Transcriptional Alteration of Gene Biomarkers in Hemocytes of Wild Ostrea edulis with Molecular Evidence of Infections with Bonamia spp. and/or Marteilia refringens Parasites
Abstract
1. Introduction
2. Results
2.1. PCR Detection of B. ostreae, B. exitiosa and M. refringens
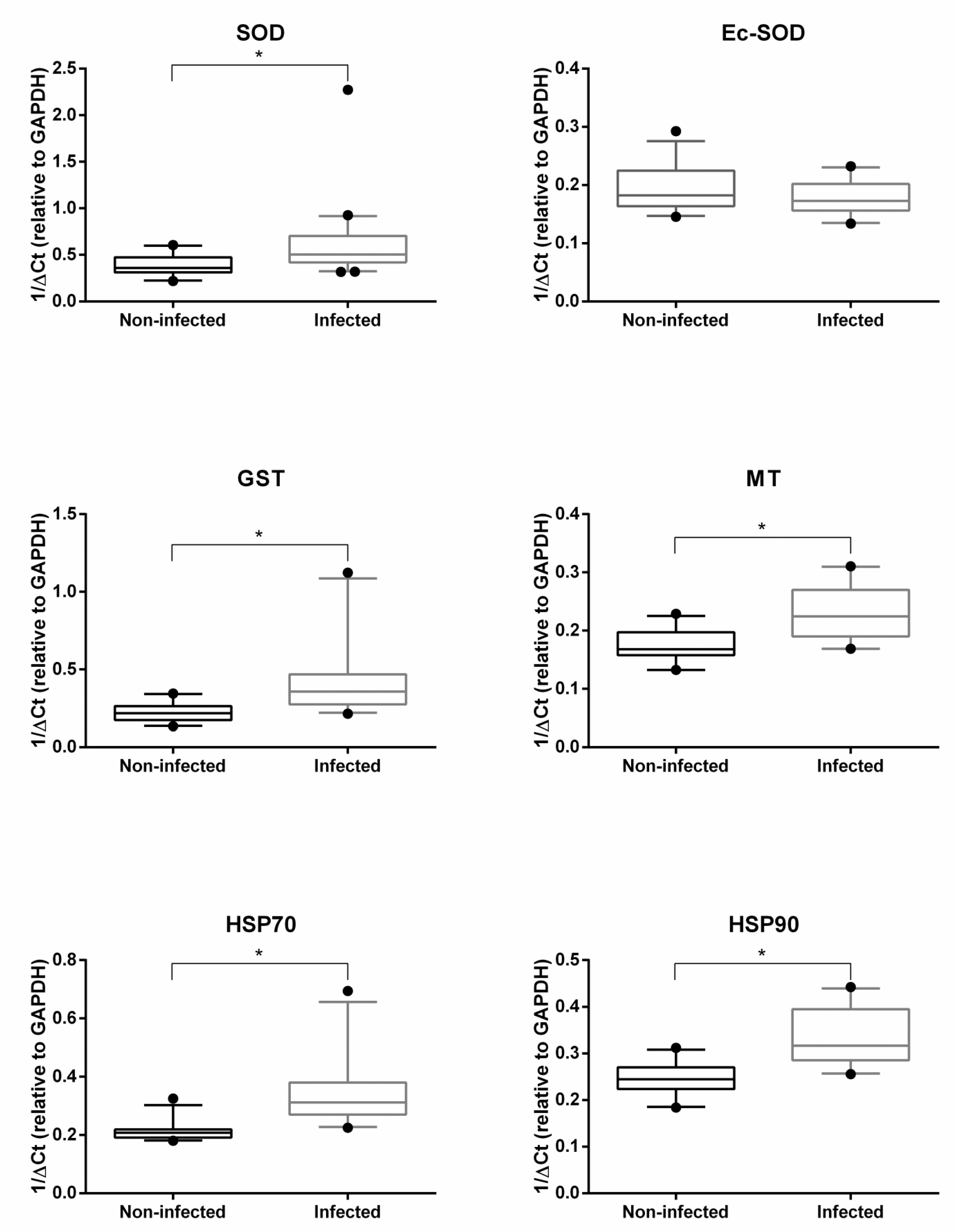
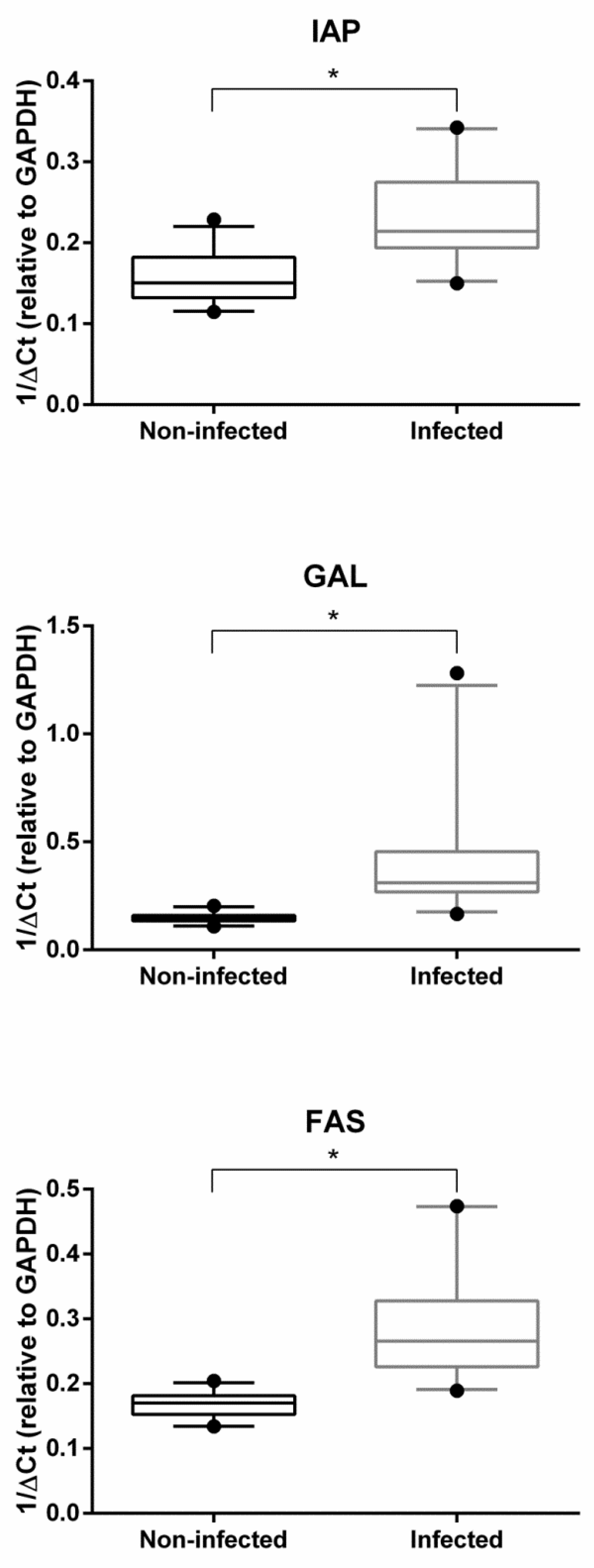
2.2. Gene Expression Analysis
2.3. Principal Component Analyses Results
3. Discussion
4. Material and methods
4.1. Sampling Sites and Oysters
4.2. Molecular Detection of Parasites: B. ostreae and M. refringens
4.3. Quantitative Real-Time PCR (qRT-PCR)
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Powered. Project of Offshore Wind Energy: Research, Experimentation, Development. In Ipa Adriatic CBC Programme 2007–2013; Code 087; Keep.eu; European Union: Brussels, Belgium, 2014. [Google Scholar]
- Petochi, T.; Gazzea, N.; Di Marco, P.; Latini, M.; Barchiesi, F.; Conte, A.; Barile, N.B.; Caruso, G.; Zaccone, R.; Cavallo, R.A.; et al. Il monitoraggio della qualità microbiologica delle aree di produzione dei molluschi bivalvi nell’ambito della Direttiva sulla Strategia Marina. In Proceedings of the II Convegno Nazionale della Società Italiana di Ricerca Applicata Alla Molluschicoltura (SIRAM) “Trasparenza tra Produttori Autorità di Controllo e Consumatori”, Cesenatico (FC), Italy, 28–29 November 2013; pp. 42–43. [Google Scholar]
- Bromley, C.; McGonigle, C.; Ashton, E.C.; Roberts, D. Restoring degraded European native oyster, Ostrea edulis, habitat: Is there a case for harrowing? Hydrobiologia 2016, 768, 151–165. [Google Scholar] [CrossRef]
- Audemard, C.; Sajus, M.C.; Barnaud, A.; Sautour, B.; Sauriau, P.G.; Berthe, F.J. Infection dynamics of Marteilia refringens in flat oyster Ostrea edulis and copepod Paracartia grani in a claire pond of Marennes-Oleron Bay. Dis. Aquat. Org. 2004, 61, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Lallias, D.; Arzul, I.; Heurtebise, S.; Ferrand, S.; Chollet, B.; Robert, M. Bonamia ostreae-induced mortalities in one-year old European flat oysters Ostrea edulis: Experimental infection by cohabitation challenge. Aquat. Living Resour. 2008, 21, 423–439. [Google Scholar] [CrossRef]
