Novel Cytoplasmic Bacteriocin Compounds Derived from Staphylococcus epidermidis Selectively Kill Staphylococcus aureus, Including Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
1. Introduction
2. Results
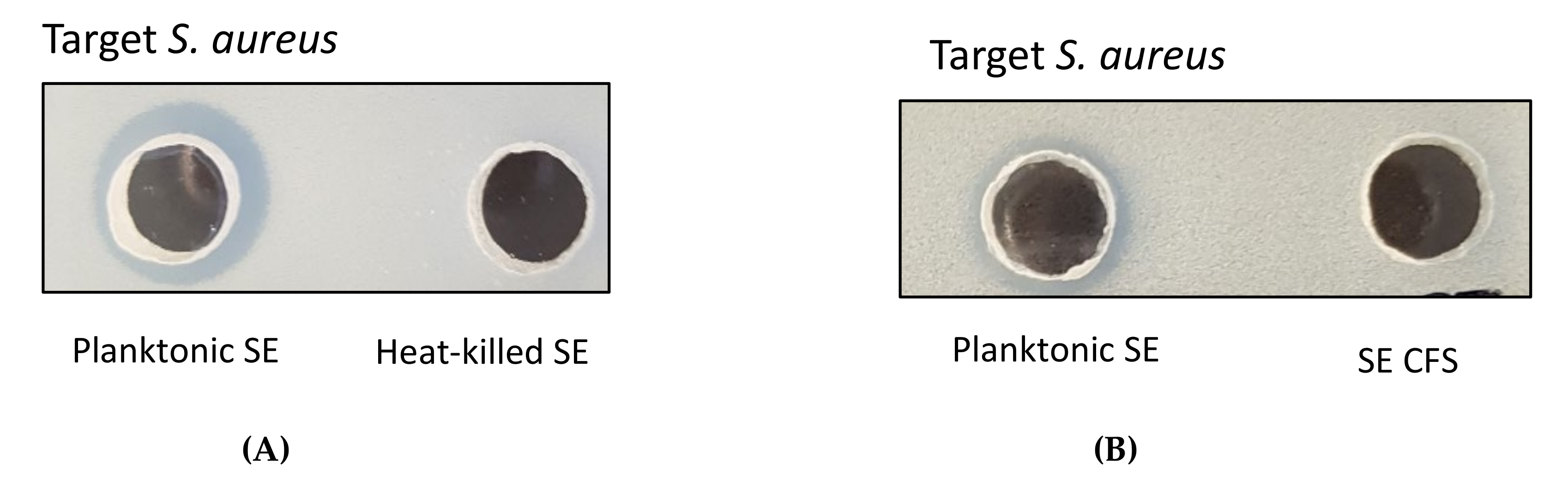
2.1. Antimicrobial Activity of Live Planktonic S. epidermidis or CFS from S. epidermidis
2.2. Antimicrobial Activity of Concentrated Proteins from CFS of S. epidermidis Strains
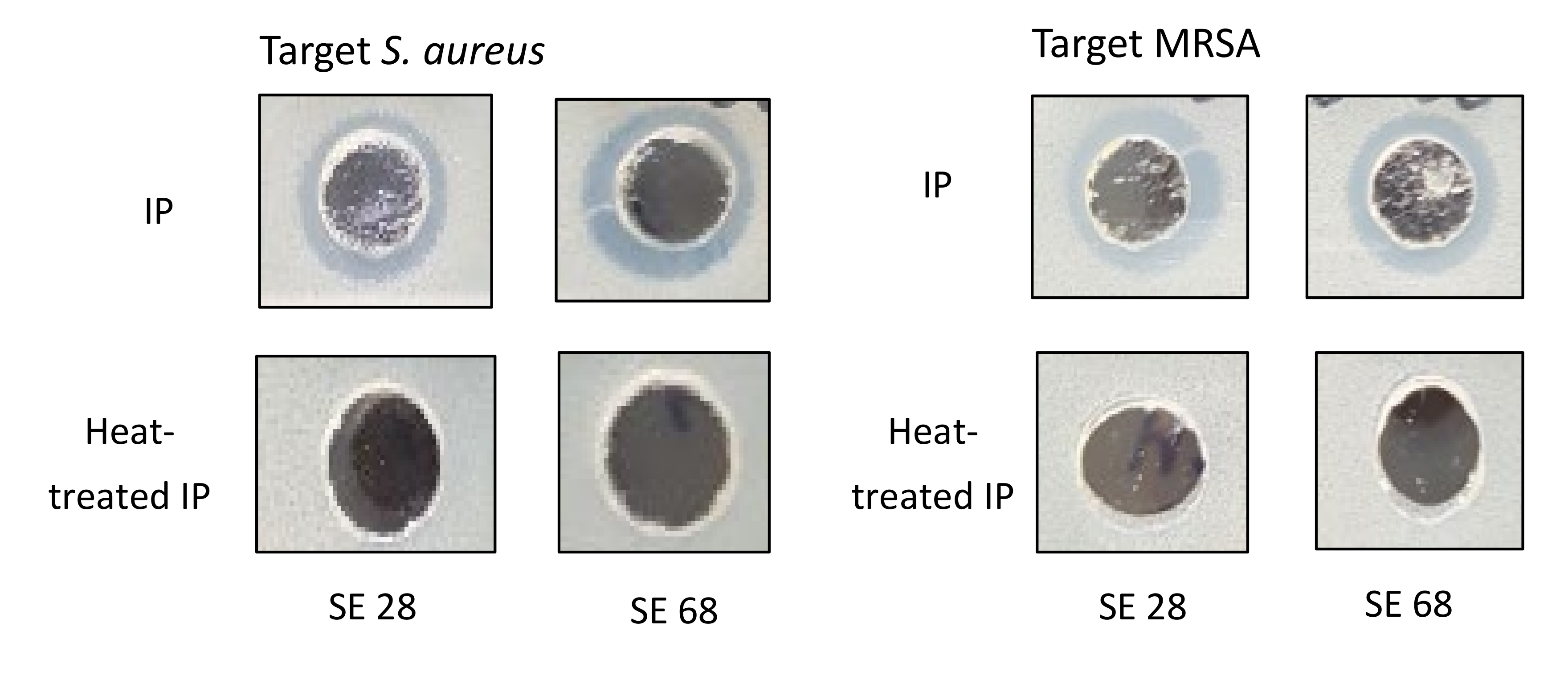
2.3. Antimicrobial Activity of Cytoplasmic Bacteriocin Compounds from S. epidermidis Strains
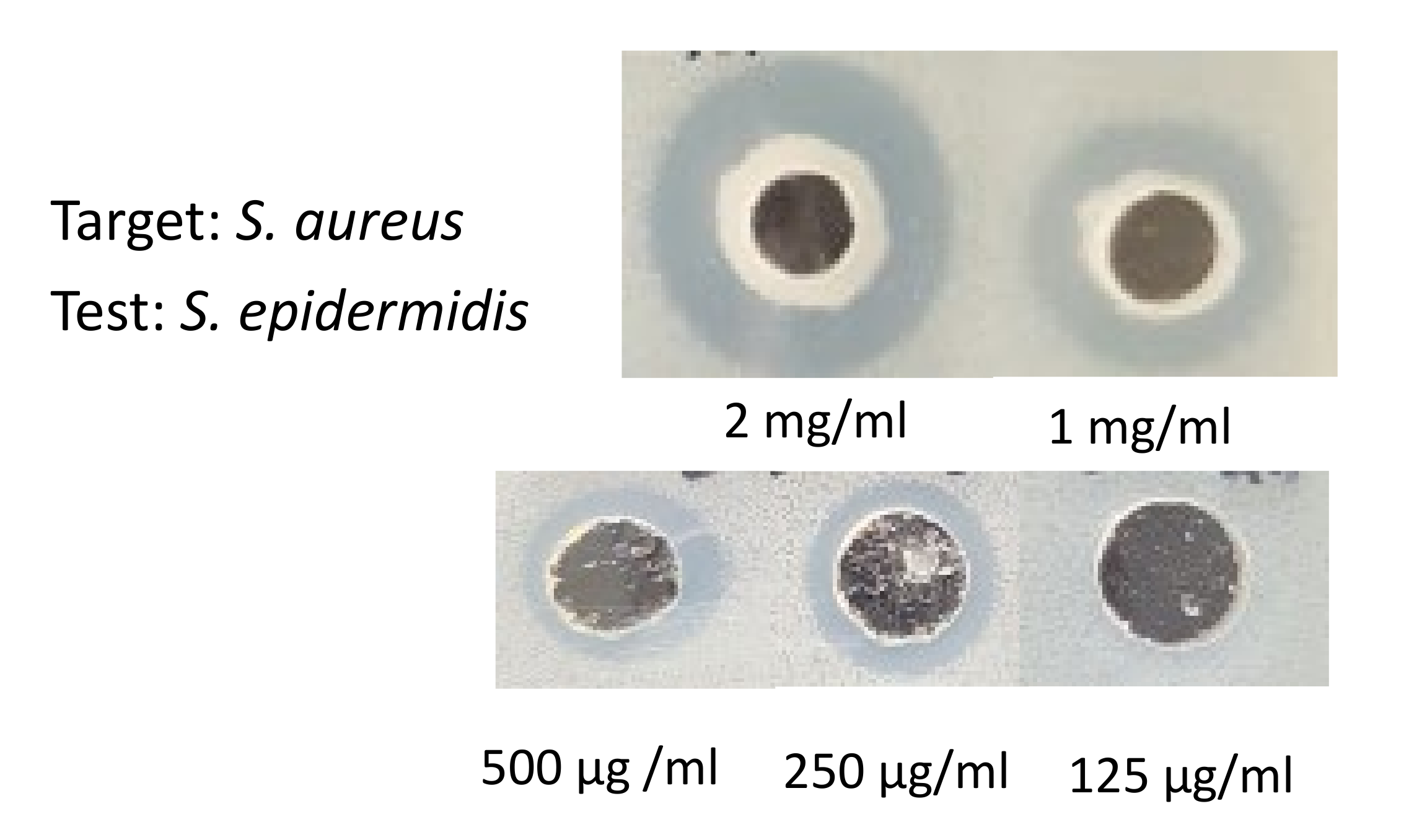
2.4. The Minimum Bactericidal Concentration of Cytoplasmic Bacteriocin Compounds
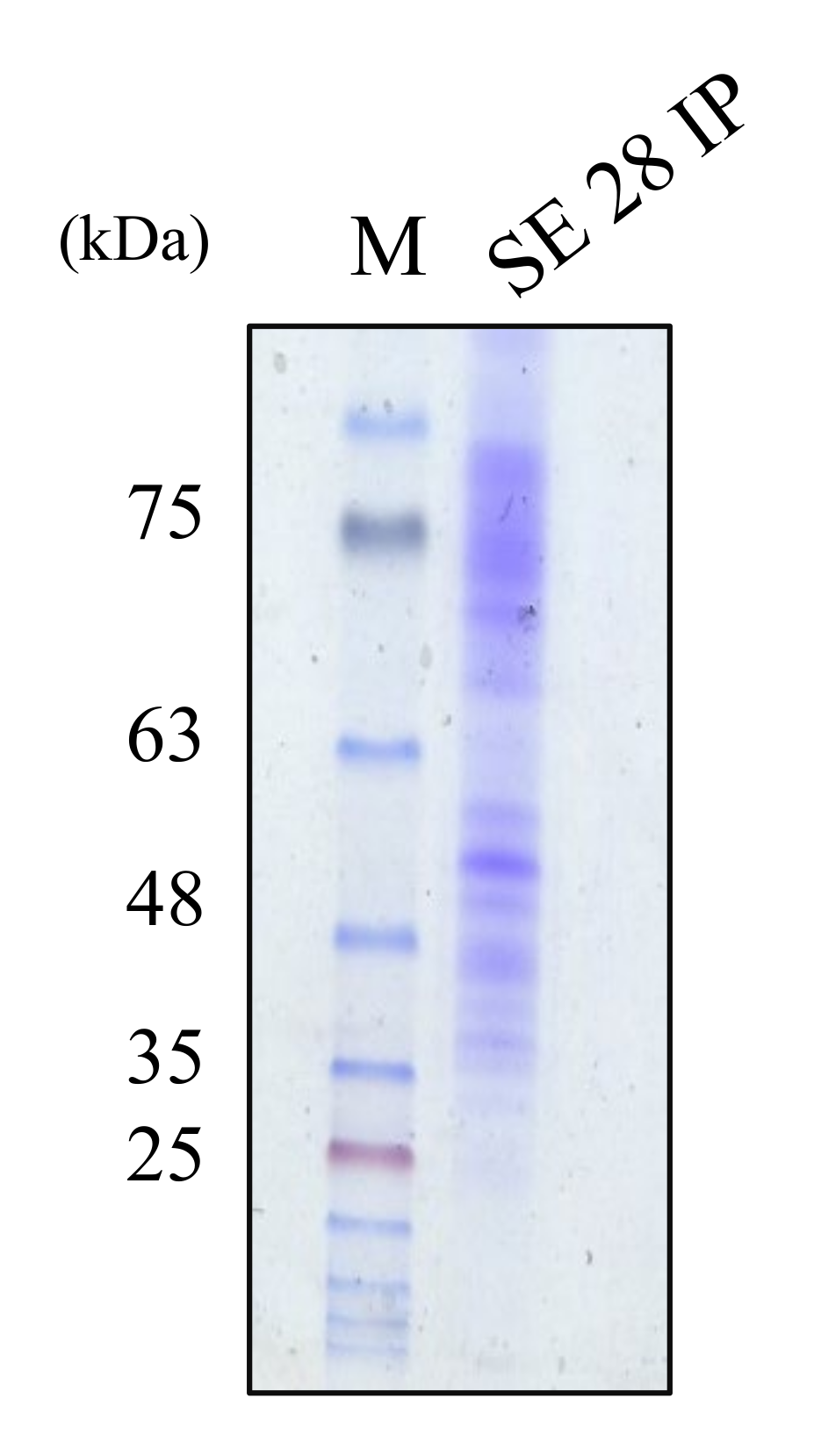
2.5. The Molecular Weight Ranges of Partially Purified Cytoplasmic Bacteriocin Compounds
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Preparation of CFS from S. Epidermidis for Antimicrobial Potentiality
4.3. Extraction of cytoplasmic bacteriocin compounds from S. epidermidis for antimicrobial potentiality
4.4. Stability Tests of Cytoplasmic Bacteriocin Compounds from S. epidermidis
4.5. Antimicrobial Activity of Bacteriocins by the Agar Well Diffusion Assay
4.6. Determination of the Minimum Bactericidal Concentration by the Agar Well Diffusion Assay
4.7. Molecular Weight Estimation with Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Author Contributions
Funding
Conflicts of Interest
References
- Kobayashi, T.; Nagao, K. Host–microbial dialogues in atopic dermatitis. Int. Immunol. 2019, 31, 449–456. [Google Scholar] [CrossRef]
