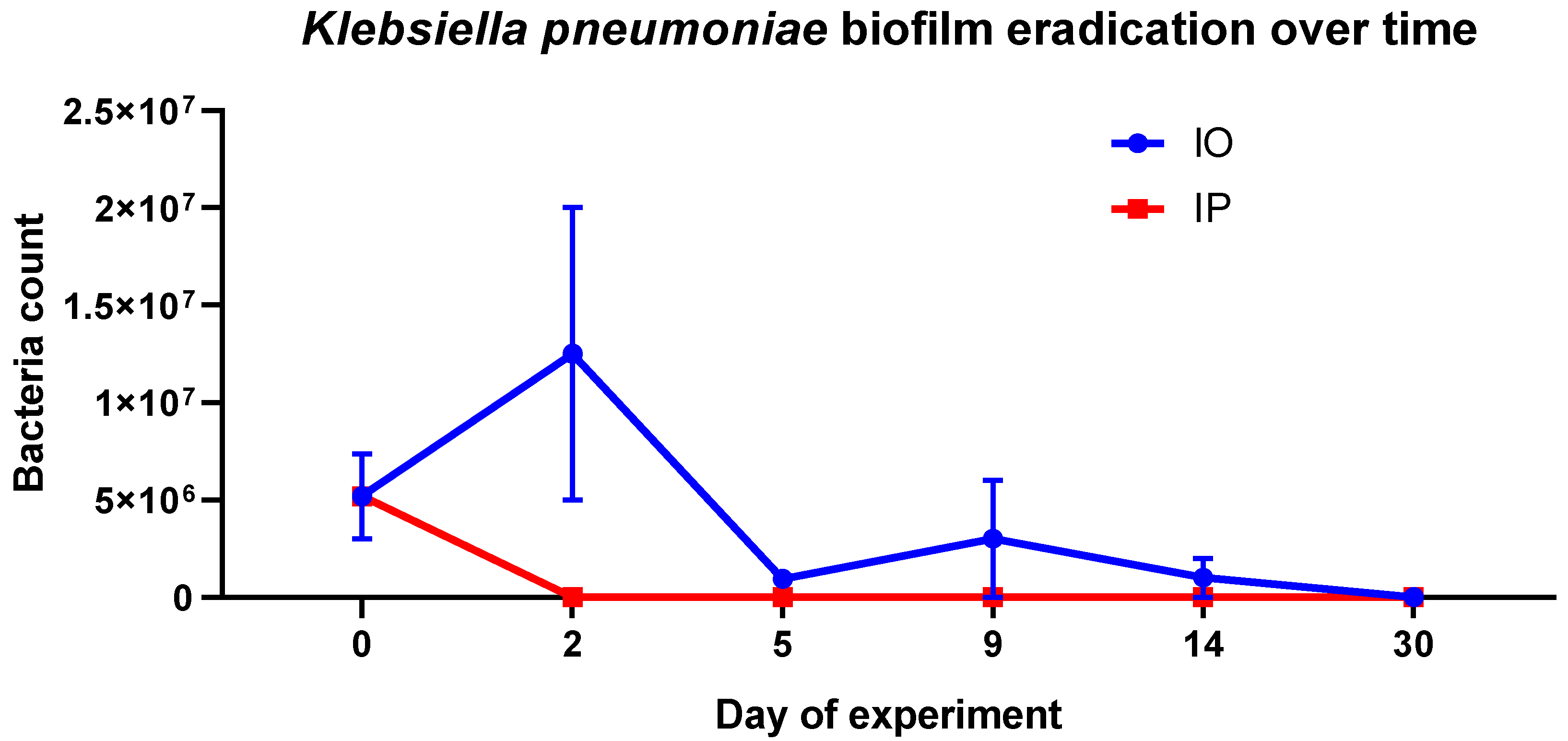
2. Results and Discussion
3. Materials and Methods
3.1. Design of the Experiment, Animal Handling and Sample Collection
3.2. LC–QTOF–MS Analysis and Data Treatment
3.2.1. Sample Preparation
3.2.2. LC–QTOF Analysis
3.2.3. LC–QTOF Data Treatment
3.3. 1H NMR Analysis and Signal Assignment
3.4. Statistical Analysis
3.5. Metabolites Identification
3.6. Quantitative Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Phu, V.D.; Wertheim, H.F.; Larsson, M.; Nadjm, B.; Dinh, Q.D.; Nilsson, L.E.; Rydell, U.; Le, T.T.; Trinh, S.H.; Pham, H.M.; et al. Burden of Hospital Acquired Infections and Antimicrobial Use in Vietnamese Adult Intensive Care Units. PLoS ONE 2016, 11, e0147544. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Sartelli, M.; McKimm, J.; Abu Bakar, M. Health care-associated infections—An overview. Infect. Drug Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Salmanov, A.; Litus, V.; Vdovychenko, S.; Litus, O.; Davtian, L.; Ubogov, S.; Bisyuk, Y.; Drozdova, A.; Vlasenko, I. Healthcare-associated infections in intensive care units. Wiad. Lek. 2019, 72, 963–969. [Google Scholar]
- Centers for Disease Control and Prevention. Healthcare-Associated Infections. HAI Data, 2018. Available online: https://www.cdc.gov/hai/data/portal/index.html (accessed on 16 September 2019).