- Narcisi, V.; Arzul, I.; Cargini, D.; Mosca, F.; Calzetta, A.; Traversa, D.; Robert, M.; Joly, J.P.; Chollet, B.; Renault, T.; et al. Detection of Bonamia ostreae and B. exitiosa (Haplosporidia) in Ostrea edulis from the Adriatic Sea (Italy). Dis. Aquat. Org. 2010, 89, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.A.; Armitage, D.V.; Coughlan, J.; Mulcahy, M.F.; Culloty, S.C. Investigating the possible role of benthic macroinvertebrates and zooplankton in the life cycle of the haplosporidian Bonamia ostreae. Exp. Parasitol. 2007, 115, 359–368. [Google Scholar] [CrossRef]
- Bucke, D. Pathology of bonamiasis. Parasitol. Today 1988, 4, 174–176. [Google Scholar] [CrossRef]
- Figueras, A.J.; Montes, J. Aber disease of edible oysters causedby Marteilia refringens. Am. Fish. Soc. Spec. Pub. 1988, 18, 38–46. [Google Scholar]
- Camacho, A.P.; Villalba, A.; Beiras, R.; Labarta, U. Absorption efficiency and condition of cultured mussels (Mytilus edulis galloprovincialis Linnaeus) of Galicia (NW Spain) infected by parasites Marteilia refringens Grizel et al. and Mytilicola intestinalis Steuer. J. Shellfish Res. 1997, 16, 77–82. [Google Scholar]
- Berthe, F.C.; Roux, F.L.; Adlard, R.D.; Figueras, A. Marteiliosis in mollusks: A review. Aquat. Living Resour. 2004, 448, 433–448. [Google Scholar] [CrossRef]
- Helmer, L.; Farrell, P.; Hendy, I.; Harding, S.; Robertson, M.; Preston, J. Active management is required to turn the tide for depleted Ostrea edulis stocks from the effects of overfishing, disease and invasive species. PeerJ 2019, 7, e6431. [Google Scholar] [CrossRef]
- Tieri, E.; Ceschia, G.; Cannone, N.; Giansante, C. Presence of Marteilia refringens in an European flat oysters (Ostrea edulis) natural bank in the Central Adriatic Sea, Italy. Ittiopatologia 2006, 3, 41–45. [Google Scholar]