- Blicharz, L.; Rudnicka, L.; Samochocki, Z. Staphylococcus aureus: an underestimated factor in the pathogenesis of atopic dermatitis? Adv. Dermatol. Allergol. 2019, 36, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome. Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Brauweiler, A.M.; Goleva, E.; Leung, N.Y. Staphylococcus aureus Lipoteichoic Acid Damages the Skin Barrier through an IL-1–Mediated Pathway. J. Investig. Dermatol. 2019, 139, 1753–1761. [Google Scholar] [CrossRef]
- Nakamura, Y.; Oscherwitz, J.; Cease, K.B.; Chan, S.M.; Muñoz-Planillo, R.; Hasegawa, M.; Villaruz, A.E.; Cheung, G.Y.; McGavin, M.J. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature 2013, 503, 397–401. [Google Scholar] [CrossRef]
- Geoghegan, J.A.; Irvine, A.D.; Foster, T.J. Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends Microbiol. 2018, 26, 484–497. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Jang, I.-T.; Yang, M.; Jo, E.-K.; Kim, H.-J.; Park, J.-K. The Effects of Staphylococci on the Degranulation of Human Mast Cell-1. J. Bacteriol. Virol. 2017, 47, 132. [Google Scholar] [CrossRef]
- Iwamoto, K.; Moriwaki, M.; Miyake, R.; Hide, M. Staphylococcus aureus in atopic dermatitis: Strain-specific cell wall proteins and skin immunity. Allergol. Int. 2019, 68, 309–315. [Google Scholar] [CrossRef]
- Williams, M.R.; Gallo, R.L. The Role of the Skin Microbiome in Atopic Dermatitis. Curr. Allergy Asthma Rep. 2015, 15, 65. [Google Scholar] [CrossRef]