- Suetens, C.; Latour, K.; Kärki, T.; Ricchizzi, E.; Kinross, P.; Moro, M.L.; Jans, B.; Hopkins, S.; Hansen, S.; Lyytikäinen, O.; et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: Results from two European point prevalence surveys, 2016 to 2017. Eurosurveillance 2018, 23, 1800516. [Google Scholar] [CrossRef]
- Cassini, A.; Plachouras, D.; Eckmanns, T.; Abu Sin, M.; Blank, H.P.; Ducomble, T.; Haller, S.; Harder, T.; Klingeberg, A.; Sixtensson, M.; et al. Burden of Six Healthcare-Associated Infections on European Population Health: Estimating Incidence-Based Disability-Adjusted Life Years through a Population Prevalence-Based Modelling Study. PLoS Med. 2016, 13, e1002150. [Google Scholar] [CrossRef]
- Veerachamy, S.; Yarlagadda, T.; Manivasagam, G.; Yarlagadda, P.K. Bacterial adherence and biofilm formation on medical implants: A review. Proc. Inst. Mech. Eng. H 2014, 228, 1083–1099. [Google Scholar] [CrossRef]
- Edmiston, C.E.; McBain, A.J.; Roberts, C.; Leaper, D. Clinical and microbiological aspects of biofilm-associated surgical site infections. Adv. Exp. Med. Biol. 2015, 830, 47–67. [Google Scholar]
- Chieng, L.O.; Hubbard, Z.; Salgado, C.J.; Levi, A.D.; Chim, H. Reconstruction of open wounds as a complication of spinal surgery with flaps: A systematic review. Neurosurg. Focus 2015, 39, E17. [Google Scholar] [CrossRef]
- Mohan, A.T.; Sur, Y.J.; Zhu, L.; Morsy, M.; Wu, P.S.; Moran, S.L.; Mardini, S.; Saint-Cyr, M. The Concepts of Propeller, Perforator, Keystone, and Other Local Flaps and Their Role in the Evolution of Reconstruction. Plast. Reconstr. Surg. 2016, 138, 710e–729e. [Google Scholar] [CrossRef]
- Mazzaferro, D.; Song, P.; Massand, S.; Mirmanesh, M.; Jaiswal, R.; Pu, L.L.Q. The Omental Free Flap-A Review of Usage and Physiology. J. Reconstr. Microsurg. 2018, 34, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Cannaday, J.E. Some uses of undetached omentum in surgery. Am. J. Surg. 1948, 76, 502–505. [Google Scholar] [CrossRef]
- Vineberg, A.M.; Kato, Y.; Pirozynski, W.J. Experimental revascularization of the entire heart. Evaluation of epicardiectomy, omental graft, and/or implantation of the internal mammary artery in preventing myocardial necrosis and death of the animal. Am. Heart J. 1966, 72, 79–93. [Google Scholar] [CrossRef]
- Vernik, J.; Singh, A.K. Omentum: Power to heal and regenerate. Int. J. Artif. Organs 2007, 30, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Lowery, E.; Braun, R.K.; Martin, A.; Huang, N.; Medina, M.; Sethupathi, P.; Seki, Y.; Takami, M.; Byrne, K.; et al. Cellular basis of tissue regeneration by omentum. PLoS ONE 2012, 7, e38368. [Google Scholar] [CrossRef] [PubMed]
- Van Wingerden, J.J.; Lapid, O.; Boonstra, P.W.; de Mol, B.A. Muscle flaps or omental flap in the management of deep sternal wound infection. Interact. Cardiovasc. Thorac. Surg. 2011, 13, 179–187. [Google Scholar] [CrossRef]
- Aquilina, D.; Darmanin, F.X.; Briffa, J.; Gatt, D. Chest wall reconstruction using an omental flap and Integra. J. Plast. Reconstr. Aesthet. Surg. 2009, 62, e200–e202. [Google Scholar] [CrossRef]
- Spindler, N.; Etz, C.D.; Misfeld, M.; Josten, C.; Mohr, F.W.; Langer, S. Omentum flap as a salvage procedure in deep sternal wound infection. Ther. Clin. Risk Manag. 2017, 13, 1077–1083. [Google Scholar] [CrossRef]
- Karasawa, J.; Touho, H.; Ohnishi, H.; Miyamoto, S.; Kikuchi, H. Cerebral revascularization using omental transplantation for childhood moyamoya disease. J. Neurosurg. 1993, 79, 192–196. [Google Scholar] [CrossRef]