- Morga, B.; Arzul, I.; Chollet, B.; Renault, T. Infection with the protozoan parasite Bonamia ostreae modifies in vitro haemocyte activities of flat oyster Ostrea edulis. Fish Shellfish Immunol. 2009, 26, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Pipe, R.K. Generation of reactive oxygen metabolites by the haemocytes of the mussel Mytilus edulis. Dev. Comp. Immunol. 1992, 16, 111–122. [Google Scholar] [CrossRef]
- Chagot, D.; Boulo, V.; Hervio, D.; Mialhe, E.; Bachere, E.; Mourton, C.; Grizel, E. Interactions between Bonamia ostreae (Protozoa: Ascetospora) and hemocytes of Ostrea edulis and Crassostrea gigas (Mollusca: Bivalvia): Entry mechanisms. J. Invertebr. Pathol. 1992, 59, 241–249. [Google Scholar] [CrossRef]
- Mourton, C.; Boulo, V.; Chagot, D.; Hervio, D.; Bachere, E.; Mialhe, E.; Grizel, H. Interactions between Bonamia ostreae (Protozoa: Ascetospora) and hemocytes of Ostrea edulis and Crassostrea gigas (Mollusca: Bivalvia): In vitro system establishment. J. Invertebr. Pathol. 1992, 59, 235–240. [Google Scholar] [CrossRef]
- Alavi, M.R.; Fernandez-Robledo, J.A.; Vasta, G.R. Development of an in vitro assay to examine intracellular survival of Perkinsus marinus trophozoites upon phagocytosis by oyster (Crassostrea virginica and Crassostrea ariakensis) hemocytes. J. Parasitol. 2009, 95, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.G.; Renault, T.; Chilmonczyk, S. Flow cytometric assessment of haemocyte sub-populations in the European flat oyster, Ostrea edulis, haemolymph. Fish Shellfish Immunol. 2001, 11, 557–567. [Google Scholar] [CrossRef]
- Cochennec-Laureau, N.; Auffret, M.; Renault, T.; Langlade, A. Changes in circulating and tissue-infiltrating hemocyte parameters of European flat oysters, Ostrea edulis, naturally infected with Bonamia ostreae. J. Invertebr. Pathol. 2003, 83, 23–30. [Google Scholar] [CrossRef]
- Morga, B.; Arzul, I.; Faury, N.; Segarra, A.; Chollet, B.; Renault, T. Molecular responses of Ostrea edulis haemocytes to an in vitro infection with Bonamia ostreae. Dev. Comp. Immunol. 2011, 35, 323–333. [Google Scholar] [CrossRef]