- Hepburn, L.; Hijnen, D.; Sellman, B.; Mustelin, T.; Sleeman, M.; May, R.; Strickland, I. The complex biology and contribution of Staphylococcus aureus in atopic dermatitis, current and future therapies. Br. J. Dermatol. 2017, 177, 63–71. [Google Scholar] [CrossRef]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.R.; Gallo, R.L. Evidence that Human Skin Microbiome Dysbiosis Promotes Atopic Dermatitis. J. Investig. Dermatol. 2017, 137, 2460–2461. [Google Scholar] [CrossRef]
- Christensen, G.J.M.; Scholz, C.F.P.; Enghild, J.J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genom. 2016, 17, 152. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis: a major player in bacterial sepsis? Futur. Microbiol. 2017, 12, 1031–1033. [Google Scholar] [CrossRef]
- Siljamäki, P.; Varmanen, P.; Kankainen, M.; Sukura, A.K.K.; Savijoki, K.; Nyman, T.A. Comparative Exoprotein Profiling of DifferentStaphylococcus epidermidisStrains Reveals Potential Link between Nonclassical Protein Export and Virulence. J. Proteome Res. 2014, 13, 3249–3261. [Google Scholar] [CrossRef]
- Götz, F.; Perconti, S.; Popella, P.; Werner, R.; Schlag, M. Epidermin and gallidermin: Staphylococcal lantibiotics. Int. J. Med Microbiol. 2014, 304, 63–71. [Google Scholar] [CrossRef]
- Bjerre, R.; Bandier, J.; Skov, L.; Engstrand, L.; Johansen, J. The role of the skin microbiome in atopic dermatitis: A systematic review. Br. J. Dermatol. 2017, 177, 1272–1278. [Google Scholar] [CrossRef]
- Lee, D.C.; Kananurak, A.; Tran, M.T.; Connolly, P.A.; Polage, C.R.; Iwase, T.; Bevins, C.L.; Underwood, M.A. Bacterial Colonization of the Hospitalized Newborn: Competition Between Staphylococcus aureus and Staphylococcus epidermidis. Pediatr. Infect. Dis. J. 2019, 38, 682–686. [Google Scholar] [CrossRef]