- Agner, C.; Yeomans, D.; Dujovny, M. The neurochemical basis for the applications of the greater omentum in neurosurgery. Neurol. Res. 2001, 23, 7–15. [Google Scholar] [CrossRef]
- Goldsmith, H.S.; Chen, W.F.; Duckett, S.W. Brain vascularization by intact omentum. Arch. Surg. 1973, 106, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Vatansev, C.; Ustün, M.E.; Oğün, C.O.; Taştekin, G.; Karabacakoğlu, A.; Yilmaz, H. Omental transposition decreases ischemic brain damage examined in a new ischemia model. Eur. Surg. Res. 2003, 35, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Maloney, C.T.; Wages, D.; Upton, J.; Lee, W.P. Free omental tissue transfer for extremity coverage and revascularization. Plast. Reconstr. Surg. 2003, 111, 1899–1904. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, H.S.; de la Torre, J.C. Axonal regeneration after spinal cord transection and reconstruction. Brain Res. 1992, 589, 217–224. [Google Scholar] [CrossRef]
- De la Torre, J.C.; Goldsmith, H.S. Coerulospinal fiber regeneration in transected feline spinal cord. Brain Res. Bull. 1994, 35, 413–417. [Google Scholar] [CrossRef]
- Goldsmith, H.S. Brain and spinal cord revascularization by omental transposition. Neurol. Res. 1994, 16, 159–162. [Google Scholar] [CrossRef]
- Rogers, S.A.; Chen, F.; Talcott, M.; Hammerman, M.R. Islet cell engraftment and control of diabetes in rats after transplantation of pig pancreatic anlagen. Am. J. Physiol. Endocrinol. Metab. 2004, 286, E502–E509. [Google Scholar] [CrossRef]
- Lee, H.; Cusick, R.A.; Utsunomiya, H.; Ma, P.X.; Langer, R.; Vacanti, J.P. Effect of implantation site on hepatocytes heterotopically transplanted on biodegradable polymer scaffolds. Tissue Eng. 2003, 9, 1227–1232. [Google Scholar] [CrossRef]
- Sigrist, S.; Mechine-Neuville, A.; Mandes, K.; Calenda, V.; Legeay, G.; Bellocq, J.P.; Pinget, M.; Kessler, L. Induction of angiogenesis in omentum with vascular endothelial growth factor: Influence on the viability of encapsulated rat pancreatic islets during transplantation. J. Vasc. Res. 2003, 40, 359–367. [Google Scholar] [CrossRef]
- Hammerman, M.R. Renal organogenesis from transplanted metanephric primordia. J. Am. Soc. Nephrol. 2004, 15, 1126–1132. [Google Scholar] [CrossRef]
- Litbarg, N.O.; Gudehithlu, K.P.; Sethupathi, P.; Arruda, J.A.; Dunea, G.; Singh, A.K. Activated omentum becomes rich in factors that promote healing and tissue regeneration. Cell Tissue Res. 2007, 328, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dudley, H.A. The metabolic response to surgery. Postgrad. Med. J. 1957, 33, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Awad, S.; Constantin-Teodosiu, D.; Macdonald, I.A.; Lobo, D.N. Short-term starvation and mitochondrial dysfunction—A possible mechanism leading to postoperative insulin resistance. Clin. Nutr. 2009, 28, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Brandi, L.S.; Frediani, M.; Oleggini, M.; Mosca, F.; Cerri, M.; Boni, C.; Pecori, N.; Buzzigoli, G.; Ferrannini, E. Insulin resistance after surgery: Normalization by insulin treatment. Clin. Sci. (Lond) 1990, 79, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Thorell, A.; Efendic, S.; Gutniak, M.; Häggmark, T.; Ljungqvist, O. Insulin resistance after abdominal surgery. Br. J. Surg. 1994, 81, 59–63. [Google Scholar] [CrossRef]
- Tewari, N.; Awad, S.; Duška, F.; Williams, J.P.; Bennett, A.; Macdonald, I.A.; Lobo, D.N. Postoperative inflammation and insulin resistance in relation to body composition, adiposity and carbohydrate treatment: A randomised controlled study. Clin. Nutr. 2019, 38, 204–212. [Google Scholar] [CrossRef]