- Morga, B.; Renault, T.; Faury, N.; Chollet, B.; Arzul, I. Cellular and molecular responses of haemocytes from Ostrea edulis during in vitro infection by the parasite Bonamia ostreae. Int. J. Parasitol. 2011, 41, 755–764. [Google Scholar] [CrossRef]
- Morga, B.; Renault, T.; Faury, N.; Lerond, S.; Garcia, C.; Chollet, B.; Joly, J.P.; Lapegue, S.; Harrang, E.; Arzul, I. Contribution of in Vivo Experimental Challenges to Understanding Flat Oyster Ostrea edulis Resistance to Bonamia ostreae. Front. Cell. Infect. Microbiol. 2017, 7, 433. [Google Scholar] [CrossRef] [PubMed]
- Mipaaf. Evolution of Italian aquaculture within the Mediterranean region. In The State of Italian Marine Fisheries and Aquaculture; Cautadella, S., Spagnolo, M., Eds.; Mipaaf: Rome, Italy, 2013; Volume Chapter 5. [Google Scholar]
- Lopez-Flores, I.; de la Herran, R.; Garrido-Ramos, M.A.; Navas, J.I.; Ruiz-Rejon, C.; Ruiz-Rejon, M. The molecular diagnosis of Marteilia refringens and differentiation between Marteilia strains infecting oysters and mussels based on the rDNA IGS sequence. Parasitology 2004, 129, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, N.; Villalba, A.; Andree, K.B.; Engelsma, M.Y.; Lacuesta, B.; Ramilo, A.; Gairin, I.; Furones, M.D. Bonamia exitiosa (Haplosporidia) observed infecting the European flat oyster Ostrea edulis cultured on the Spanish Mediterranean coast. J. Invertebr. Pathol. 2012, 110, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Comps, M. Observations sur les causes d’une mortalité abnormal des huîtres plates dans le bassin de Marennes. Rev. Trav. Inst. Pech. Marit. 1970, 34, 317–326. [Google Scholar]
- Grizel, H.; Comps, M.; Bonami, J.R.; Cousserans, F.; Duthoit, J.L.; Pennec, M. Epizooty of the common oyster Ostrea edulis. Part 1. Study of the agent of digestive gland disease in Ostrea edulis (Linne). Sci. Pech. 1974, 240, 1–30. [Google Scholar]
- Bower, S. Synopsis of Infectious Diseases and Parasites of Commercially Exploited Shellfish: Marteiliosis (Aber Disease) of Oysters. 2011. Available online: https://www.dfo-mpo.gc.ca/science/aah-saa/diseases-maladies/madoy-eng.html (accessed on 4 December 2018).