- Lambers, H.; Piessens, S.; Bloem, A.; Pronk, H.; Finkel, P. Natural skin surface pH is on average below 5, which is beneficial for its resident flora. Int. J. Cosmet. Sci. 2006, 28, 359–370. [Google Scholar] [CrossRef]
- Sandiford, S.; Upton, M. Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against Staphylococci. Antimicrob Agents Ch. 2012, 56, 1539–1547. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Genet. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Cogen, A.L.; Yamasaki, K.; Sanchez, K.M.; Dorschner, R.A.; Lai, Y.; MacLeod, D.T.; Torpey, J.W.; Otto, M.; Nizet, V.; Kim, J.E.; et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010, 130, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Claudel, J.-P.; Auffret, N.; Leccia, M.-T.; Poli, F.; Corvec, S.; Dréno, B. Staphylococcus epidermidis: A Potential New Player in the Physiopathology of Acne? Dermatology 2019, 235, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. NPG 2010, 465, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Bürgers, R.; Morsczeck, C.; Felthaus, O.; Gosau, M.; Beck, H.; Reichert, T.E. Induced surface proteins of Staphylococcus [corrected] epidermidis adhering to titanium implant substrata. Clin. Oral Investig. 2018, 22, 2663–2668. [Google Scholar] [CrossRef] [PubMed]
- Alreshidi, M.M.; Dunstan, R.H.; Gottfries, J.; Macdonald, M.M.; Crompton, M.J.; Ang, C.