- Thorell, A.; Efendic, S.; Gutniak, M.; Häggmark, T.; Ljungqvist, O. Development of postoperative insulin resistance is associated with the magnitude of operation. Eur. J. Surg. 1993, 159, 593–599. [Google Scholar]
- Lehrke, M.; Broedl, U.C.; Biller-Friedmann, I.M.; Vogeser, M.; Henschel, V.; Nassau, K.; Göke, B.; Kilger, E.; Parhofer, K.G. Serum concentrations of cortisol, interleukin 6, leptin and adiponectin predict stress induced insulin resistance in acute inflammatory reactions. Crit. Care 2008, 12, R157. [Google Scholar] [CrossRef]
- Steenburg, R.W.; Ganong, W.F. Observations on the influence of extra-adrenal factors on circulating 17-hydroxycorticoids in the surgically stressed adrenalectomized animal. Surgery 1955, 38, 92–104. [Google Scholar]
- Friedrich, M.; Rixecker, D.; Friedrich, G. Evaluation of stress-related hormones after surgery. Clin. Exp. Obstet. Gynecol. 1999, 26, 71–75. [Google Scholar] [PubMed]
- Şimşek, T.; Şimşek, H.U.; Cantürk, N.Z. Response to trauma and metabolic changes: Posttraumatic metabolism. Ulus Cerrahi Derg 2014, 30, 153–159. [Google Scholar] [PubMed]
- Conlay, L.A.; Wurtman, R.J.; Blusztajn, K.; Coviella, I.L.; Maher, T.J.; Evoniuk, G.E. Decreased plasma choline concentrations in marathon runners. N. Engl. J. Med. 1986, 315, 892. [Google Scholar]
- Conlay, L.A.; Sabounjian, L.A.; Wurtman, R.J. Exercise and neuromodulators: Choline and acetylcholine in marathon runners. Int. J. Sports Med. 1992, 13 (Suppl. 1), S141–S142. [Google Scholar] [CrossRef]
- Buchman, A.L.; Jenden, D.; Roch, M. Plasma free, phospholipid-bound and urinary free choline all decrease during a marathon run and may be associated with impaired performance. J. Am. Coll. Nutr. 1999, 18, 598–601. [Google Scholar] [CrossRef]
- Ulus, I.H.; Ozyurt, G.; Korfali, E. Decreased serum choline concentrations in humans after surgery, childbirth, and traumatic head injury. Neurochem. Res. 1998, 23, 727–732. [Google Scholar] [CrossRef]
- Ilçöl, Y.O.; Ozyurt, G.; Kilicturgay, S.; Uncu, G.; Ulis, I.H. The decline in serum choline concentration in humans during and after surgery is associated with the elevation of cortisol, adrenocorticotropic hormone, prolactin and beta-endorphin concentrations. Neurosci. Lett. 2002, 324, 41–44. [Google Scholar] [CrossRef]
- Garcia, M.; Mamedova, L.K.; Barton, B.; Bradford, B.J. Choline Regulates the Function of Bovine Immune Cells and Alters the mRNA Abundance of Enzymes and Receptors Involved in Its Metabolism. Front. Immunol. 2018, 9, 2448. [Google Scholar] [CrossRef]
- Dong, F.; Wang, B.; Zhang, L.; Tang, H.; Li, J.; Wang, Y. Metabolic response to Klebsiella pneumoniae infection in an experimental rat model. PLoS ONE 2012, 7, e51060. [Google Scholar] [CrossRef][Green Version]
- Bartoszewicz, M.; Mączyńska, B.; Smutnicka, D.; Junka, A.; Domanasiewicz, A.; Barc, P.; Secewicz, A.; Rzeszutko, M.; Nowicka, J.; Janczura, A.; et al. Ability of greater omentum (omentum majus) to eradicate Klebsiella biofilm from surgical biomaterials implanted to Wistar female rats—Initial evaluation. In Proceedings of the Special Issue CMI: Abstracts of the 22nd European Congress of Clinical Microbiology and Infectious Diseases, London, UK, 31 March–3 April 2012; Volume R2414, pp. 718–719. [Google Scholar]