- Carrasco, N.; Arzul, I.; Berthe, F.C.; Furones, M.D. In situ hybridization detection of initial infective stages of Marteilia refringens (Paramyxea) in its host Mytilus galloprovincialis. J. Fish Dis. 2008, 31, 153–157. [Google Scholar] [CrossRef]
- Guillou, F.; Mitta, G.; Galinier, R.; Coustau, C. Identification and expression of gene transcripts generated during an anti-parasitic response in Biomphalaria glabrata. Dev. Comp. Immunol. 2007, 31, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Brophy, P.M.; Pritchard, D.I. Immunity to helminthes:Ready totip the biochemical balance. Parasitol. Today 1992, 8, 419–420. [Google Scholar] [CrossRef]
- Brophy, P.M.; Pritchard, D.I. Parasitic helminth glutathione S-transferases: An update on their potential as targets for immuno- and chemotherapy. Exp. Parasitol. 1994, 79, 89–96. [Google Scholar] [CrossRef]
- Ruttkay-Nedecky, B.; Nejdl, L.; Gumulec, J.; Zitka, O.; Masarik, M.; Eckschlager, T.; Stiborova, M.; Adam, V.; Kizek, R. The role of metallothionein in oxidative stress. Int. J. Mol. Sci. 2013, 14, 6044–6066. [Google Scholar] [CrossRef]
- Desclaux-Marchand, C.; Paul-Pont, I.; Gonzalez, P.; Baudrimont, M.; de Montaudouin, X. Metallothionein gene identification and expression in the cockle (Cerastoderma edule) under parasitism (Trematodes) and cadmium contaminations. Aquat. Living Resour. 2007, 20, 43–49. [Google Scholar] [CrossRef]
- Encomio, V.G.; Chu, F.L. Heat shock protein (hsp70) expression and thermal tolerance in sublethally heat-shocked eastern oysters Crassostrea virginica infected with the parasite Perkinsus marinus. Dis. Aquat. Org. 2007, 76, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, R.I.; Sarge, K.D.; Abravaya, K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J. Biol. Chem. 1992, 267, 21987–21990. [Google Scholar]
- Morimoto, R.I. Cells in stress: Transcriptional activation of heat shock genes. Science 1993, 259, 1409–1410. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, R.I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998, 12, 3788–3796. [Google Scholar] [CrossRef]
- Silar, P.; Butler, G.; Thiele, D.J. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Mol. Cell. Biol. 1991, 11, 1232–1238. [Google Scholar] [CrossRef]
- Sorger, P.K. Heat shock factor and the heat shock response. Cell 1991, 65, 363–366. [Google Scholar] [CrossRef]
- Morga, B.; Renault, T.; Faury, N.; Arzul, I. New insights in flat oyster Ostrea edulis resistance against the parasite Bonamia ostreae. Fish Shellfish Immunol. 2012, 32, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Prado-Alvarez, M.; Gestal, C.; Novoa, B.; Figueras, A. Differentially expressed genes of the carpet shell clam Ruditapes decussatus against Perkinsus olseni. Fish Shellfish Immunol. 2009, 26, 72–83. [Google Scholar] [CrossRef]
- Hughes, F.M.; Foster, B.; Grewal, S.; Sokolova, I.M. Apoptosis as a host defense mechanism in Crassostrea virginica and its modulation by Perkinsus marinus. Fish Shellfish Immunol. 2010, 29, 247–257. [Google Scholar] [CrossRef]
- Prado-Alvarez, M.; Chollet, B.; Faury, N.; Robert, M.; Morga, B.; Ibara, D.J.; Lupo, C.; Renault, T.; Arzul, I. Interactions between Ostrea edulis galectin (OE-GAL) and the protozoan parasite Bonamia ostreae. Fish Shellfish Immunol. 2013, 34, 1674. [Google Scholar] [CrossRef]
- OIE. Manual of Diagnostic Tests for Aquatic Animals. Available online: http://www.oie.int/en/standard-setting/aquatic-manual/access-online/ (accessed on 19 August 2019).