-S.; Williamson, N.A.; Roberts, T.K. Changes in the Cytoplasmic Composition of Amino Acids and Proteins Observed in Staphylococcus aureus during Growth under Variable Growth Conditions Representative of the Human Wound Site. PLOS ONE 2016, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, S.A.; Medina, L.M.P.; Glasner, C.; Tsompanidou, E.; De Jong, A.; Grasso, S.; Schaffer, M.; Mäder, U.; Larsen, A.R.; Gumpert, H.; et al. Signatures of cytoplasmic proteins in the exoproteome distinguish community- and hospital-associated methicillin-resistantStaphylococcus aureusUSA300 lineages. Virulence 2017, 8, 891–907. [Google Scholar] [CrossRef]
- García-Pérez, A.N.; De Jong, A.; Junker, S.; Becher, D.; Chlebowicz, M.A.; Duipmans, J.C.; Jonkman, M.F.; Van Dijl, J.M. From the wound to the bench: exoproteome interplay between wound-colonizing Staphylococcus aureus strains and co-existing bacteria. Virulence 2018, 9, 363–378. [Google Scholar] [CrossRef]
- Solis, N.; Cain, J.A.; Cordwell, S.J. Comparative analysis of Staphylococcus epidermidis strains utilizing quantitative and cell surface shaving proteomics. J. Proteom. 2016, 130, 190–199. [Google Scholar] [CrossRef]
- García-Gómez, E.; Miranda-Ozuna, J.F.T.; Díaz-Cedillo, F.; Vázquez-Sánchez, E.A.; Rodríguez-Martínez, S.; Jan-Roblero, J.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Staphylococcus epidermidis lipoteichoic acid: exocellular release and ltaS gene expression in clinical and commensal isolates. J. Med Microbiol. 2017, 66, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Pidutti, P.; Federici, F.; Brandi, J.; Manna, L.; Rizzi, E.; Marini, U.; Cecconi, D. Purification and characterization of ribosomal proteins L27 and L30 having antimicrobial activity produced by theLactobacillus salivariusSGL 03. J. Appl. Microbiol. 2018, 124, 398–407. [Google Scholar] [CrossRef] [PubMed]