- Mączyńska, B.; Smutnicka, D.; Przondo-Mordarska, A.; Bartoszewicz, M.; Junka, A.; Janczura, A.; Nowicka, J. Biofilm Formation by Clinical Klebsiella Strains Expressing Various Types of Adhesins on CathetersMade of Different Materials. Adv. Clin. Exp. Med. 2010, 19, 443–453. [Google Scholar]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Lizewska, B.; Teul, J.; Kuc, P.; Lemancewicz, A.; Charkiewicz, K.; Goscik, J.; Kacerovsky, M.; Menon, R.; Miltyk, W.; Laudanski, P. Maternal Plasma Metabolomic Profiles in Spontaneous Preterm Birth: Preliminary Results. Mediat. Inflamm. 2018, 2018, 9362820. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. J. Magn. Reson. 2010, 202, 190–202. [Google Scholar] [CrossRef]
- Dawiskiba, T.; Deja, S.; Mulak, A.; Ząbek, A.; Jawień, E.; Pawełka, D.; Banasik, M.; Mastalerz-Migas, A.; Balcerzak, W.; Kaliszewski, K.; et al. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J. Gastroenterol. 2014, 20, 163–174. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS (R) models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
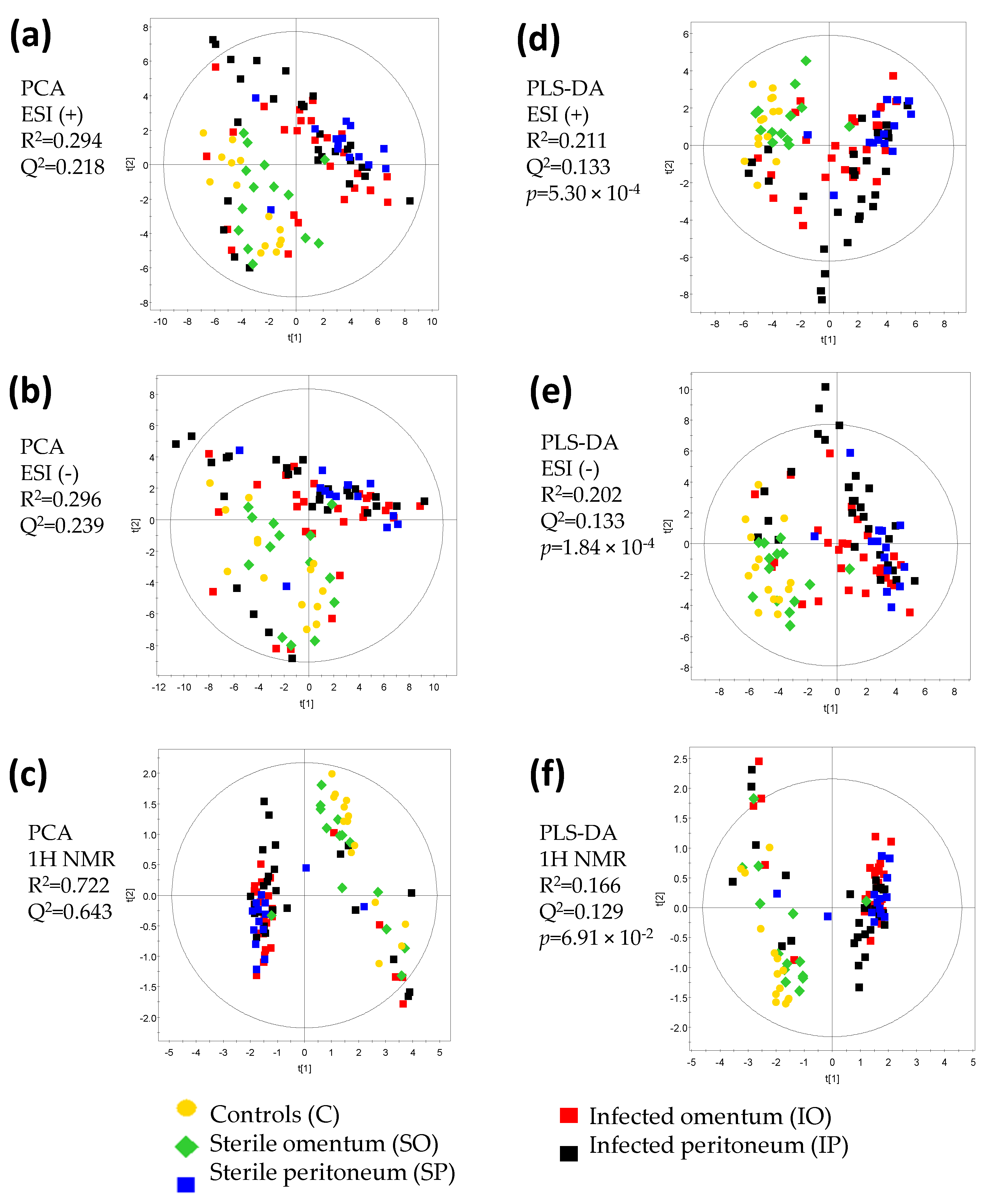
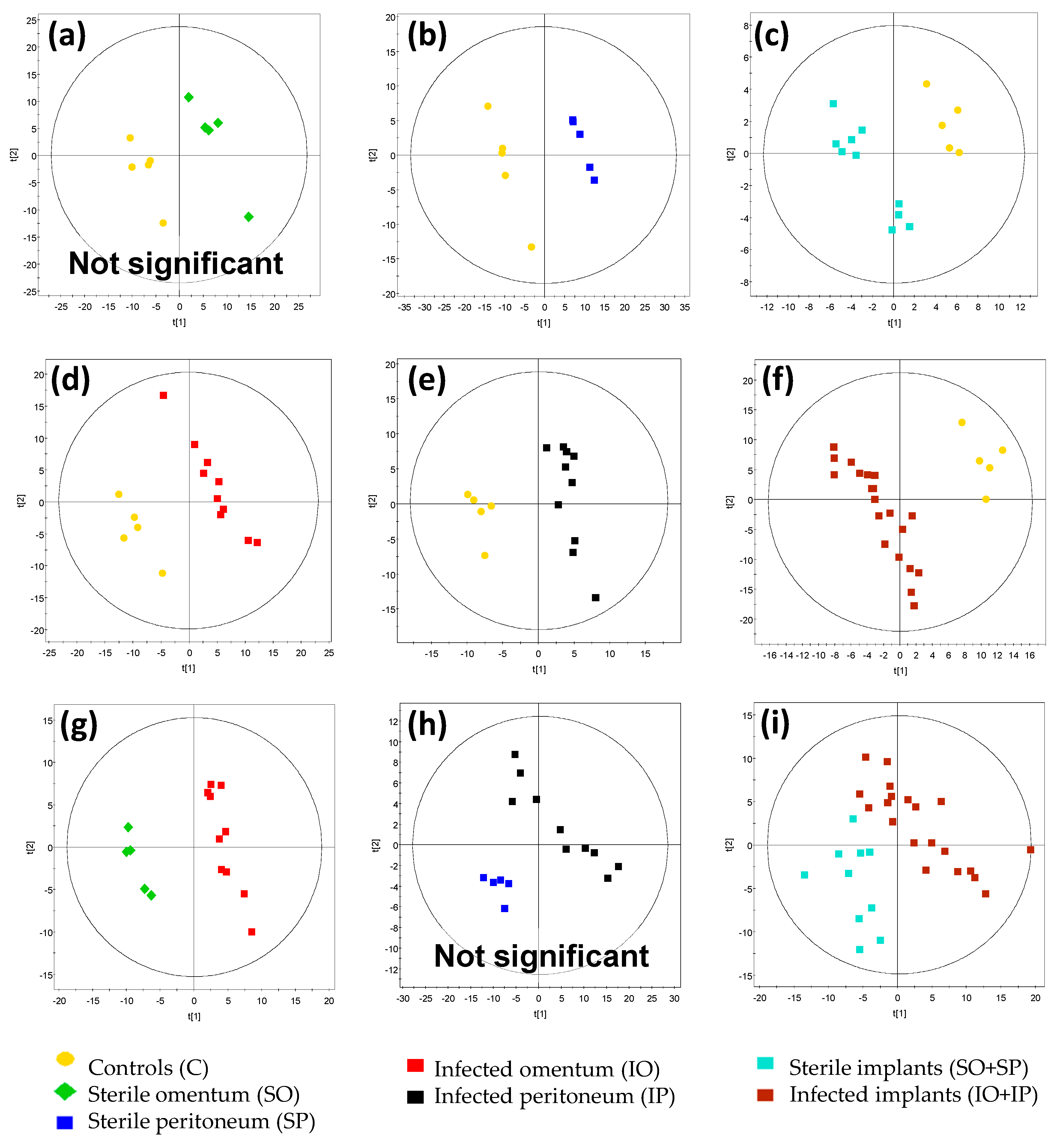

| Figure | Comparison | R2 Value | Q2 Value | CV ANOVA p-Value | Model Significance |
|---|---|---|---|---|---|
| Figure 4a | SO vs. C | 0.976 | 0.804 | 2.40 × 10−1 | No |
| Figure 4b | SP vs. C | 0.999 | 0.93 | 1.20 × 10−2 | Yes |
| Figure 4c | SO + SP vs. C | 0.955 | 0.739 | 1.17 × 10−2 | Yes |
| Figure 4d | IO vs. C | 0.984 | 0.903 | 9.04 × 10−4 | Yes |
| Figure 4e | IP vs. C | 0.974 | 0.792 | 1.69 × 10−3 | Yes |
| Figure 4f | IO + IP vs. C | 0.963 | 0.913 | 6.44 × 10−9 | Yes |
| Figure 4g | IO vs. SO | 0.975 | 0.773 | 1.84 × 10−2 | Yes |
| Figure 4h | IP vs. SP | 0.954 | 0.354 | 3.57 × 10−1 | No |
| Figure 4i | IO + IP vs. SO + SP | 0.790 | 0.382 | 3.50 × 10−2 | Yes |
| Day | d0 | d2 | d5 | d0 | d2 | d5 | d0 | d2 | d5 | d0 | d2 | d5 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Groups | IO_SO | IO_SO | IO_SO | IP_SP | IP_SP | IP_SP | IO_IP | IO_IP | IO_IP | SO_SP | SO_SP | SO_SP | ||||||||||||
| TCA CYCLE, CoA Biosynthesis | ||||||||||||||||||||||||
| N-acetylaspartate (p, a) | 5% | - | 33% | - | −33% | - | 12% | - | 7% | - | 18% | - | −22% | - | −12% | - | −14% | - | −17% | - | −30% | - | 52% | - |
| L−Lactic acid (m) | −88% | ** | −83% | - | 138% | - | 89% | * | 30% | - | 98% | - | −12% | - | 302% | - | 16% | - | 1259% | ** | 1245% | ** | 209% | - |
| Formic acid (m) | −38% | - | −83% | ** | −79% | - | −63% | ** | −22% | - | 166% | - | 13% | - | −5% | - | 35% | - | −55% | - | 36% | - | 325% | * |
| Acetic acid (m) | −82% | ** | −93% | - | −62% | - | 18% | * | −7% | - | 271% | - | 11% | - | 208% | - | 40% | - | 575% | ** | 424% | * | 569% | * |
| Pyruvic acid (m) | −15% | * | −38% | - | −14% | - | 8% | - | 2% | - | 42% | - | 15% | - | 26% | * | 58% | - | 62% | ** | 66% | * | 70% | * |
| Citric acid (m) | 14% | - | 474% | - | 313% | - | 1% | - | 1% | - | −36% | - | 20% | - | 18% | - | 4% | - | 36% | * | 30% | - | −77% | * |
| alpha-ketoglutaric acid (m) | 60% | - | 68% | - | 53% | - | 8% | - | 19% | - | −2% | - | 45% | - | 52% | - | 84% | * | 76% | ** | 60% | - | 19% | - |
| cis-Aconitic acid (m) | 7% | - | 6% | - | 159% | - | −23% | - | 4% | - | −7% | - | 26% | - | 6% | - | 30% | - | −19% | - | −3% | - | 12% | - |
| trans-Aconitic acid (m) | 18% | * | 33% | - | −27% | - | 8% | - | −3% | - | 7% | - | 0% | - | 3% | - | −7% | - | −15% | * | −10% | - | −3% | - |
| Succinic acid (m) | −38% | - | −66% | - | −16% | - | −24% | - | −9% | - | 27% | - | 52% | * | 50% | * | 136% | * | 65% | - | 61% | - | 263% | * |