- Pernas, M.; Novoa, B.; Berthe, F.; Tafalla, C.; Figueras, A. Molecular methods for the diagnosis of Marteilia refringens. Bull. Eur. Assoc. Fish Pathol. 2001, 21, 200. [Google Scholar]
- Robert, M.; Garcia, C.; Chollet, B.; Lopez-Flores, I.; Ferrand, S.; Francois, C.; Joly, J.P.; Arzul, I. Molecular detection and quantification of the protozoan Bonamia ostreae in the flat oyster, Ostrea edulis. Mol. Cell. Probes 2009, 23, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Ramilo, A.; Navas, J.I.; Villalba, A.; Abollo, E. Species-specific diagnostic assays for Bonamia ostreae and B. exitiosa in European flat oyster Ostrea edulis: Conventional, real-time and multiplex PCR. Dis. Aquat. Org. 2013, 104, 149–161. [Google Scholar] [CrossRef]
- Lane, H.S.; Webb, S.C.; Duncan, J. Bonamia ostreae in the New Zealand oyster Ostrea chilensis: A new host and geographic record for this haplosporidian parasite. Dis. Aquat. Org. 2016, 118, 55–63. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Morga, B.; Arzul, I.; Faury, N.; Renault, T. Identification of genes from flat oyster Ostrea edulis as suitable housekeeping genes for quantitative real time PCR. Fish Shellfish Immunol. 2010, 29, 937–945. [Google Scholar] [CrossRef]
- Tanguy, A.; Boutet, I.; Riso, R.; Boudry, P.; Auffret, M.; Moraga, D. Metallothionein genes in the European flat oyster Ostrea edulis: A potential ecological tool forenvironmental monitoring? Mar. Ecol. Prog. Ser. 2003, 257, 87–97. [Google Scholar] [CrossRef]
- Farcy, E.; Voiseux, C.; Lebel, J.M.; Fievet, B. Transcriptional expression levels of cell stress marker genes in the Pacific oyster Crassostrea gigas exposed to acute thermal stress. Cell Stress Chaperones 2009, 14, 371–380. [Google Scholar] [CrossRef]

| Gene | Forward Sequence (5′–3′) | Reverse Sequence (5′–3′) | References | Efficiency (%) |
|---|---|---|---|---|
| MT | CTAATTTTACTCCTTCCAAC | CAGGCGACCATTAATTCAC | [53] | 99.52 |
| SOD | TCGTCAATGTCAGCGTGAA | AAATGTTGGGGCTGGTGA | [21] | 102.87 |
| GST | GGTCGTCAGGGGTCAGTTT | GGTTCCCGTTCTTGAGCA | [21] | 102.52 |
| HSP70 | AGCAAGCCAGCACAGCA | GCGATGATTTCCACCTTC | [54] | 98.27 |
| HSP90 | TTTGTGGAACGGGTCAAAA | AACGTCGAGCACAGTCGAG | [21] | 95.76 |
| IAP | CTACCTCCCCAGGATTGTCA | CACCACTCTCCTCCATGTCA | [42] | 97.56 |
| FAS | TTTGGGCAGTGGTGTAAGTG | TAGCCCTGTTTCTCCACCAG | [42] | 98.50 |
| GAL | TCGGAGGTCGCCCTTAAT | TTGCCGTGAACAATCAACA | [22] | 99.30 |
| EcSOD | GAGGAGGAAGAGGACCATCC | ATTTTCCTCCGCTTTGTGTG | [42] | 97.67 |
| GAPDH | TCCCGCTAGCATTCCTTG | TTGGCGCCTCCTTTCATA | [52] | 98.50 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cocci, P.; Roncarati, A.; Capriotti, M.; Mosconi, G.; Palermo, F.A. Transcriptional Alteration of Gene Biomarkers in Hemocytes of Wild Ostrea edulis with Molecular Evidence of Infections with Bonamia spp. and/or Marteilia refringens Parasites. Pathogens 2020, 9, 323. https://doi.org/10.3390/pathogens9050323
Cocci P, Roncarati A, Capriotti M, Mosconi G, Palermo FA. Transcriptional Alteration of Gene Biomarkers in Hemocytes of Wild Ostrea edulis with Molecular Evidence of Infections with Bonamia spp. and/or Marteilia refringens Parasites. Pathogens. 2020; 9(5):323. https://doi.org/10.3390/pathogens9050323
Chicago/Turabian StyleCocci, Paolo, Alessandra Roncarati, Martina Capriotti, Gilberto Mosconi, and Francesco Alessandro Palermo. 2020. "Transcriptional Alteration of Gene Biomarkers in Hemocytes of Wild Ostrea edulis with Molecular Evidence of Infections with Bonamia spp. and/or Marteilia refringens Parasites" Pathogens 9, no. 5: 323. https://doi.org/10.3390/pathogens9050323
APA StyleCocci, P., Roncarati, A., Capriotti, M., Mosconi, G., & Palermo, F. A. (2020). Transcriptional Alteration of Gene Biomarkers in Hemocytes of Wild Ostrea edulis with Molecular Evidence of Infections with Bonamia spp. and/or Marteilia refringens Parasites. Pathogens, 9(5), 323. https://doi.org/10.3390/pathogens9050323