| Bacterial indicators | PCSE | CFS |
|---|---|---|
| S. aureus (ATCC 25923) S. aureus (NCCP 14780) | 11.1 ± 0.1 11.4 ± 0.2 | ― ― |
| MRSA (ATCC 33591) | 10.6 ± 0.1 | ― |
| S. epidermidis (ATCC 12228) | ― | ― |
| E. coli (NCCP 14762) | ― | ― |
| Salmonella Typhimurium (NCCP 10438) | ― | ― |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, I.-T.; Yang, M.; Kim, H.-J.; Park, J.-K. Novel Cytoplasmic Bacteriocin Compounds Derived from Staphylococcus epidermidis Selectively Kill Staphylococcus aureus, Including Methicillin-Resistant Staphylococcus aureus (MRSA). Pathogens 2020, 9, 87. https://doi.org/10.3390/pathogens9020087
Jang I-T, Yang M, Kim H-J, Park J-K. Novel Cytoplasmic Bacteriocin Compounds Derived from Staphylococcus epidermidis Selectively Kill Staphylococcus aureus, Including Methicillin-Resistant Staphylococcus aureus (MRSA). Pathogens. 2020; 9(2):87. https://doi.org/10.3390/pathogens9020087
Chicago/Turabian StyleJang, In-Taek, Miso Yang, Hwa-Jung Kim, and Jeong-Kyu Park. 2020. "Novel Cytoplasmic Bacteriocin Compounds Derived from Staphylococcus epidermidis Selectively Kill Staphylococcus aureus, Including Methicillin-Resistant Staphylococcus aureus (MRSA)" Pathogens 9, no. 2: 87. https://doi.org/10.3390/pathogens9020087
APA StyleJang, I.-T., Yang, M., Kim, H.-J., & Park, J.-K. (2020). Novel Cytoplasmic Bacteriocin Compounds Derived from Staphylococcus epidermidis Selectively Kill Staphylococcus aureus, Including Methicillin-Resistant Staphylococcus aureus (MRSA). Pathogens, 9(2), 87. https://doi.org/10.3390/pathogens9020087