| Succinic acid semialdehyde (n, a) | −20% | - | −10% | - | 178% | * | 69% | - | 45% | - | 10% | - | 21% | - | 43% | - | 81% | - | 155% | * | 128% | - | −29% | - |
| Fumaric acid (m) | 159% | - | 331% | - | 387% | - | −14% | - | −6% | - | −27% | - | 40% | - | 57% | - | −23% | - | −39% | - | −22% | - | −69% | * |
| Malic acid (n, a) | 42% | - | 64% | * | 25% | - | −2% | - | 2% | - | −29% | - | 15% | - | 58% | * | 53% | - | −21% | - | −2% | - | −12% | - |
| AMINO ACIDS | ||||||||||||||||||||||||
| Pyroglutamic acid (n, b) | −11% | - | 38% | - | 21% | - | 4% | - | −10% | - | −13% | - | −26% | - | 14% | - | 51% | - | −14% | - | −25% | - | 9% | - |
| L-Alanine (m) | −49% | ** | −89% | - | −63% | ** | −9% | - | 9% | - | 138% | - | 8% | - | 40% | - | 0% | - | 59% | * | 152% | * | 514% | * |
| TRYPTOPHAN METABOLISM | ||||||||||||||||||||||||
| Tryptophyl-Glutamate (n, b) | 14% | - | −12% | - | −22% | - | −3% | - | 29% | - | −4% | - | 46% | - | 31% | - | 42% | - | 25% | - | 92% | * | 75% | - |
| Indoxyl sulfate (n, a) | −18% | - | −17% | - | 5% | - | −30% | - | 68% | - | 1% | - | 18% | - | −16% | - | 37% | - | 1% | - | 71% | - | 33% | - |
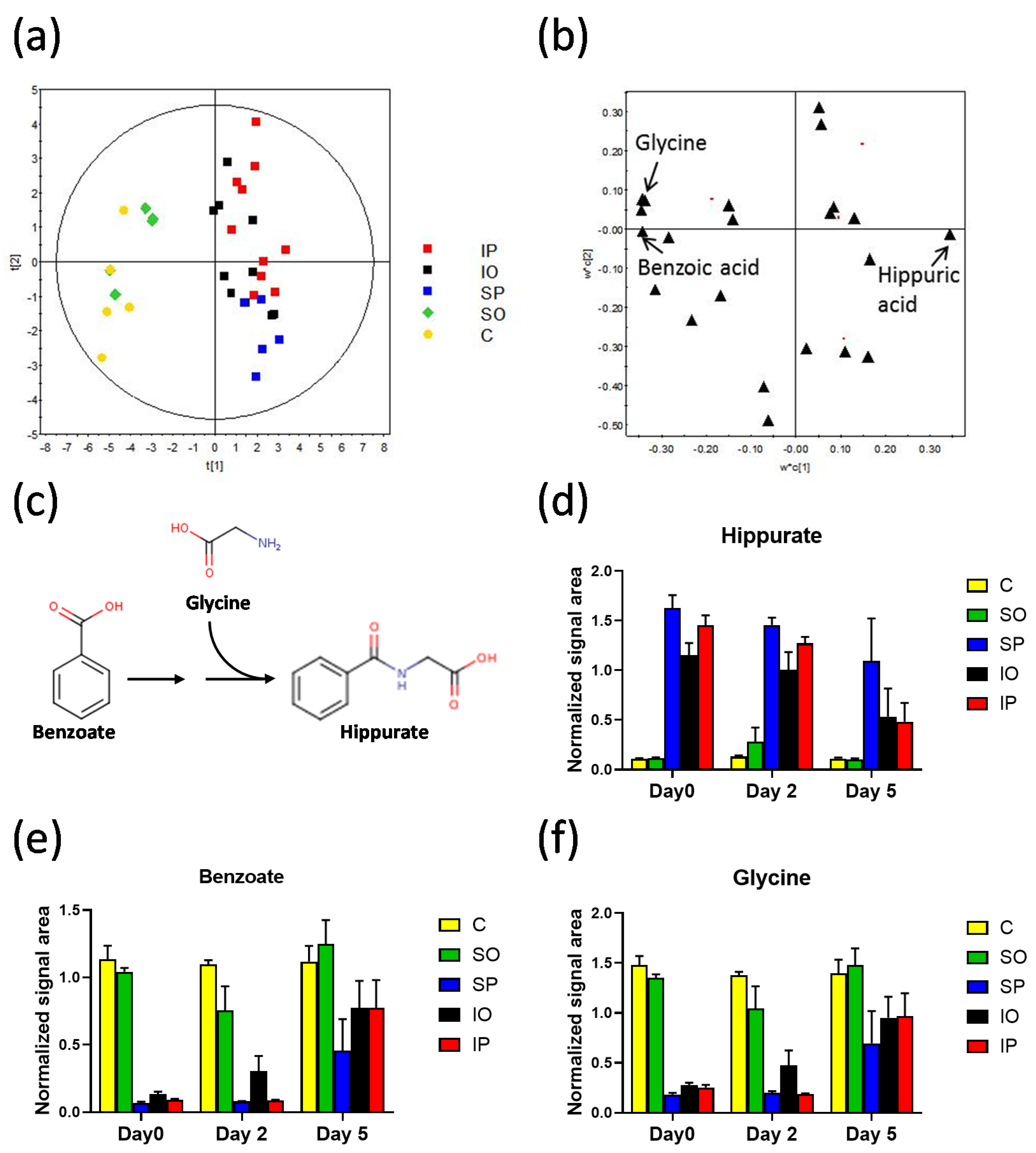
| HIPPURATE BIOSYNTHESIS | ||||||||||||||||||||||||
| Benzoic acid (m) | −83% | ** | −76% | - | −21% | - | 33% | - | 8% | - | 70% | - | 44% | - | 258% | * | 0% | - | 1419% | ** | 857% | ** | 173% | * |
| Glycine (m) | −74% | ** | −68% | - | 38% | - | 39% | * | −7% | - | 39% | - | 8% | - | 155% | - | −2% | - | 639% | ** | 426% | * | 113% | - |
| Hippuric acid (m) | 310% | ** | 877% | - | 442% | - | −11% | - | −13% | - | −56% | - | −21% | - | −21% | - | 10% | - | −93% | ** | −81% | ** | −91% | * |
| CHOLINE, LIPID METABOLISM | ||||||||||||||||||||||||
| Choline (p, a) | −8% | - | −67% | * | −7% | - | 54% | - | 4% | - | 228% | - | 45% | - | −24% | * | 22% | - | 145% | * | 139% | * | 333% | - |
| Betaine (m) | 7% | - | −34% | - | −19% | - | −18% | - | −8% | - | −5% | - | 15% | - | −4% | - | 46% | - | 21% | - | 24% | * | 5% | - |
| Creatinine (p, a) | 48% | - | 5% | - | 82% | - | 4% | - | −20% | - | 8% | - | −3% | - | 18% | - | −16% | - | −32% | - | −11% | - | −50% | * |
| Phosphorylcholine (p, b) | 52% | - | 36% | - | 243% | - | −32% | - | −27% | - | −49% | - | 2% | - | 14% | - | 27% | - | −55% | - | −39% | - | −81% | - |
| Tetrahydrofolic acid (n, b) | 1% | - | 1% | - | 66% | - | 139% | * | −4% | - | 10% | - | −34% | - | −9% | - | 48% | - | 56% | - | −14% | - | −2% | - |
| ACYL GLYCINES | ||||||||||||||||||||||||
| Isonicotinylglycine (p, a) | 126% | ** | 75% | - | na | - | −41% | * | −16% | - | −54% | - | 33% | - | 8% | - | −51% | - | −65% | * | −48% | - | na | - |
| Phenylacetylglycine (p, a) | 55% | - | 38% | - | 1041% | - | −35% | - | 40% | - | −41% | - | 17% | - | −23% | - | −1% | - | −51% | - | −22% | - | −95% | * |
| 3-Hydroxyhippuric acid (n, a) | 78% | - | 129% | * | 683% | - | 12% | - | −1% | - | 65% | - | −2% | - | 4% | * | 21% | - | −39% | - | −55% | - | −74% | - |
| HORMONES | ||||||||||||||||||||||||
| Tetrahydrocortisone (p, b) | 16% | - | 18% | - | −8% | - | 33% | - | 44% | * | −7% | - | −18% | - | −13% | - | −4% | - | −6% | - | 6% | - | −2% | - |
| Dihydrocortisol (p, b) | 56% | - | 34% | - | −47% | - | 5% | - | 20% | - | −46% | - | 9% | - | −18% | - | −6% | - | −27% | - | −27% | - | −4% | - |
| DIETARY METABOLITES | ||||||||||||||||||||||||
| Taurine (p, a) | 12% | - | −23% | - | 80% | - | −30% | - | 26% | - | −12% | - | −1% | - | −24% | - | −22% | - | −38% | - | 24% | - | −62% | * |
| Indolylacryloylglycine (n, a) | -4% | - | 7% | - | 103% | - | −18% | - | −17% | - | −10% | - | 46% | - | 41% | - | 36% | - | 25% | - | 9% | - | −40% | - |
| kamlolenic acid (p, b) | 143% | - | −8% | - | 25% | - | 35% | - | 34% | - | 160% | - | 19% | - | 7% | - | −4% | - | −34% | - | 56% | - | 99% | - |
| Xylitol (+Ribitol) (n, a) | 16% | - | −1% | - | 73% | - | 15% | - | 35% | - | 5% | - | −1% | - | −19% | - | 11% | - | −2% | - | 11% | - | −32% | - |
| Stachyose (n, a) | 13% | - | −40% | * | −61% | - | 648% | - | 17% | - | −1% | - | −58% | - | −27% | * | −32% | - | 176% | * | 42% | - | 73% | - |
| POLYPHENOLS INTAKE | ||||||||||||||||||||||||
| Phenol sulphate (n, a) | 10% | - | −8% | - | 60% | - | −46% | * | 6% | - | −52% | * | 34% | - | 0% | - | 11% | - | −34% | - | 16% | - | −66% | * |
| Tyrosol 4-sulfate (n, b) | 5% | - | −15% | - | −12% | - | −10% | - | −32% | - | −40% | - | −17% | - | 12% | - | 73% | - | −28% | - | −11% | - | 17% | - |
| Pyrocatechol sulphate (n, a) | −5% | - | 37% | * | −14% | - | −36% | ** | −8% | - | −14% | - | 2% | - | 0% | * | −11% | - | −31% | * | −33% | * | −11% | - |
| URIC ACID | ||||||||||||||||||||||||
| Allantoin (n, a) | 29% | - | 5% | - | 58% | - | 45% | * | 12% | - | −20% | - | 5% | - | 1% | - | 14% | - | 19% | - | 8% | - | −42% | - |
| CAFFEIC ACID FERRULIC ACID | ||||||||||||||||||||||||
| Caffeic acid 3-sulfate / Caffeic acid 4-sulfate (p, a) | −13% | - | −1% | - | −15% | - | −24% | * | −15% | - | 36% | - | 10% | - | 30% | - | 5% | - | −4% | - | 11% | - | 69% | - |
| Dihydrocaffeic acid 3-sulfate (n, a) | 56% | - | 51% | - | −4% | - | −35% | * | −24% | - | −44% | * | 15% | - | 4% | - | 22% | - | −52% | * | −48% | * | −28% | - |
| Ferulic acid (n, a) | 71% | * | 19% | - | −4% | - | −40% | ** | −4% | - | −34% | - | 0% | - | 0% | - | 35% | - | −65% | * | −20% | - | −7% | - |
| Ferulic acid 4-O-sulfate (n, b) | 39% | - | 31% | - | 0% | - | −44% | * | −24% | - | −35% | - | −10% | - | 28% | - | 29% | - | −64% | * | −26% | - | −17% | - |
| Dihydroferulic acid (n, a) | −54% | - | 60% | - | 56% | - | 56% | - | 6% | - | −27% | - | 9% | - | 480% | - | 134% | - | 270% | - | 284% | * | 10% | - |
| Ferrulic acid 4-O-glucuronide (n, a) | 175% | ** | 31% | - | 159% | - | −18% | - | 10% | - | −75% | - | 17% | - | 4% | - | 41% | - | −65% | - | −13% | - | −86% | - |
| Dihydrocaffeic acid 3-O-glucuronide (n, b) | 88% | - | 34% | - | 8274% | - | −27% | - | −26% | * | −31% | - | 21% | - | −27% | - | 0% | - | −52% | * | −60% | * | −99% | - |
| Dihydroferulic acid 4-O-glucuronide (n, b) | 96% | - | 60% | - | 642% | - | −27% | * | −11% | - | −41% | - | 21% | - | −14% | - | 90% | - | −55% | * | −52% | * | −85% | - |
| Dihydroferuloylglycine (p, b) | 229% | - | −5% | - | 144% | - | −48% | - | 5% | - | −29% | - | 57% | - | 29% | - | 130% | - | −75% | * | 43% | - | −33% | - |
| OTHER METABOLITES | ||||||||||||||||||||||||
| Urea (m) | 15% | - | −14% | - | 3065% | - | −25% | * | 7% | - | −28% | - | 10% | - | −7% | - | 53% | - | −32% | ** | −12% | - | 15% | - |
| Histamine (p, a) | 36% | - | 27% | - | 52% | - | −30% | - | 24% | - | −39% | * | 16% | - | 9% | - | 13% | - | −40% | - | 7% | - | −55% | - |
| 17,21-Dihydroxypregnenolone (p, b) | 78% | - | 91% | - | −3% | - | −9% | - | 121% | - | 11% | - | −7% | - | −24% | - | −25% | - | −53% | - | −13% | - | −14% | - |
| Isohomovanillic acid (n, a) | 11% | - | 30% | - | −18% | - | −14% | - | 18% | - | −20% | - | 15% | - | 79% | - | 31% | - | −11% | - | 63% | - | 28% | - |
| 4-Hydroxybenzaldehyde (n, a) | −30% | - | 0% | - | −20% | - | −21% | - | 21% | - | −16% | - | 44% | * | 15% | - | −11% | - | 62% | - | 39% | - | −7% | - |
| 3-Hydroxydodecanedioic acid (n, b) | −6% | - | −27% | - | −35% | - | −3% | - | 1% | - | −2% | - | 47% | - | 25% | - | 24% | - | 51% | * | 73% | - | 86% | * |
| 3-Hydroxyisoheptanoic acid / Ethyl 2-hydroxyisovalerate (n, b) | 15% | - | −13% | - | −14% | - | 66% | - | 51% | - | 61% | - | −12% | - | 24% | - | 2% | - | 27% | - | 116% | * | 90% | - |
| O-methoxycatechol-O-sulphate (n, b) | 10% | - | 7% | - | −17% | - | −47% | ** | 10% | - | −50% | - | 18% | - | −13% | - | −5% | - | −43% | * | −11% | - | −42% | - |
| Trigonelline (m) | 30% | * | −9% | - | 8% | - | −16% | - | −22% | - | −11% | - | 0% | - | 8% | - | 10% | - | −34% | ** | −29% | ** | 0% | - |
| Oxolan-3-one (n, a) | 35% | * | 20% | * | 3% | - | −10% | - | 9% | - | −6% | - | 11% | - | 8% | * | 10% | - | −26% | * | −2% | - | 1% | - |
| N-Acetyl-7-O-acetylneuraminic acid (n, a) | 100% | - | −49% | - | 7% | - | −8% | - | −1% | - | 46% | - | 136% | - | −21% | - | 33% | - | 9% | - | 53% | - | 81% | - |
| Ethanol (m) | −2% | - | −32% | - | −9% | - | 19% | - | 3% | - | 65% | - | −10% | - | 2% | - | 38% | - | 3% | - | 1% | - | 42% | * |
| 2-Octenedioic acid / cis-4-Octenedioic acid / trans-3-Octenedioic acid (n, b) | −7% | - | 40% | - | 74% | - | 152% | * | 19% | - | 24% | - | −17% | - | 2% | - | 44% | - | 126% | * | −14% | - | 2% | - |
| beta-D-Mannosylphosphodecaprenol (n, b) | −68% | ** | −45% | * | −5% | - | 141% | * | 37% | - | 101% | * | −41% | - | 22% | * | −11% | - | 345% | * | 202% | * | 89% | - |
| Number of Urine Samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Implant | Place of Implant Grafting | Name of the Group | Initial Number of Rats | Day 0 | Day 2 | Day 5 | Day 9 | Day 14 | Day 30 |
| Sterile | Peritoneum | SP | 5 | 5 | 5 | 5 | 3 | 2 | 1 |
| Sterile | Omentum | SO | 5 | 5 | 5 | 4 | 2 | 1 | 1 |
| Colonized | Peritoneum | IP | 10 | 10 | 10 | 8 | 6 | 4 | 2 |
| Colonized | Omentum | IO | 10 | 10 | 10 | 7 | 6 | 4 | 2 |
| - | No surgery | C | 5 | 5 | 5 | 4 | 3 | 2 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teul, J.; Deja, S.; Celińska-Janowicz, K.; Ząbek, A.; Młynarz, P.; Barć, P.; Junka, A.; Smutnicka, D.; Bartoszewicz, M.; Pałka, J.; et al. LC-QTOF-MS and 1H NMR Metabolomics Verifies Potential Use of Greater Omentum for Klebsiella pneumoniae Biofilm Eradication in Rats. Pathogens 2020, 9, 399. https://doi.org/10.3390/pathogens9050399
Teul J, Deja S, Celińska-Janowicz K, Ząbek A, Młynarz P, Barć P, Junka A, Smutnicka D, Bartoszewicz M, Pałka J, et al. LC-QTOF-MS and 1H NMR Metabolomics Verifies Potential Use of Greater Omentum for Klebsiella pneumoniae Biofilm Eradication in Rats. Pathogens. 2020; 9(5):399. https://doi.org/10.3390/pathogens9050399
Chicago/Turabian StyleTeul, Joanna, Stanisław Deja, Katarzyna Celińska-Janowicz, Adam Ząbek, Piotr Młynarz, Piotr Barć, Adam Junka, Danuta Smutnicka, Marzenna Bartoszewicz, Jerzy Pałka, and et al. 2020. "LC-QTOF-MS and 1H NMR Metabolomics Verifies Potential Use of Greater Omentum for Klebsiella pneumoniae Biofilm Eradication in Rats" Pathogens 9, no. 5: 399. https://doi.org/10.3390/pathogens9050399
APA StyleTeul, J., Deja, S., Celińska-Janowicz, K., Ząbek, A., Młynarz, P., Barć, P., Junka, A., Smutnicka, D., Bartoszewicz, M., Pałka, J., & Miltyk, W. (2020). LC-QTOF-MS and 1H NMR Metabolomics Verifies Potential Use of Greater Omentum for Klebsiella pneumoniae Biofilm Eradication in Rats. Pathogens, 9(5), 399. https://doi.org/10.3390/pathogens9050399